Abstract
Reduced levonorgestrel concentrations from the levonorgestrel contraceptive implant was previously seen when given concomitantly with efavirenz. We sought to assess whether single nucleotide polymorphisms (SNPs) in genes involved in efavirenz and nevirapine metabolism were linked to these changes in levonorgestrel concentration. SNPs in CYP2B6, CYP2A6, NR1I2 and NR1I3 were analysed. Associations of participant demographics and genotype with levonorgestrel pharmacokinetics were evaluated in HIV-positive women using the levonorgestrel implant plus efavirenz- or nevirapine-based ART, in comparison to ART-naïve women using multivariate linear regression. Efavirenz group: CYP2B6 516G>T was associated with lower levonorgestrel log10 Cmax and log10 AUC. CYP2B6 15582C>T was associated with lower log10 AUC. Nevirapine group: CYP2B6 516G>T was associated with higher log10 Cmax and lower log10 Cmin. Pharmacogenetic variations influenced subdermal levonorgestrel pharmacokinetics in HIV-positive women, indicating that the magnitude of the interaction with non-nucleoside reverse transcriptase inhibitors (NNRTIs) is influenced by host genetics.
Keywords: Levonorgestrel, Efavirenz, Nevirapine, Pharmacokinetics, Pharmacogenetics, HIV, CYP2A6, CYP2B6, NR1I2, NR1I3, Single nucleotide polymorphisms
Introduction
Hormonal contraceptives and antiretroviral therapy (ART) are critical components of healthcare for the estimated 17 million women living with HIV and are key pillars in the effort to globally reduce mother-to-child HIV transmission.(1–3) The subdermal levonorgestrel (LNG) implant is a highly effective and desirable method of contraception.(4, 5) LNG is released from the implant initially at 100mcg/day, decreasing to 40mcg/day within one year and to 30mcg/day within three years, providing relatively stable daily drug concentrations.(6) Notably, the use of contraceptive implants is rapidly expanding in low- and middle-income countries.(7) In these same settings, the World Health Organization (WHO) recommends ART-containing two nucleos(t)ide reverse transcriptase inhibitors (NRTI) in combination with efavirenz (EFV) as a first-line regimen for HIV treatment.(8) The use of EFV-based ART in women of childbearing potential has expanded in the past decade, first following characterization of the lower than expected level of fetal risk during in-utero EFV exposure (9–11), and subsequently due to recent WHO recommendations for universal use of ART irrespective of CD4+ cell count.(12) Considered an alternate first-line option, nevirapine (NVP), is still used by many women in low- and middle-income countries.(8) In light of this, identifying safe and efficacious combinations of both therapies with hormonal contraceptives in HIV-positive women is a critical public health priority.
We previously described a significant drug-drug interaction between the LNG implant and EFV-based ART in HIV-positive Ugandan women, which resulted in 47% lower LNG concentrations compared to control participants, and a high rate of unintended pregnancy, in women receiving EFV-based ART [3 (15%) of 20 participants].(13) Induction of CYP3A4- and CYP3A5-mediated metabolism of LNG is the proposed mechanism of the observed interaction with EFV. No pharmacokinetic interaction or unintended pregnancies were observed in women receiving NVP-based ART within the same study. These findings are supported by a retrospective cohort study of Kenyan women that found a three-fold higher adjusted pregnancy incidence in HIV-positive women receiving the implant plus EFV-based ART compared to those receiving NVP-based ART.(14)
LNG, EFV and NVP are substrates of cytochrome P450 (CYP) enzymes; LNG is metabolized by CYP3A4 and CYP3A5, (15) EFV is metabolized by CYP2B6 with minor contributions from CYP2A6, CYP3A4, and UGT2B7, (16, 17) while NVP is metabolized by CYP2B6, CYP3A4 and partially by CYP2D6 (18, 19). The nuclear receptors NR1I2 and NR1I3 are known to regulate the expression and activity of several CYPs, including CYP2B6 and CYP3A4. Multiple SNPs in both NR1I2 and NR1I3 genes have been linked to variation in expression of CYP3A4 and CYP2B6 (20–22), which may alter the pharmacokinetics (PK) of substrate drugs. Activation of NR1I2 and NR1I3 results in up regulation of target CYPs and alters the PK of their substrates. Both phenomenon’ have been demonstrated for EFV and NVP. (23, 24) CYP2B6 516G>T has previously been associated with higher EFV and NVP plasma concentrations. (25–31) Notably, several polymorphisms associated with EFV and NVP PK are found more commonly within African populations compared to Caucasians or Asians.(1, 28, 32–35)
The primary objective of this secondary study analysis was to ascertain the contributory effect of SNPs in key genes of interest in LNG, EFV, and NVP metabolism (CYP2B6, CYP2A6, NR1I2 and NR1I3) on plasma LNG PK when the subdermal implant is given in combination with EFV- or NVP-based ART (EFV and NVP groups, respectively) in HIV-positive women. The control group of the primary study was not included in this pharmacogenetics analysis because the purpose of the study was to investigate the genetic basis for the interaction between NNRTIs and LNG. Since these SNPs have been shown to influence non-nucleoside reverse transcriptase inhibitors (NNRTI) PK, our underlying hypothesis was that there would be consequences of different NNRTI concentrations for the degree of CYP3A induction, and therefore a genetic basis for the subsequent magnitude of the interaction with LNG.
Results
All women in the EFV and NVP group from the primary study were included in this analysis (n=40). (13) Patient characteristics and genotype frequencies are summarized in Table 1. The median (interquartile range [IQR]) age and weight of all patients was 31 years (29–34 years) and 59kg (53–68kg). All SNPs were in Hardy-Weinberg equilibrium, except CYP2B6 15582C>T (rs4803419) (X2=20.62, P=<0.001) and NR1I3 540C>T (rs2307424) (X2=12.36, P=<0.001), which compromises their interpretation. Univariate and multiple regression analysis for LNG PK parameters based on EFV and NVP groups are presented in Table 2. All regression results can be found in Supplementary Table 1.
Table 1.
Characteristics of the study participants at entry
| Characteristics | Total (n=40) | Efavirenz group (n=20) | Nevirapine group (n=20) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 31.0 (29.0–34.0) | 31.0 (28.3–34.0) | 32.5 (31.0–35.8) | ||||||
| Weight (kg) | 59.0 (53.0–68.0) | 59.5 (52.3–63.8) | 59.5 (54.3–69.8) | ||||||
| CD4 count (cells/mm3) | 579.0 (464.0–731.0) | 556.5 (477.3–663.5) | 626.0 (399.8–857.3) | ||||||
| Albumin concentration at baseline (g/L) | 43.7 (41.5–46.5) | 43.3 (40.7–45.1) | 43.1 (41.8–45.6) | ||||||
| SHBG at baseline (nmol/L) | 105.0 (82.3–136.0) | 100.0 (79.4–125.9) | 100.0 (100.0–125.9) | ||||||
| Genotype frequencies | |||||||||
| CYP2B6 516G>T (rs3745274) (%) | GG | GT | TT | GG | GT | TT | GG | GT | TT |
| 43 | 55 | 2 | 40 | 55 | 5 | 45 | 55 | 0 | |
| CYP2B6 983T>C (rs28399499) (%) | TT | CT | CC | TT | CT | CC | TT | CT | CC |
| 80 | 18 | 2 | 85 | 15 | 0 | 75 | 20 | 5 | |
| CYP2B6 15582C>T (rs4803419) (%) | CC | CT | TT | CC | CT | TT | CC | CT | TT |
| 28 | 70 | 2 | 25 | 75 | 0 | 30 | 65 | 5 | |
| CYP2A6*9B 1836G>T (rs8192726) (%) | GG | GT | TT | GG | GT | TT | GG | GT | TT |
| 82 | 18 | 0 | 80 | 20 | 0 | 75 | 25 | 0 | |
| CYP2A6 48A>C (rs28399433) (%) | AA | AC | CC | AA | AC | CC | AA | AC | CC |
| 78 | 22 | 0 | 75 | 25 | 0 | 70 | 30 | 0 | |
| NR1I2 63396C>T (rs2472677) (%) | CC | CT | TT | CC | CT | TT | CC | CT | TT |
| 63 | 30 | 7 | 65 | 30 | 5 | 60 | 30 | 10 | |
| NR1I3 540C>T (rs2307424) (%) | CC | CT | TT | CC | CT | TT | CC | CT | TT |
| 25 | 75 | 0 | 30 | 70 | 0 | 20 | 80 | 0 | |
| NR1I3 1089T>C (rs3003596) (%) | TT | CT | CC | TT | CT | CC | TT | CT | CC |
| 35 | 55 | 10 | 30 | 60 | 10 | 40 | 50 | 10 | |
Values shown as median (interquartile range) and percentage of population.
Table 2.
Univariate and multivariate linear regression analysis from each study group
| Efavirenz group | ||||||
|---|---|---|---|---|---|---|
| log10 Cmax | Univariate linear regression | Multivariate linear regression | ||||
| P value | β value (95% CI) | r2 | P value | β value (95% CI) | r2 | |
| Age (years) | 0.049 | −0.027 (−0.01,0.0) | 0.199 | |||
| CYP2B6 516G>T (rs3745274) | 0.021 | −0.209 (−0.4,0.0) | 0.263 | 0.021 | −0.209 (−0.4,0.0) | 0.263 |
| log10 AUC0–24 weeks | P value | β (pg/mL) (95% CI) | r2 | P value | β (pg/mL) (95% CI) | r2 |
| Age (years) | 0.028 | −0.022 (0.0,0.0) | 0.242 | 0.039 | −0.018 (−0.0,0.0) | 0.739 |
| CD4 (log10cells/mm3) | 0.085 | 0.520 (−0.1,1.1) | 0.156 | |||
| CYP2B6 516G>T (rs3745274) | <0.000 | −0.216 (−0.3,−0.1) | 0.512 | 0.023 | −0.132 (−0.2,0.0) | 0.739 |
| CYP2B6 15582C>T (rs4803419) | 0.058 | −0.172 (−0.3,0.0) | 0.186 | 0.021 | −0.163 (−0.3,0.0) | 0.756 |
| Nevirapine Group | ||||||
| log10 Cmax | Univariate linear regression | Multivariate linear regression | ||||
| P value | β value (95% CI) | r2 | P value | β value (95% CI) | r2 | |
| Age (years) | 0.056 | 0.015 (0.0,0.0) | 0.189 | 0.044 | 0.013 (0.0,0.3) | 0.569 |
| Albumin concentration at baseline (g/L) | 0.076 | 0.019 (0.0,0.0) | 0.164 | |||
| SHBG at baseline (log10 pg/mL) | 0.017 | 0.480 (0.1,0.9) | 0.280 | 0.007 | 0.469 (0.0,0.8) | 0.569 |
| CYP2B6 516G>T (rs3745274) | 0.115 | 0.098 (0.0,0.2) | 0.133 | 0.034 | 0.103 (0.0,0.2) | 0.569 |
| Tmax | P value | β value (95% CI) | r2 | P value | β value (95% CI) | r2 |
| Age (years) | 0.043 | −0.259 (−0.5,0.0) | 0.209 | |||
| Weight (log10 kg) | 0.029 | −16.956 (−31.9,−0.2) | 0.239 | 0.002 | −15.910 (−24.9,−6.9) | 0.771 |
| CD4 (log10 cells/mm3) | 0.157 | −3.489 (−8.5,1.5) | 0.108 | |||
| Albumin concentration at baseline (g/L) | 0.001 | −0.499 (−0.8,0.2) | 0.446 | 0.003 | −0.340 (−0.5,−0.1) | 0.771 |
| NR1I2 63396C>T (rs2472677) | 0.013 | 2.000 (0.5,3.5) | 0.300 | 0.003 | 1.623 (0.6,2.6) | 0.771 |
| log10 Cmin | P value | β value (95% CI) | r2 | P value | β value (95% CI) | r2 |
| Weight (log10kg) | 0.168 | −0.751 (−1.8,0.3) | 0.103 | |||
| SHBG at baseline (log10 nmol/L) | 0.022 | 0.496 (0.1,0.9) | 0.258 | 0.011 | 0.522 (0.1,0.9) | 0.561 |
| CYP2B6 516G>T (rs3745274) | 0.118 | 0.104 (0.0,0.2) | 0.130 | 0.048 | 0.115 (0.0,0.2) | 0.561 |
| log10 AUC0–24 weeks | P value | β value (95% CI) | r2 | P value | β value (95% CI) | r2 |
| CD4 (log10cells/mm3) | 0.034 | −0.282 (−0.5,0.0) | 0.227 | |||
| SHBG at baseline (log10 pg/mL) | 0.008 | 0.465 (0.1,0.8) | 0.333 | 0.004 | 0.484 (0.2,0.8) | 0.457 |
| CYP2B6 516G>T (rs3745274) | 0.179 | 0.075 (0.0,0.0) | 0.098 | |||
Univariate linear regression (P=≤0.2) completed, all statistically significant results then carried through to multivariate linear regression analysis (P=≤0.05). All statistically significant variables from multivariate linear regression shown in bold type.
β is the regression coefficient and represents incremental change in the log10 LNG pharmacokinetic parameter per unit change in a patient characteristic (e.g. per kg body weight or per allele carried). So if β=0.5 an increase per unit in the patient characteristic results in the log10 LNG pharmacokinetic parameter increasing by a factor of 0.5.
Levonorgestrel, efavirenz and nevirapine pharmacokinetics
LNG Cmax, Tmax, Cmin, and AUC0–24week are summarized by study group and SNP in Table 3. EFV C12–14h (mg/L) was 76% higher in homozygous CYP2B6 516 TT participants compared to those who were homozygous for the G allele. Within the NVP group NVP C11–13h (mg/L) was 5% lower in heterozygous CYP2B6 516 CT participants compared to homozygotes for the G allele.
Table 3.
Levonorgestrel (LNG), Efavirenz (EFV) and Nevirapine (NVP) pharmacokinetic parameters shown as median (interquartile range), summarized by associated CYP2B6, NR1I2 or CYP2A6 genotype.
| LNG Cmax (pg/mL) | CYP2B6 516G>T (rs3745274) | CYP2B6 15582C>T (rs4803419) | CYP2A6*9B 1836G>T (rs8192726) | NR1I2 63396C>T (rs2472677) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GT | TT | CC | CT | TT | GG | GT | TT | CC | CT | TT | |
| EFV group | 692.57 (486.82–887.95) | 402.40 (223.07–1027.65) | 200.63 | 692.57 (501.09–706.56) | 486.82 (347.10–775.28) | - | 486.82 (359.66–706.56) | 585.19 (223.07–1405.43) | - | 486.82 (347.10–775.28) | 463.03 (419.79–1027.65) | 585.19 |
| NVP group | 1310.78 (1069.99–2530.06) | 1449.53 (1053.42–1836.94) | - | 1097.94 (814.90–2157.00) | 1449.53 (1069.99–1836.94) | 1267.16 | 1449.53 (1267.16–1836.94) | 1097.94 (1069.99–1543.09) | - | 1436.43 (1054.42–1877.35) | 1310.78 (1303.20–1818.52) | 1466.49 (775.98–2157.00) |
| LNG Tmax (wk) | ||||||||||||
| EFV group | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 4.00 | 1.00 (1.00–1.00) | 1.00 (1.00–12.00) | - | 1.00 (1.00–4.00) | 1.00 (1.00–36.00) | - | 1.00 (1.00–12.00) | 1.00 (1.00–1.00) | 1.00 |
| NVP group | 1.00 (1.00–1.00) | 1.00 (1.00–12.00) | - | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | - | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 6.50 (1.00–12.00) |
| LNG Cmin (pg/mL) | ||||||||||||
| EFV group | 272.21 (189.45–377.85) | 202.23 (175.86–359.66) | 121.82 | 298.99 (272.98–307.27) | 208.13 (188.23–274.21) | - | 224.66 (188.23–307.27) | 202.23 (189.45–1405.43) | - | 223.07 (188.23–359.66) | 202.23 (189.13–298.99) | 189.45 |
| NVP group | 553.65 (365.16–1094.09) | 562.62 (504.42–807.67) | - | 504.42 (343.67–713.96) | 562.38 (512.46–986.40) | 526.38 | 562.38 (504.42–807.67) | 724.96 (540.18–986.40) | - | 540.18 (501.00–956.95) | 562.62 (562.38–986.40) | 599.33 (512.46–686.21) |
| LNG AUC0–24weeks (pg*wk/mL) | ||||||||||||
| EFV group | 10114.61 (8069.30–12862.18) | 6311.80 (6569.71–14937.71) | 3622.06 | 10114.61 (8499.98–12798.81) | 7754.35 (5676.92–9435.56) | - | 7754.35 (6311.80–11399.63) | 8069.30 (4321.25–11070.02) | - | 6830.55 (5676.92–11399.63) | 7754.35 (7371.57–10114.61) | 8069.30 |
| NVP group | 19970.24 (15259.29–30339.86) | 15965.35 (15729.55–26301.13) | - | 15761.90 (11338.65–22478.35) | 20167.18 (15729.55–27442.02) | 19970.24 | 19970.24 (15761.90–26301.13) | 16948.72 (15258.29–21199.91) | - | 16948.72 (15729.55–26301.13) | 16759.11 (15258.29–27442.02) | 19189.39 (15900.44–22478.35) |
| EFV C12–14h (mg/L) | 2.06 (1.39–2.61) | 2.63 (2.05–6.89) | 8.71 | 2.38 (2.20–2.63) | 2.35 (1.75–5.06) | - | 2.38 (1.75–3.03) | 2.35 (1.99–6.64) | - | 2.61 (2.05–6.64) | 2.21 (2.01–2.38) | 1.62 |
| NVP C11–13h (mg/L) | 6.51 (4.40–7.36) | 6.16 (5.10–10.03) | - | 6.75 (5.85–8.33) | 6.10 (4.95–8.34) | 8.01 | 6.10 (5.10–8.42) | 6.63 (5.45–7.14) | - | 6.75 (4.81–8.42) | 6.16 (5.50–6.86) | 7.06 (5.85–8.27) |
EFV C12–14h (mg/L) and NVP C11–13h (mg/L) determined from individual participant’s geometric mean value calculated from concentration measured at study entry, week 1, 4, 12, 24, 36 and 48 and summarized for the group as median (interquartile range).
Efavirenz group
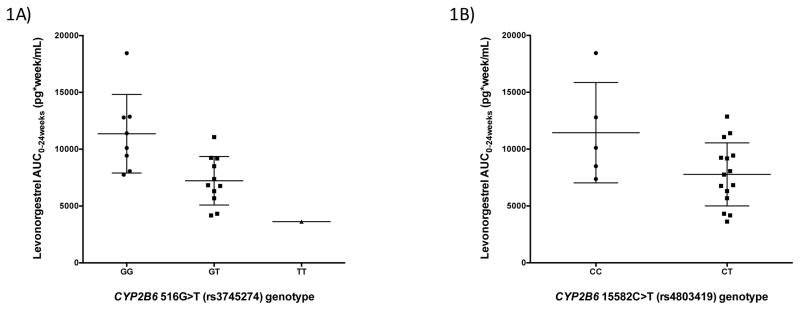
All patients in the EFV group (n=20) received EFV 600mg daily plus 2 NRTIs for a median (IQR) of 10.5 months (6.3–37.8) prior to study entry. After multivariate analysis, CYP2B6 516G>T (rs3745274) was associated with lower log10 LNG Cmax (β=−0.209, P=0.021) and log10 LNG AUC0–24weeks (β=−0.132, P=0.023). This meant that LNG Cmax median values were 692.8, 402.4 and 200.6 pg/mL in the GG, GT and TT genotype groups, respectively (71% difference between homozygote groups) and LNG AUC0–24weeks median values were 10,114.6, 6311.8 and 3622.1 pg*wk/mL in the GG, GT and TT genotype groups, respectively (64% difference between homozygote groups). CYP2B6 15582C>T (rs4803419) was associated with lower log10 LNG AUC0–24weeks (β=−0.163, P=0.021), resulting in LNG AUC0–24weeks median values of 10,114.6 and 7754.4 pg*wk/mL in the CC and CT genotype groups, respectively (23% difference between groups). Figure 1 illustrates the LNG AUC0–24weeks within the EFV group according to these genotypes. In addition to these genetic associations, age was inversely associated with log10 LNG AUC0–24weeks (β=−0.018 per 1 year, P=0.039) (Table 2).
Figure 1. Levonorgestrel AUC0–24weeks within the efavirenz group shown by statistically significant genotype.
Levonorgestrel AUC0–24weeks compared by statistically significant genotype.
Data is represented by median (interquartile range) and compared by genotype for each of the single nucleotide polymorphisms (SNPs) significantly associated levonorgestrel log10 AUC0–24weeks found through multivariate analysis (P=0.05) within the efavirenz group.
Consistent with previous reports, through univariate linear regression analysis, CYP2B6 516G>T was associated with higher EFV concentration (β=0.269, P=0.011), meaning that C12–14h values were 2.1, 2.6 and 8.7 mg/L in GG, GT and TT genotype groups, respectively (76% difference between homozygote groups). Additionally, higher EFV C12–14h (log10 mg/L) was associated with lower LNG log10 AUC0–24week (β=−0.030, P=0.003).
Nevirapine group
Patients in the NVP group (n=20) received NVP 200mg twice daily plus 2 NRTIs for a median (IQR) of 30.5 months (13.5–80.3) prior to study entry. CYP2B6 516G>T (rs3745274) was associated with higher log10 LNG Cmax (β=0.103, P=0.034) and log10 LNG Cmin (β=0.115, P=0.048). Resulting in LNG Cmax median values of 1310.8 and 1449.5 pg/mL in the GG and GT genotype groups, respectively (10% difference between homozygote groups) and LNG Cmin median values of 553.7 and 562.6 pg/mL in the GG and GT genotype groups (2% difference between groups). NR1I2 63396C>T was associated with delayed LNG Tmax (β=1.623, P=0.003) with a median value of 1.0, 1.0 and 6.5 weeks in the CC, CT and TT genotype groups. Additionally, log10 weight (β=−15.910, P=0.002) and albumin concentration at baseline (β=−0.340, P=0.003) were associated with shorter LNG Tmax. Log10 SHBG was associated with higher log10 LNG AUC0–24weeks (β=0.484, P=0.004), log10 LNG Cmin (β=0.522, P=0.011) and log10 LNG Cmax (β=0.469, P=0.007). Age was associated with higher log10 LNG Cmax (β=0.013, P=0.044) (Table 2).
No other genetic associations were observed in any of the groups; however, the sample size was insufficient to robustly assess the CYP2B6 983T>C variant (rs28399499).
Pharmacodynamic outcome in the EFV group
Three women in the EFV group became pregnant between study weeks 36 and 48 (3 (15%) of 20 participants). Pharmacogenetic characteristics of these three patients were as follows for the statistically significant genotypes within the EFV group: One woman was both homozygous TT for CYP2B6 516G>T (rs3745274) and heterozygous CT for CYP2B6 15582C>T (rs4803419). She was the only patient who possessed both of these genotypes in the study group, and her LNG concentration at the study visit prior to pregnancy (week 36) was 122 pg/mL (LNG AUC0–24weeks 3622 pg*wk/mL). The other two women were heterozygous CT for CYP2B6 516G>T and homozygous CC for CYP2B6 1582 C>T. They were the only patients in the EFV group who possessed these genotypes, and their LNG concentration prior to pregnancy (week 36) were 299 and 303 pg/mL (LNG AUC0–24weeks 10,115 and 8500 pg*wk/mL), respectively.
Discussion
The impact of this investigation is greatest for women receiving a combination of EFV-based ART with a LNG subdermal implant. Women receiving this drug combination are at risk for suboptimal LNG exposure, which results in a higher rate of contraceptive failures.(13, 14) For women who are heterozygous or homozygous for CYP2B6 516G>T or CYP2B6 15582C>T, which was associated with lower LNG AUC when combined with EFV, this drug-drug interaction will be more pronounced. This was evidenced by the 3 patients who became pregnant in the EFV group: all possessed at least one of these genotypes and the patient who possessed both significant genotypes had the lowest LNG exposure of any study participant. Importantly, nine additional women in the EFV group were heterozygous for either CYP2B6 516G>T or CYP2B6 15582C>T, and had LNG exposure (median LNG AUC0–24weeks 6831 pg*wk/mL), consistent with those women who became pregnant, indicating that they were also at risk for contraceptive failure. We hypothesize that high EFV plasma concentrations associated with these SNPs may result in greater EFV induction of CYP3A4, resulting in increased LNG metabolism and lower LNG exposure in the patients who were heterozygous or homozygous for CYP2B6 516G>T or CYP2B6 15582C>T. Further contributing to this effect may be the modest inhibitory effect of LNG on CYP2B6.(36, 37) It is possible that LNG may inhibit CYP2B6 activity, resulting in high EFV concentrations and further inhibition of CYP3A4.
Within the NVP group, the significant association between CYP2B6 516G>T and higher LNG log10 Cmax and LNG log10 Cmin is surprising, given that NVP is an inducer of CYP3A4, the enzyme believed to be primarily responsible for LNG metabolism. In a previous study of three HIV-positive Malawian women receiving 30 μg ethinyl estradiol/300 μg norgestrel (a racemic mixture of LNG and dextronorgestrel) and NVP-based ART, higher LNG exposure was also observed compared to HIV-uninfected women.(38) Notably, two of the three women were heterozygous for CYP2B6 516G>T. Furthermore, CYP2B6 516G>T has been associated with elevated NVP concentrations in previous studies.(1, 39) Therefore, the association between CYP2B6 516G>T and LNG PK parameters observed in our study could be attributed to NVP-mediated inhibition of LNG metabolism by CYP3A4, or potential inhibition of CYP3A5. However, this result must be interpreted with caution, given that overall exposure (as measured by AUC) was not associated with the same SNP. Further investigation is required to determine the mechanism underpinning this finding.
We observed an association between NR1I2 63396C>T (rs2472677) and delayed LNG Tmax in patients on NVP, which has the potential to result in suboptimal LNG concentrations. Previously, NR1I2 63396C>T (in linkage disequilibrium (LD) with 63704G>A, 63813(CAAA)n/(CA)n, and 65104T>C) has been associated with higher NR1I2 expression, elevated CYP3A4 expression, and unboosted atazanavir plasma concentrations.(23, 40, 41) NR1I2 63396C>T may be directly or indirectly associated with inducibility of CYP3A4 and hence reduced LNG metabolism. This is supported by observations in primary human hepatocytes where the 63396T allele was associated with higher basal activity but lower rifampicin-mediated inducibility of CYP3A4.(23)
LNG pharmacokinetics demonstrate high inter-individual variability, often attributed to variations in protein binding(42) and bodyweight.(43–45) Although we did not observe a consistent association with weight and LNG PK, this may be related to the lower median body weight of HIV-infected Ugandan women compared to those in other studied populations. LNG is highly protein-bound to sex hormone-binding globulin (SHBG) and albumin. (42) Elevated log10 SHBG concentration has previously been associated with higher serum LNG concentration.(46) However, the observed association of log10 SHBG concentration at baseline with higher LNG log10 Cmax, LNG log10 Cmin and LNG log10 AUC0–24weeks, and of albumin concentration at baseline with lower Tmax within the NVP group, may reflect some influence of baseline protein binding on overall LNG exposure.
Limitations of this study include a relatively small sample size, particularly related to CYP2B6 15582C>T (rs4803419) and NR1I3 540C>T (rs2307424), as both SNPs were not in Hardy Weinberg equilibrium. In addition, the consequences of the Tmax associations are unclear, but should in any case be interpreted cautiously given the intermittent measurement of LNG PK during the study period.
Our study demonstrates pharmacogenetic associations with LNG PK when administered as a LNG subdermal implant in HIV-positive women receiving EFV- or NVP-based ART. Even with the EFV interaction with LNG, effectiveness of the LNG subdermal implant may still be higher than that for oral contraception due to improved medication adherence.(14, 47) Notwithstanding, any increased risk of pregnancy during contraception is important, given the impact that unintended pregnancy has for women living with HIV. Based upon our findings, screening for SNPs associated with low LNG exposure in patients receiving EFV represents a novel approach to identify women at the highest risk for an unintended pregnancy. Added pharmacogenetic knowledge would allow clinicians to personalize counseling of women on their choice of a contraceptive method based on individualized risks and benefits. Overall, this study highlights the potential role of affordable, accessible and clinically-relevant pharmacogenetic screening in resource-constrained settings. Specifically, future studies with larger populations and a more diverse range of ethnicities, should focus upon prospective evaluation of the relationship between pharmacogenetics and the effectiveness of the LNG implant in the presence of EFV-related drug-drug interactions.
Methods
Aim of the study
To assess the association between LNG PK and SNPs in CYP2B6, CYP2A6, NR1I3 and NR1I2 in Ugandan HIV-positive women participating in a drug-drug interaction PK study. (13)
Ethical approval
All study procedures followed the Helsinki Declaration of 1975 and were approved by ethics boards at the Joint Clinical Research Centre Kampala, Uganda National Council for Science and Technology, and the University of Nebraska Medical Centre. The study was registered on clinicaltrials.gov (NCT01789879).
Study design and cohort
This was a prospective PK evaluation of HIV-positive Ugandan women receiving EFV-based ART (EFV group, n=20), NVP-based ART (NVP group, n=20), or ART-naïve (n=17). For the pharmacogenetic study, all participants in the EFV and NVP group were included (n=40) and the ART-naïve group was excluded. The two-rod (75mg/rod) LNG sub-dermal implant was inserted in all patients upon study entry. Inclusion criteria for the EFV and NVP groups included prescription of EFV or NVP plus 2 NRTIs for 30 days or longer and a HIV-RNA of <400 copies/mL. (48)
Sample and data collection
Over a total of 48 weeks, a plasma sample was collected at weeks 1, 4, 12, 24, 36 and 48 to assess LNG, NVP and EFV PK. Follow-up was interrupted for 9 subjects between weeks 36–44 after 3 pregnancies were identified in the EFV group. The primary endpoint for the PK analysis was 24 weeks. Timing of blood sample collection for the EFV group was based on EFV mid-dosing interval at 12–14 hours post dose and for the NVP group based on the end dosing interval of 11–13 hours post dose. PK parameters included in this analysis were area under the concentration time curve (AUC) from entry to week 24 (AUC0–24 weeks), maximum concentration (Cmax), time to Cmax (Tmax) and minimum concentration (Cmin). Cmax and Cmin represent the highest and lowest concentrations observed over the entire study period. AUC was calculated using the trapezoidal rule (Phoenix WinNonlin, Certara®). LNG concentrations were analyzed by a validated LC-MS/MS method; EFV and NVP plasma concentrations were determined using HPLC assays with ultraviolet detection as previously outlined by Scarsi et al. (13)
Genotyping
Genomic DNA was extracted from whole blood through use of the manufacturers protocol (E.Z.N.A Blood DNA Mini Kit; Omega bio-tek; Norcross, GA). Extracted DNA was quantified using NanoDrop (Thermo Fisher Scientific, Wilmington, DE). Genotyping was completed using real-time allelic discrimination polymerase chain reaction (PCR) assay on a DNA Engine Chromo4 system (Bio-Rad Laboratories, Hercules, CA). The PCR protocol followed denaturation at 95°C for 10 minutes, followed by 50 cycles of amplification at 92°C for 15 seconds and annealing at 60°C for 1 minute 30 seconds. Taqman Genotyping Master mix and assays CYP2B6 516G>T (rs3745274, catalogue number C___7817765_60), CYP2B6 983T>C (rs28399499, catalogue number C__60732328_20), CYP2B6 15582C>T (rs4803419, catalogue number C___7817764_10), CYP2A6 -48A>C (rs28399433, catalogue number C__30634332_10) CYP2A6*9B 1836G>T (rs8192726, catalogue number C__29560333_20) NR1I2 63396C>T (rs2472677, catalogue number C_2607845), NR1I3 540C>T (rs2307424, catalogue number C__25746794_20) and NR1I3 1089T>C (rs3003596, catalogue number C__16194070_10) were purchased from Life Technologies (Paisley, Renfrewshire, UK). Opticon Monitor v.3.1 software (Bio-Rad Laboratories) was used to obtain allelic discrimination plots and identify genotypes.
Statistical analysis
Compliance with Hardy Weinberg equilibrium was tested using previously outlined methods.(49) Genotypes were coded for regression analyses as 0=homozygous common allele, 1=heterozygous and 2=homozygous variant allele. Categorical variables were described using relative frequencies, whilst continuous variables were described using median and IQR. The Shapiro-Wilk Test was used to test for normality, with a P value ≤0.05 considered as statistically significant. A univariate analysis through linear enter regression was carried out in order to identify independent variables associated with LNG PK parameters within all patients, the EFV group, and the NVP group. Variables with a P value ≤0.2 for the univariate analysis were carried through to a linear backwards multivariate analysis were a P value ≤0.05 was classed as statistically significant. All statistical analyses were carried out using IBM SPSS Statistics v.22 (IBM Armonk, NY). All charts were produced using GraphPad Prism 6 (GraphPad Software, La Jolla, CA).
Supplementary Material
Study highlights.
1. What is the current knowledge on the topic?
Use of the subdermal levonorgestrel implant with efavirenz-based antiretroviral therapy (ART) results in suboptimal concentrations of levonorgestrel, a result that was not observed for patients receiving nevirapine. CYP2B6, CYP2A6, NR1I3 and NR1I2 single nucleotide polymorphisms (SNPs) have previously been associated, through multiple studies, with alterations in efavirenz and nevirapine concentrations.
2. What question did this study address?
Are SNPs in genes involved in efavirenz and nevirapine metabolism linked to the reduced LNG concentrations observed from a drug-drug interaction between levonorgestrel and ART?
3. How this might change clinical pharmacology or translation science
This is the first study to demonstrate an association between pharmacogenetic variations in CYP2B6 and the extent of the drug-drug interaction observed when sub-dermal levonorgestrel was combined with ART. In total, this manuscript adds to the current understanding of the mechanism by which efavirenz- and nevirapine-based ART interacts with the LNG sub-dermal implant.
Acknowledgments
This project was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the National Institutes of Health [grant numbers 5R21HD074462 and 1R01HD085887 (Scarsi)]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank the patients, staff, and administration of the Infectious Diseases Institute, Makerere University, Kampla, Uganda, for their collaboration and support.
Footnotes
Author responsibility
The corresponding author accepts final responsibility and confirms access to all data in the study.
Conflict of interest/Disclosure
Megan Neary, Kimberly Scarsi have no conflict of interest/disclosures.
Author contributions:
M.N., M.S., A.O., and K.S. wrote the manuscript; M.L., K.D., C.M., P.B., D.B., A.O., and K.S. designed the research; M.N., M.L., A.O., K.D., C.M., P.B., D.B., and K.S. performed the research, M.N. analyzed the data; M.S. and A.O. contributed new reagents/analytical tools.
Contributor Information
Megan Neary, Molecular and Clinical Pharmacology, Institute of Translational Medicine, University of Liverpool, Liverpool, UK.
Mohammed Lamorde, Infectious Diseases Institute, Makerere University College of Health Sciences, Kampala, Uganda.
Adeniyi Olagunju, Faculty of Pharmacy, Obafemi Awolowo University, Ile-Ife, Nigeria.
Kristin M. Darin, Center for Global Health, Northwestern University Feinberg School of Medicine, Chicago, USA
Concepta Merry, Department of Medicine, Trinity College Dublin, Dublin, Ireland.
Pauline Byakika-Kibwika, Infectious Diseases Institute, Makerere University College of Health Sciences, Kampala, Uganda.
David J. Back, Molecular and Clinical Pharmacology, Institute of Translational Medicine, University of Liverpool, Liverpool, UK
Marco Siccardi, Molecular and Clinical Pharmacology, Institute of Translational Medicine, University of Liverpool, Liverpool, UK.
Andrew Owen, Molecular and Clinical Pharmacology, Institute of Translational Medicine, University of Liverpool, Liverpool, UK.
Kimberly K. Scarsi, Department of Pharmacy Practice, College of Pharmacy, University of Nebraska Medical Center, Omaha, USA
References
- 1.Penzak S, Kabuye G, Mugyenyi P, Mbamanya F, Natarajan V, Alfaro R, et al. Cytochrome P450 2B6 (CYP2B6) G516T influences nevirapine plasma concentrations in HIV-infected patients in Uganda. HIV medicine. 2007;8(2):86–91. doi: 10.1111/j.1468-1293.2007.00432.x. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. Geneva: World Health Organization; Sep, 2015. [PubMed] [Google Scholar]
- 3.Panel on Treatment of HIV-Infected Pregnant Women and Prevention of Perinatal Transmission. [Accessed: 2 May 2016];Recommendations for Use of Antiretroviral Drugs in Pregnant HIV-1- Infected Women for Maternal Health and Interventions to Reduce Perinatal HIV Transmission in the United States. Available at http://aidsinfo.nih.gov/contentfiles/lvguidelines/PerinatalGL.pdf.
- 4.Lehtovirta P, Paavonen J, Heikinheimo O. Experience with the levonorgestrel-releasing intrauterine system among HIV-infected women. Contraception. 2007;75(1):37–9. doi: 10.1016/j.contraception.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Heikinheimo O, Lehtovirta P, Aho I, Ristola M, Paavonen J. The levonorgestrel-releasing intrauterine system in human immunodeficiency virus–infected women: a 5-year follow-up study. American Journal of Obstetrics and Gynecology. 2011;204(2):126e1–e4. doi: 10.1016/j.ajog.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Kopp JB, Winkler C. HIV-associated nephropathy in African Americans1. Kidney international. 2003;63:S43–S9. doi: 10.1046/j.1523-1755.63.s83.10.x. [DOI] [PubMed] [Google Scholar]
- 7.Phillips SJ, Curtis KM, Polis CB. Effect of hormonal contraceptive methods on HIV disease progression: a systematic review. Aids. 2013;27(5):787–94. doi: 10.1097/QAD.0b013e32835bb672. [DOI] [PubMed] [Google Scholar]
- 8.Organization WH. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. 2016. [PubMed] [Google Scholar]
- 9.Ford N, Calmy A, Mofenson L. Safety of efavirenz in the first trimester of pregnancy: an updated systematic review and meta-analysis. Aids. 2011;25(18):2301–4. doi: 10.1097/QAD.0b013e32834cdb71. [DOI] [PubMed] [Google Scholar]
- 10.Organization WH. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Geneva: World Health Organization; 2013. [PubMed] [Google Scholar]
- 11.Organization WH. Use of Efavirenz During Pregnancy: A Public Health Perspective: Technical Update on Treatment Optimization. World Health Organization; 2012. [Google Scholar]
- 12.Health WHOR. Medical eligibility criteria for contraceptive use. World Health Organization; 2010. [Google Scholar]
- 13.Scarsi KK, Darin KM, Nakalema S, Back DJ, Byakika-Kibwika P, Else LJ, et al. Unintended pregnancies observed with combined use of the levonorgestrel contraceptive implant and efavirenz-based antiretroviral therapy: a three-arm pharmacokinetic evaluation over 48 weeks. Clinical Infectious Diseases. 2016;62(6):675–82. doi: 10.1093/cid/civ1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel RC, Onono M, Gandhi M, Blat C, Hagey J, Shade SB, et al. Pregnancy rates in HIV-positive women using contraceptives and efavirenz-based or nevirapine-based antiretroviral therapy in Kenya: a retrospective cohort study. The Lancet HIV. 2015;2(11):e474–e82. doi: 10.1016/S2352-3018(15)00184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno I, Quiñones L, Catalán J, Miranda C, Roco Á, Sasso J, et al. Influence of CYP3A4/5 polymorphisms in the pharmacokinetics of levonorgestrel: a pilot study. Biomédica. 2012;32(4):570–7. doi: 10.1590/S0120-41572012000400012. [DOI] [PubMed] [Google Scholar]
- 16.Bélanger A-S, Caron P, Harvey M, Zimmerman PA, Mehlotra RK, Guillemette C. Glucuronidation of the antiretroviral drug efavirenz by UGT2B7 and an in vitro investigation of drug-drug interaction with zidovudine. Drug Metabolism and Disposition. 2009;37(9):1793–6. doi: 10.1124/dmd.109.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogburn ET, Jones DR, Masters AR, Xu C, Guo Y, Desta Z. Efavirenz primary and secondary metabolism in vitro and in vivo: identification of novel metabolic pathways and cytochrome P450 2A6 as the principal catalyst of efavirenz 7-hydroxylation. Drug Metabolism and Disposition. 2010;38(7):1218–29. doi: 10.1124/dmd.109.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan-Havard P, Liu Z, Chou M, Ling Y, Barrail-Tran A, Haas DW, et al. Pharmacokinetics of phase I nevirapine metabolites following a single dose and at steady state. Antimicrobial agents and chemotherapy. 2013;57(5):2154–60. doi: 10.1128/AAC.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cammett AM, MacGregor TR, Wruck JM, Felizarta F, Miailhes P, Mallolas J, et al. Pharmacokinetic assessment of nevirapine and metabolites in human immunodeficiency virus type 1-infected patients with hepatic fibrosis. Antimicrobial agents and chemotherapy. 2009;53(10):4147–52. doi: 10.1128/AAC.00460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyen C, Hendra H, Siccardi M, Platten M, Jaeger H, Harrer T, et al. Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. Journal of antimicrobial chemotherapy. 2011:dkr272. doi: 10.1093/jac/dkr272. [DOI] [PubMed]
- 21.Zhang J, Kuehl P, Green ED, Touchman JW, Watkins PB, Daly A, et al. The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics and Genomics. 2001;11(7):555–72. doi: 10.1097/00008571-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Swart M, Whitehorn H, Ren Y, Smith P, Ramesar RS, Dandara C. PXR and CAR single nucleotide polymorphisms influence plasma efavirenz levels in South African HIV/AIDS patients. BMC medical genetics. 2012;13(1):1. doi: 10.1186/1471-2350-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metabolism and Disposition. 2008;36(1):169–81. doi: 10.1124/dmd.107.016600. [DOI] [PubMed] [Google Scholar]
- 24.Lamba J, Lamba V, Schuetz E. Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Current drug metabolism. 2005;6(4):369–83. doi: 10.2174/1389200054633880. [DOI] [PubMed] [Google Scholar]
- 25.Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH. CYP2B6 (c. 516G→ T) and CYP2A6 (* 9B and/or* 17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. British journal of clinical pharmacology. 2009;67(4):427–36. doi: 10.1111/j.1365-2125.2009.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas DW, Smeaton LM, Shafer RW, Robbins GK, Morse GD, Labbé L, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing Efavirenz and/or Nelfinavir: an Adult Aids Clinical Trials Group Study. Journal of Infectious Diseases. 2005;192(11):1931–42. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 27.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. Aids. 2004;18(18):2391–400. [PubMed] [Google Scholar]
- 28.Wang J, Sönnerborg A, Rane A, Josephson F, Lundgren S, Ståhle L, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenetics and genomics. 2006;16(3):191–8. doi: 10.1097/01.fpc.0000189797.03845.90. [DOI] [PubMed] [Google Scholar]
- 29.Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. 2007 doi: 10.2217/14622416.8.6.547. [DOI] [PubMed] [Google Scholar]
- 30.Sarfo FS, Zhang Y, Egan D, Tetteh LA, Phillips R, Bedu-Addo G, et al. Pharmacogenetic associations with plasma efavirenz concentrations and clinical correlates in a retrospective cohort of Ghanaian HIV-infected patients. Journal of Antimicrobial Chemotherapy. 2013:dkt372. doi: 10.1093/jac/dkt372. [DOI] [PubMed] [Google Scholar]
- 31.Oluka MN, Okalebo FA, Guantai AN, McClelland RS, Graham SM. Cytochrome P450 2B6 genetic variants are associated with plasma nevirapine levels and clinical response in HIV-1 infected Kenyan women: a prospective cohort study. AIDS research and therapy. 2015;12(1):1. doi: 10.1186/s12981-015-0052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehlotra RK, Bockarie MJ, Zimmerman PA. CYP2B6 983T> C polymorphism is prevalent in West Africa but absent in Papua New Guinea: implications for HIV/AIDS treatment. British journal of clinical pharmacology. 2007;64(3):391–5. doi: 10.1111/j.1365-2125.2007.02884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solus JF, Arietta BJ, Harris JR, Sexton DP, Steward JQ, McMunn C, et al. Genetic variation in eleven phase I drug metabolism genes in an ethnically diverse population. Pharmacogenomics. 2004;5(7):895–931. doi: 10.1517/14622416.5.7.895. [DOI] [PubMed] [Google Scholar]
- 34.Bertrand J, Chou M, Richardson DM, Verstuyft C, Leger PD, Mentré F, et al. Multiple genetic variants predict steady-state nevirapine clearance in HIV-infected Cambodians. Pharmacogenetics and genomics. 2012;22(12):868. doi: 10.1097/FPC.0b013e32835a5af2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gounden V, Van Niekerk C, Snyman T, George JA. Presence of the CYP 2 B 6 516 G> T polymorphism, increased plasma Efavirenz concentrations and early neuropsychiatric side effects in South African HIV-infected patients. AIDS research and therapy. 2010;7:32. doi: 10.1186/1742-6405-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsky RL, Astuccio AV, Obach RS. Evaluation of 227 drugs for in vitro inhibition of cytochrome P450 2B6. The Journal of Clinical Pharmacology. 2006;46(12):1426–38. doi: 10.1177/0091270006293753. [DOI] [PubMed] [Google Scholar]
- 37.Palovaara S, Pelkonen O, Uusitalo J, Lundgren S, Laine K. Inhibition of cytochrome P450 2B6 activity by hormone replacement therapy and oral contraceptive as measured by bupropion hydroxylation. Clinical Pharmacology & Therapeutics. 2003;74(4):326–33. doi: 10.1016/S0009-9236(03)00202-9. [DOI] [PubMed] [Google Scholar]
- 38.Stuart GS, Moses A, Corbett A, Phiri G, Kumwenda W, Mkandawire N, et al. Combined oral contraceptives and antiretroviral PK/PD in Malawian women: pharmacokinetics and pharmacodynamics of a combined oral contraceptive and a generic combined formulation antiretroviral in Malawi. Journal of acquired immune deficiency syndromes (1999) 2011;58(2):e40–3. doi: 10.1097/QAI.0b013e31822b8bf8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenetics and genomics. 2005;15(1):1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 40.Schipani A, Siccardi M, D’Avolio A, Baietto L, Simiele M, Bonora S, et al. Population pharmacokinetic modeling of the association between 63396C→ T pregnane X receptor polymorphism and unboosted atazanavir clearance. Antimicrobial agents and chemotherapy. 2010;54(12):5242–50. doi: 10.1128/AAC.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siccardi M, D’Avolio A, Baietto L, Gibbons S, Sciandra M, Colucci D, et al. Association of a single-nucleotide polymorphism in the pregnane X receptor (PXR 63396C→ T) with reduced concentrations of unboosted atazanavir. Clinical infectious diseases. 2008;47(9):1222–5. doi: 10.1086/592304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coukell AJ, Balfour JA. Levonorgestrel subdermal implants. Drugs. 1998;55(6):861–87. doi: 10.2165/00003495-199855060-00019. [DOI] [PubMed] [Google Scholar]
- 43.Sivin I, Lahteenmaki P, Mishell DR, Jr, Alvarez F, Diaz S, Ranta S, et al. First week drug concentrations in women with levonorgestrel rod or Norplant capsule implants. Contraception. 1997;56(5):317–21. doi: 10.1016/s0010-7824(97)00153-4. [DOI] [PubMed] [Google Scholar]
- 44.Sivin I, Lahteenmaki P, Ranta S, Darney P, Klaisle C, Wan L, et al. Levonorgestrel concentrations during use of levonorgestrel rod (LNG ROD) implants. Contraception. 1997;55(2):81–5. doi: 10.1016/s0010-7824(96)00276-4. [DOI] [PubMed] [Google Scholar]
- 45.Sivin I, Wan L, Ranta S, Alvarez F, Brache V, Mishell DR, Jr, et al. Levonorgestrel concentrations during 7 years of continuous use of Jadelle contraceptive implants. Contraception. 2001;64(1):43–9. doi: 10.1016/s0010-7824(01)00226-8. [DOI] [PubMed] [Google Scholar]
- 46.Abdalla K, Shabaan M, Stanczyk F. Interrelationship of serum levonorgestrel and sex hormone-binding globulin levels following vaginal and oral administration of combined steroid contraceptive tablets. Contraception. 1992;45(2):111–8. doi: 10.1016/0010-7824(92)90045-u. [DOI] [PubMed] [Google Scholar]
- 47.Shelton JD. Reduced Effectiveness of Contraceptive Implants for Women Taking the Antiretroviral Efavirenz (EFV): Still Good Enough and for How Long? Global Health: Science and Practice. 2015;3(4):528–31. doi: 10.9745/GHSP-D-15-00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uganda Ministry of Health. Addendum to the national antiretroviral treatment guidelines. Dec, 2013. [Google Scholar]
- 49.Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. American journal of epidemiology. 2009;169(4):505–14. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.