Abstract
Pre-mRNA splicing is executed in mammalian cell nuclei within a huge (21 MDa) and highly dynamic molecular machine – the supraspliceosome - that individually package pre-mRNA transcripts of different sizes and number of introns into complexes of a unique structure, indicating their universal nature. Detailed structural analysis of this huge and complex structure requires a stepwise approach using hybrid methods. Structural studies of the supraspliceosome by room temperature electron tomography, cryo-electron tomography, and scanning transmission electron microscope mass measurements revealed that it is composed of four native spliceosomes, each resembling an in vitro assembled spliceosome, which are connected by the pre-mRNA. It also elucidated the arrangement of the native spliceosomes within the intact supraspliceosome. Native spliceosomes and supraspliceosomes contain all five spliceosomal U snRNPs together with other splicing factors, and are active in splicing. The structure of the native spliceosome, at a resolution of 20 Å, was determined by cryo-electron microscopy, and a unique spatial arrangement of the spliceosomal U snRNPs within the native spliceosome emerged from in-silico studies. The supraspliceosome also harbor components for all pre-mRNA processing activities. Thus the supraspliceosome - the endogenous spliceosome - is a stand-alone complete macromolecular machine capable of performing splicing, alternative splicing, and encompass all nuclear pre-mRNA processing activities that the pre-mRNA has to undergo before it can exit from the nucleus to the cytoplasm to encode for protein. Further high-resolution cryo-electron microscopy studies of the endogenous spliceosome are required to decipher the regulation of alternative splicing, and elucidate the network of processing activities within it.
Keywords: pre-mRNA splicing, supraspliceosome, electron tomography, scanning transmission electron microscopy mass measurements, cryo-electron microscopy, cryo-electron microscopy single particle reconstruction
1. Introduction
Eukaryotic pre-mRNAs must undergo several post-transcriptional modifications such as 5’-end capping, 3’-end processing, editing, and splicing before their export to the cytoplasm as functional mRNAs. The accurate splicing reaction involves cis elements (5’ and 3’ splice sites (SSs) consensus sequences, a branch site, a polypirimidine sequence and exonic and intronic splicing enhancers and silencers), which are recognized by trans factors, (the five-spliceosomal U small nuclear ribonucleoproteins (snRNPs): U1, U2, U4, U5 and U6 snRNPs, and non-snRNP protein splicing factors, such as the Serine/Arginine (SR) rich protein family and heterogeneous nuclear ribonucleoproteins (hnRNPs) (reviewed in Refs [1–3]).
Major effort in the field of splicing was invested in developing in vitro systems for the analysis of the mechanism of the splicing reaction, its components, and the structure of the splicing intermediate complexes (reviewed in Refs [1–3]). These studies revealed that the two-step splicing transesterification reactions take place in the spliceosome - a huge mega Dalton dynamic RNP complex - in which the pre-mRNA is assembled with the spliceosomal U snRNPs and protein splicing factors. The assembly of the spliceosome in vitro occurs in a step-wise manner, and involves an intricate series of dynamic interactions between the five major U snRNPs, which play a major role in splicing accuracy and catalysis, as well as with a number of non-snRNP splicing factors, which are recruited to the spliceosome when an exogenous pre-mRNA is added to a nuclear extract (reviewed in refs [2, 4]). Binding of U1 snRNP to the 5’SS of the pre-mRNA and of U2 snRNP to the branch-point, followed by the assembly of U4/U6.U5 tri-snRNP forms the inactive pre-catalytic B complex, that harbors the five-spliceosomal U snRNPs. This step is followed by major rearrangements in RNA:RNA and RNA:protein interactions, destabilization of U1 and U4 snRNPs and formation of activated complex Bact. This is followed by generation of complex B*, which catalyzes the first step of splicing, generating complex C. Complex C is converted to complex C*, which catalyzes the second step of splicing, resulting in the ligation of the two exons resulting in complex P, release of the intron lariat spliceosome (ILS), and recycling of the U snRNPs and protein components. Thus, the in vitro assembled splicing active complex contains U2/U6.U5 snRNPs [2]. Studies of the structure and function of the in vitro assembled spliceosome have been summarized in a number of reviews [2, 3].
For understanding the mechanism of a molecular machine as complex as the spliceosome, structural, biochemical and genetic studies are required. In view of its huge dimensions, complexity, and dynamics, electron microscopy (EM) has been the method of choice for the structural analysis of the spliceosome. Structural analysis of splicing intermediates assembled in vitro or splicing complexes isolated from cells’ nuclei have been performed and until recently were limited to resolutions lower than 20-Å (reviewed in Refs [5, 6]). The recent advancements in cryo-EM, with the development of a new generation of direct electron detectors, novel image processing tools, and improvements in microscope optic and stage stability, enabling structure determination at subnanometric resolution [7] have revolutionized the field of structural molecular biology, including the splicing field. The recent subnanometric structures of the tri-snRNP [8–10] and of the Bact [11, 12], C [13, 14], C* [15, 16] and ILS [17] splicing complexes assembled on a one intron transcript provide insight into the catalytic center of the spliceosome, comprised of Prp8, helix I of the U2/U6 snRNA duplex, the intramolecular stem loom (ISL) of U6 snRNA, Loop I of U5 snRNA and Mg2+ ions. These studies also highlight the dynamic changes that occur during the two steps of the splicing reaction [8–19].
Most RNA polymerase II (Pol II) transcripts are multi-intronic, and undergo alternative splicing (AS). This process is a major source for the diversity of the human proteome and plays a major role in the regulation of gene expression (reviewed in Refs [20–23]). Yet, the regulation of AS is at present not well understood. Importantly, defects in AS and misregulation of AS factors were shown to be involved in several human diseases including cancer [24–26]. Because most transcripts are multi-intronic, the endogenous spliceosome should be able to account for the regulation of constitutive and alternative splicing and for all the pre-mRNA processing activities. This regulation requires coordination between spliceosomes to select the correct splice junctions and generate a “meaningful” mRNA.
We took a different and unique top-down, approach, and have studied the splicing complex in its intact form, isolated from nuclei of living mammalian cells under native conditions, using novel methods we developed [27–29]. The complexes we have isolated from cell nuclei are much larger than the splicing complexes assembled in vitro, and are thus termed supraspliceosomes (Fig. 1). This 21 MDa RNP complex [30] is composed of four active native spliceosomes, each resembling the in vitro assembled spliceosome, which are connected via the pre-mRNA (reviewed in Refs [6, 31, 32]).
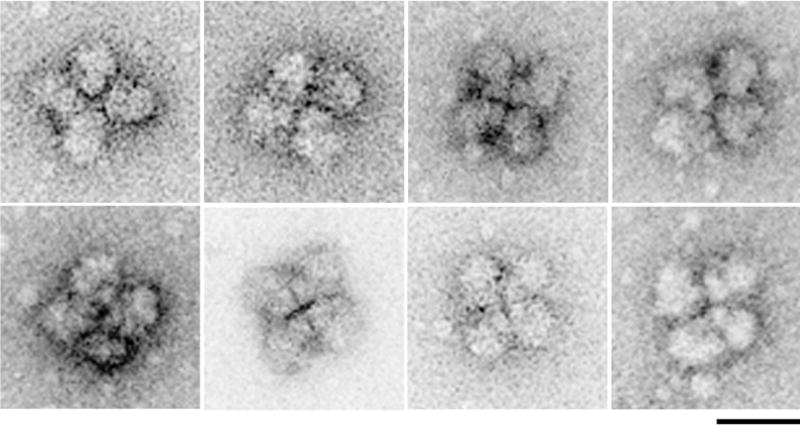
Fig. 1. EM visualization of supraspliceosomes.
A gallery of supraspliceosomes, each composed of four native spliceosomes. Supraspliceosomes were isolated from HeLa cell nuclei and fractionated on a glycerol gradient in 10 mM Tris-HCl, 0.1 M NaCl, 2 mM MgCl2, pH 8.0 [27, 37]. They were stained with uranyl acetate (1%) and applied to copper grids that were coated with carbon and glow discharged. Images were recorded by TEM (Tecnai T12, 120kV, LaB6 filament) at a magnification of 30,000. Scale bar represents 50 nm. Adapted from [80].
Here we review the structural studies of the huge (21 MDa) and dynamic supraspliceosomes, with EM as the preferred method. The complexity of the system also requires that the structural analysis will be carried out in steps and through an interdisciplinary approach, using hybrid methods, and combining structural biology and molecular biology.
Based on our finding that all nuclear pre-mRNAs, independent of their length or number of introns are individually packaged with all known splicing components in a 50×50 nm [33], 21 MDa supraspliceosome [27, 30, 33], we initiated our structural studies by room temperature electron tomography, followed by cryo-electron tomography [34–36]. These studies were accompanied by mass measurements by Scanning Transmission EM (STEM) [30] that confirmed the generality of the supraspliceosome particle and the similarity of its four substructures - the native spliceosomes - that are active in splicing[37]. The similarity of the native spliceosome, as confirmed by the mass measurements, as well as the flexibility of the individual subunits within the supraspliceosomes, as revealed by the EM studies, led to the isolation of the native spliceosome by specific cleavage of the pre-mRNA without cleaving the U snRNAs and to the determination of its structure by cryo-EM single particle techniques, at 20-Å resolution [29]. These experiments also confirmed that the supraspliceosome is composed of four native spliceosomes interconnected by the pre-mRNA [29, 37]. Native spliceosomes contain all five spliceosomal U snRNPs together with other splicing factors, and are functional in splicing (hence termed “native spliceosomes”) [29, 37, 38].
The supraspliceosome composed of four active native spliceosomes connected by the pre-mRNA provides a unique and general machine that can account for the extensive network of interactions and offers coordination and regulation of the different splicing events that a multiintronic pre-mRNA has to undergo. We have demonstrated that AS factors are predominantly found in supraspliceosomes, and that AS is regulated there [39, 40]. Notably, a quality control mechanism of pre-mRNA processing was identified in supraspliceosomes [31, 41, 42], as well as all the additional processing activities, such as 5’-end and 3’-end processing, and RNA editing [35, 43]. Furthermore, intronic pre-microRNAs were found in supraspliceosomes and their processing occurs there [44, 45]. Also, a sub-fraction of C/D BOX small nucleolar RNAs (SNORDs) assembled in non-canonical RNPs were found in supraspliceosomes likely playing a role in alternative splicing [46]. Support for the functional significance of supraspliceosomes was provided from a study showing that U2/U6 snRNA base pairing, which characterizes active spliceosomes assembled in vitro, was found in complexes sedimenting between 150–300S but not in 60S spliceosome complexes [47]. Supraspliceosomes were also visualized in the cell nucleus from the site of transcription to the nuclear pore [48]. Thus, the supraspliceosome – the endogenous spliceosome - is functional in all nuclear processing activities that the pre-mRNA has to undergo [6, 31, 32].
2. The endogenous spliceosome – the supraspliceosome
Our approach for the study of the endogenous spliceosome was through analysis of complexes isolated from cell nuclei under physiological conditions and having intact RNA (reviewed in Refs [6, 31, 32]).
2.1 Isolation and biochemical characterization of supraspliceosomes from live cells
To develop a protocol for the isolation of the endogenous spliceosome from mammalian cell nuclei under physiological conditions, we optimized conditions whereby over 85% of 3H-labeled Pol II transcripts could be released to the nuclear supernatant after mild sonication of the nuclei and removal of the chromatin. Subsequent fractionation of such nuclear supernatants in sucrose or glycerol gradients revealed that the labeled pol II transcripts sedimented at the 200S region of the gradient [27, 28, 37]. We next used this protocol for the release of complexes of CAD (for carbamoyl-phosphate synthetase, aspartate transcarbamylase, dihydro-orotase) RNA expressed from a cell line in which this RNA overaccumulated due to amplification (200-fold) of the CAD gene [49, 50], and in which CAD RNA was highly abundant in the cell nucleus [27]. Analysis of the distribution of CAD RNP in sucrose gradients revealed that intact CAD RNA sedimented as 200S complexes [27]. Further studies showed that the general population of nuclear polyadenylated RNAs, as well as additional specific Pol II transcripts (dihydrofolate reductase (DHFR), β-actin, hnRNP AB, survival motor neuron (SMN), adenosine deaminase that acts on RNA 2 (ADAR2), and Adenovirus major late (AdML)), all sedimented at the 200S region of the gradient [33, 38, 40, 51]. Visualization by EM of aliquots from fractions across the gradient revealed a peak of a well defined novel structure, a large tetrameric complex, having overall dimensions of 50 × 50 nm (Fig. 1), that sedimented at 200S and coincided with the peak of the Pol II transcripts [33]. These 200S complexes, initially called large nuclear RNP (lnRNP) complexes, were termed supraspliceosomes (reviewed in Refs [6, 31, 32]). Immunoprecipitation experiments, using antibodies against splicing factors, showed that all five spliceosomal U snRNPs are integral components of the supraspliceosome [28, 37, 51–53], as well as all known protein splicing factors, including the non-snRNP protein splicing factor U2AF and all members of the SR protein family [28, 38, 39, 51–55]. Notably, the phosphorylated SR proteins, which are required for spliceosome assembly and alternative splicing, are predominantly associated within the supraspliceosome [52]. A remarkable feature of supraspliceosomes is that they package pre-mRNA transcripts of different sizes and of different number of introns into complexes of a unique size, hydrodynamic properties and structure, indicating their universal nature (reviewed in Refs [6, 31, 32]).
2.2 Electron Tomography at room temperature (ET)
The huge size of the supraspliceosome required structural analysis in steps. We initiated the analysis by performing automated computed room temperature electron tomography (defined here as ET) for the three-dimensional (3D) image reconstruction of individual supraspliceosomes isolated from mammalian cells nuclei and negatively stained [34, 56]. We have chosen this approach rather than single particle analysis because the four substructures of the supraspliceosome are interconnected in a flexible way and may thus adopt different angular settings, which imposed a significant restriction on reaching high resolution in EM image analyses at that time. For each particle, a tilt series of 71 images (±75°) was collected by direct digital recording of the images on a CCD camera attached to a computer controlled Transmission electron microscope (TEM) facility. The 3D image was reconstructed according to the back projection principle. For rendering, real time display and comparison of the reconstructed particles, interactive computer graphics was employed. The reconstructed 3D images (Fig. 2) show a compact structure, 50×50×35 nm, composed of four major subunits connected to each other. Comparison of the reconstructed supraspliceosomes revealed morphological similarity of the individual particles, as well as similarity among the substructures. The uniformity of the supraspliceosomes was further substantiated by measurement of the volume engulfed by their surface [56]. Based on these findings, we proposed a model for the packaging of nuclear pre-mRNAs in supraspliceosomes, where each substructure represents a functional unit. This model is compatible with the requirements for alternative splicing of multi-intronic pre-mRNAs, and with the fact that the splicing of multi-intronic pre-mRNAs does not occur in a sequential manner.
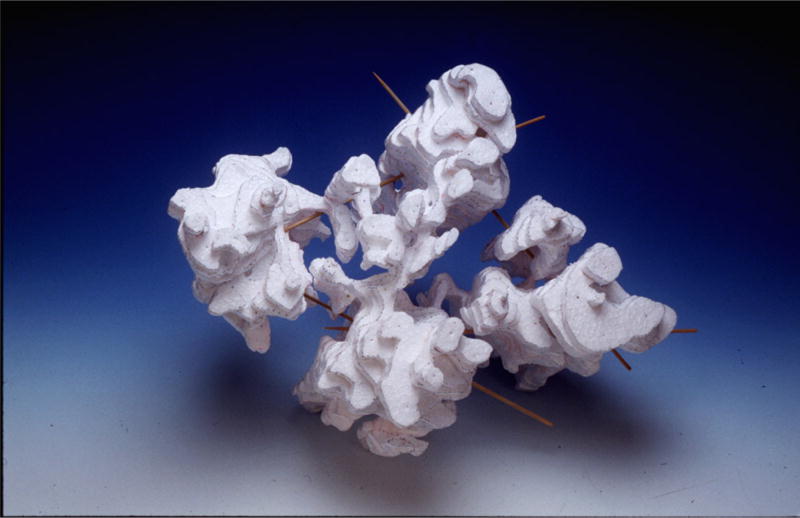
Fig. 2. A physical model of a reconstructed supraspliceosome.
Automated ET was employed for the 3D image reconstruction of individual supraspliceosomes isolated from mammalian cells nuclei and negatively stained. For each particle, a tilt series of 71 images was collected by direct digital recording of the images on a CCD camera attached to a computer controlled TEM facility. The model was built from the density map (density sections of ~2 nm thickness parallel to the XY plane of the grid). Adapted from Ref [34].
2.3. Cryo-electron tomography of supraspliceosomes
Cryo-EM of frozen-hydrated specimens is considered as an EM technique that preserves the sample in its native conformation like in solution [57], unlike negative staining, where distortions due to flattening might occur. Since neither staining nor any other fixative is used, this technique is almost artifact-free and therefore is highly suitable to study the complex and dynamic structure of supraspliceosomes.
2.3.1. Concentration of supraspliceosomes on a charged lipid monolayer
ET of ice-embedded biological complexes is hampered by the relatively low contrast of the specimen and its high sensitivity to the electron beam, which necessitates low-dose conditions. This disadvantage can be compensated by collecting data sets from samples containing high concentrations of evenly distributed ice-embedded particles. The concentration of supraspliceosomes isolated from gradients turned out not to be sufficient for cryo-ET. Typically, at the desired magnification only one or two particles were present in the field of view [36], which would make ET very inefficient in time and effort. Most attempts to concentrate the particles by standard methods (e.g. gel filtration, ultrafiltration) led to aggregates of particles, which were not useful for structural analysis. We therefore tried a different approach of concentrating the particles on charged lipid monolayer films. This methodology has been successfully used in electron crystallography studies of biologically active complexes [58, 59]. We adapted this method to reproducibly concentrate and generate, on a positively charged monolayer, a layer with a high density of supraspliceosomes that can be picked up on holey carbon grids, blotted and plunged into liquid ethane, and visualized in the frozen-hydrated state [36]. These preparations gave a relatively high concentration of particles, evenly distributed in the ice films, which enabled us to obtain good quality images of the supraspliceosome (Fig. 3). We attributed the concentrating effect of the positively charged lipid to the negative charge that is conferred on the supraspliceosome by the partially exposed pre-mRNA they package [30]. The quality of the ice films was high, they exhibited good contrast, and the concentration of the particles in the ice film was increased by at least an order of magnitude, while preserving their supraspliceosomal configuration, which makes it possible to efficiently investigate the particles by cryo-EM. In addition, the areas containing the lipid layer can be easily identified at low magnification, enabling searching for ice-embedded particles on such a grid a straightforward task [29, 36, 37]. This method may also be applicable to generate concentrated frozen-hydrated preparations of other biological complexes.
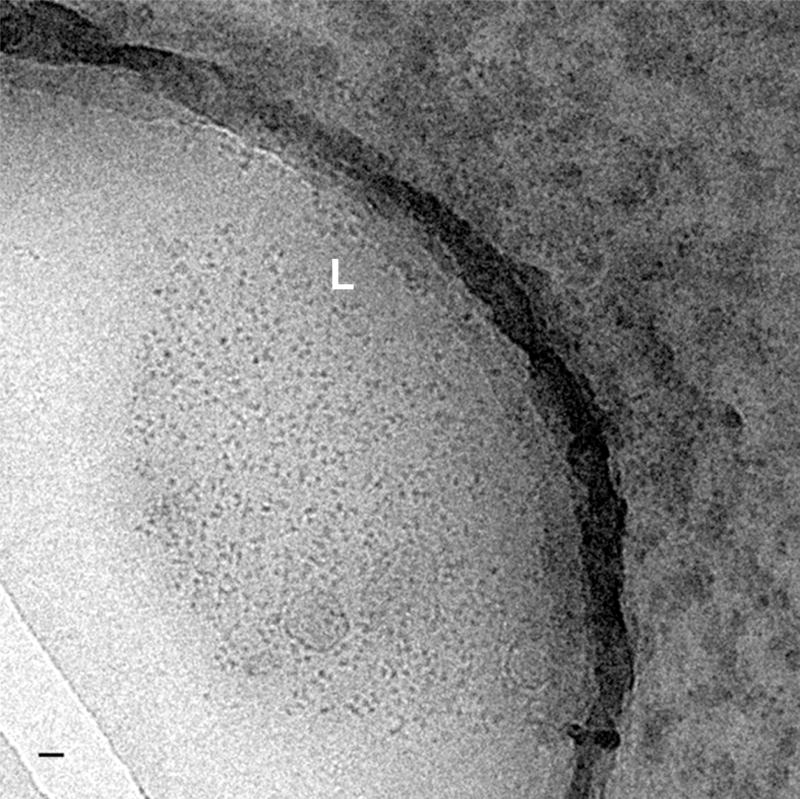
Fig. 3. Cryo-images of supraspliceosomes concentrated on a charged lipid monolayer.
A low magnification image of frozen-hydrated supraspliceosomes concentrated on a positively charged lipid monolayer (phosphatidylcholine and stearylamine) viewed inside a hole in the carbon-coated grid. The edges of the lipid film (L) are clearly visible and only very few particles are found outside the lipid layer. The bar represents 200 nm. Adapted from Ref [36].
2.3.2. Cryo-ET
We employed automated cryo-ET for the structural analysis of supraspliceosomes. Tilt series (±70°) of frozen-hydrated preparations of supraspliceosomes adsorbed to charged lipid film, were recorded and evaluated, and 22 particles were reconstructed. 3D image reconstruction showed the supraspliceosome as forming a closed structure, 50×50×35 nm in dimensions, composed mainly of four similar native spliceosomes, which appear in contact with each other (Fig. 4A) [60]. In the cryo-EM and cryo-ET studies [60] a fifth substructure appears sometimes, and it appears to be larger than a smaller fifth substructure, sometimes observed in negative stain (see Fig. 4. See also Fig. 2 where a smaller fifth substructure can be observed at the top of the tetrameric structure). This additional domain is correlated with a 1.9 MDa substructure determined by STEM mass measurements (see section 3.1), and is different from the 4.8 MDa mass of native spliceosomes. This substructure was attributed to 5’end and 3’end processing components that are present once in a supraspliceosome [30]. The difference in dimensions between the cryo and negative stain preparations and its non-frequent appearance may be explained by assuming that the fifth substructures is biochemically different and loosely bound, which causes it to be frequently lost during sample preparation. The additional domain, unlike the four similar spliceosomal subunits, might be more sensitive to the negative staining procedure or interacts differently with the carbon film, while it is better preserved in the frozen-hydrated state. This is consistent with the idea that the four-spliceosomal subunits are composed mainly of spliceosomal factors, whereas the fifth domain harbors factors involved in the other pre-mRNA processing activities [60].
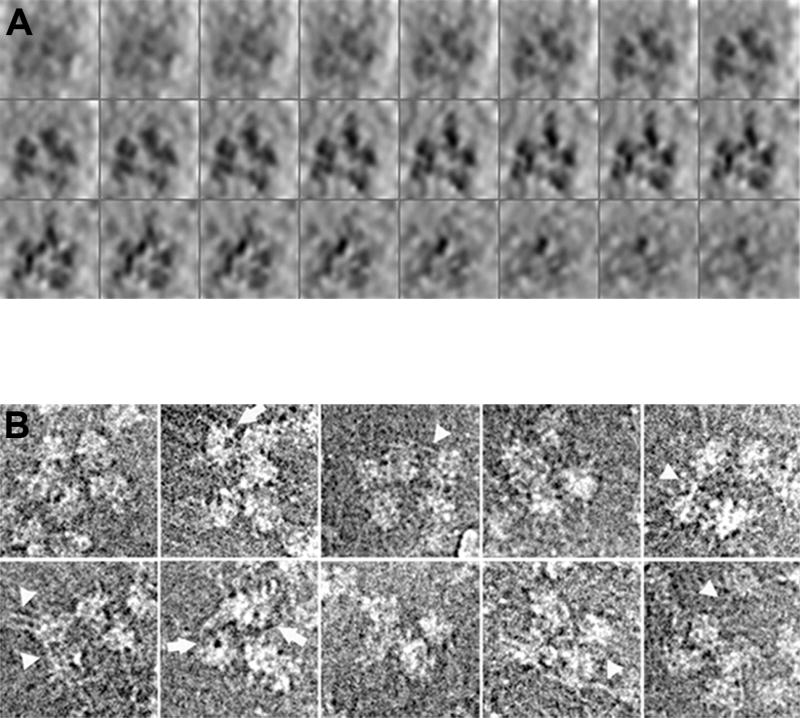
Fig. 4. Structural analysis of supraspliceosomes by cryo-EM.
(A) 3D image reconstruction of a supraspliceosome by automated cryo-ET. The reconstructed model of an ice-embedded supraspliceosome is presented as a series of x-y sections, 1.6 nm in thickness, along the z axis (top left to bottom right), enhanced by denoising. (B) Image restoration of selected ice-embedded supraspliceosomes. Particles displaying four or five substructures are presented. Some of the images display partially disassembled particles with an “opened-up” configuration. Fibrillary structures, presumably RNA covered with proteins, connecting the subunits are visualized particularly in the more open configurations (arrow-heads). Also, low-density areas, representing holes in the native spliceosomes, are visualized in several substructures (arrows). Adapted from Ref [36].
2.3.3. Cryo image restoration
To improve the information obtained by 2D imaging, we also performed image restoration from focus series [61, 62] of the ice-embedded supraspliceosomes [60]. These results were a first glance into the high-resolution structure of the supraspliceosome. Most of the restored images revealed a novel feature of fibrillary elongated structures, 1–2 nm in width, emanating from the subunits and connecting them (Fig. 4B), presumably representing pre-mRNA coated with proteins, consistent with protein-coated pre-mRNA fibrils, seen as strands and loops that emanate from the subunits detected by STEM in positively stained supraspliceosomes (see Section 3.1 and Fig. 5) [30]. Another novel feature of the spliceosomal subunits that has come out of this analysis is the presence of low-density regions that can be visualized in some of the substructures, depending on their relative orientation in the ice film. These regions represent holes in the substructures (Fig. 4B). Although holes or tunnels in the substructures were also indicated by the 3D reconstruction of negatively stained particles [34, 56], we could not determine whether such areas represent stain-filled regions or regions where (positively stained) RNA is present. The image restoration of unstained frozen-hydrated supraspliceosomes resolved this issue, providing the first demonstration of low-density regions within the native spliceosome [60].
Fig. 5. Strands and loops of RNA covered with proteins emanate from the supraspliceosome.
Positively stained supraspliceosomes recorded in the STEM dark-field mode. Protein and RNA are displayed in light shades. Strands and loops can be seen spreading out from the supraspliceosome subunits – the native spliceosomes. The scale bar represents 50 nm. Adapted from Ref [30].
3. The supraspliceosome is composed of four native spliceosomes connected by the pre-mRNA
3.1. STEM mass measurements
To substantiate the apparent generality of the supraspliceosome structure and the similarity between it’s subunits, we performed mass measurements by STEM [30]. The advantage of this technology is that it enables measurements of the mass of each individual particle visualized, and also measurements of the mass of hundreds of particles and thus providing statistically significant numbers. Aliquots from the 200S supraspliceosome peak fractions of HeLa cells were analyzed, after glutaraldehyde or UV stabilization and washing with volatile ammonium acetate. During isolation and grid preparation the tetrameric complex partially dissociated to yield a heterogeneous mixture of particles containing one, two, three or four subunits. The mass uniformity of the 28 nm × 19 nm monomeric substructure - that we termed native spliceosome [4.8± 0.5 MDa, SE = 0.022 MDa, (n=506)] indicates the presence of a single particle population, and confirms the subunit similarity of the oligomers. The data clearly show that the mass of the tetrameric supraspliceosome is 21.1 MDa (n ≈ 396), while that of the dimeric and trimeric complexes being 10.6 MDa (n ≈ 281) and 16.3 MDa (n ≈ 540), respectively. Furthermore, both visual inspection and the mass values indicate that the monomeric 4.8 MDa native spliceosome dissociates into two even smaller particles, with masses of 1.5 MDa and 3.1 MDa [30].
The mass measurements by STEM show that the trimeric and tetrameric particles are 1.9 MDa heavier than three and four subunits, respectively. The significance of these mass differences is shown by the corresponding SEs. We attributed the 1.9 MDa surplus to an additional domain often visualized on the micrographs, and better visualized by cryo-EM (see Section 2.3.2). Although other possibilities cannot be excluded, the 1.9 MDa additional mass may be attributed to the 5'- and 3'-end processing components that we have shown to be associated with the supraspliceosome [35, 38]. As each supraspliceosome is assembled on a single pre-mRNA molecule (see Section 5.2), these components are present as a single copy per tetrameric supraspliceosome.
We have made a rough estimate of the mass of a 60S in vitro assembled spliceosome from its known sedimentation coefficient, assuming similar sedimentation behavior to that of an icosahedral virus. Using the density, partial specific volume, and diffusion coefficient of 132S bushy stunt virus (BSV), which has a mass of 10.7 MDa [63], the mass of the 60S spliceosome is estimated as 4.9 MDa. Interestingly, the mass of the yeast Bact complex, lacking U1 and U4, whose structure was recently solved at 5.8 Å resolution, was reported as 3.8 MDa [12]. Thus, the reported mass of the Bact complex is getting closer to the measured mass of the HeLa native spliceosome of 4.8 MDa [30], providing further support for the connection between the native spliceosome and the in vitro assembled spliceosome.
Another observation was of strands and loops of material that could sometimes be seen protruding from the unstained supraspliceosomes [30]. These were more clearly revealed using a positive staining protocol, which allows the visualization of nucleic acids [64]. Representative tetrameric supraspliceosomes are shown in Fig. 5, with filamentous material generally appearing radiating from the outermost regions of the subunits. These strands and loops are RNA covered with proteins as they disappeared upon RNase treatment.
The similarity of the 4.8 MDa monomers, the mass of the tetrameric supraspliceosomes (21.1 MDa), and the biochemical data showing that the supraspliceosome harbors all components of the spliceosome [6, 31, 32] supports the notion that the endogenous spliceosome – the supraspliceosome - is composed of four native spliceosomes.
3.2. Isolation and characterization of native spliceosomes
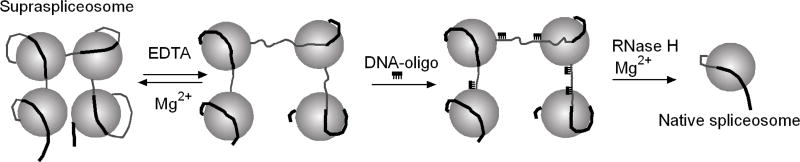
To examine the hypothesis that the four native spliceosomes are connected by the pre-mRNA within the supraspliceosome, we have chosen to cleave the pre-mRNA without cleaving the U snRNAs. This approach is also beneficial to our structural studies for the following reasons. The four substructures of the supraspliceosome are interconnected in a flexible way and may thus adopt different angular settings, which impose a significant restriction on reaching high resolution in EM image analyses. We have therefore used the methodology we developed to cleave the pre-mRNA and isolate the native spliceosomes from supraspliceosomes, for cryo-EM single particle reconstruction of native spliceosomes.
The strategy developed to specifically cleave the general population of pre-mRNAs, while keeping the U snRNAs within the supraspliceosome intact was based on the premise that the consensus sequence of the 5’ splice site (5'SS) in U2 introns (AG/GTRAGT; where R denotes purine and / denotes the splice junction) is highly abundant in such introns (~9 5’SSs/intron that remain latent under normal growth conditions)[65, 66]. These latent 5’SSs, not involved in splicing, are thus likely available for base pairing with complementary oligodeoxynucleotides and to digestion by RNase H. To facilitate cleavage at the intronic latent 5’SSs, we took advantage of the fact that upon chelating the magnesium ions, which help stabilize supraspliceosomes, these complexes adopt an open-up conformation that presumably exposes the introns [28]. We have thus treated purified supraspliceosomes with EDTA, hybridized them to the redundant DNA oligonucleotide NCTYACCV (where N=A+C+G+T; Y=C+T; V=A+C+G) and subjected them to RNase H digestion (Fig. 6). The resulting complexes were purified by centrifugation in a 10–45% glycerol gradient, where they sedimented at the 60–70S region, revealing monomeric substructures. Fig. 7 shows cryo-EM images of the monomeric substructures, sedimenting at 60–70S, spread on a thin carbon film [37]. We termed these complexes as native spliceosomes as we have shown that they are active in splicing [37], and because they are similar to the in vitro assembled spliceosome with respect to dimensions and estimated mass. The mass of the native spliceosome as determined by STEM is 4.8 MDA [30] (see section 3.1).
Fig. 6. A scheme of the isolation of native spliceosomes.
Treatment of supraspliceosomes with EDTA induces a conformational change (please note that this step is reversible) [28]. This change makes the pre-mRNA available for base pairing with an oligodeoxynucleotide complementary to abundant intronic sequences that conform to the 5’SS consensus sequence. This part of the scheme is oversimplified because some components dissociate upon EDTA treatment but reassociate upon addition of magnesium cations. Subsequent treatment with RNase H and readdition of Mg2+ yield native spliceosomes (please note that this step is irreversible). Adapted from Ref. [37].
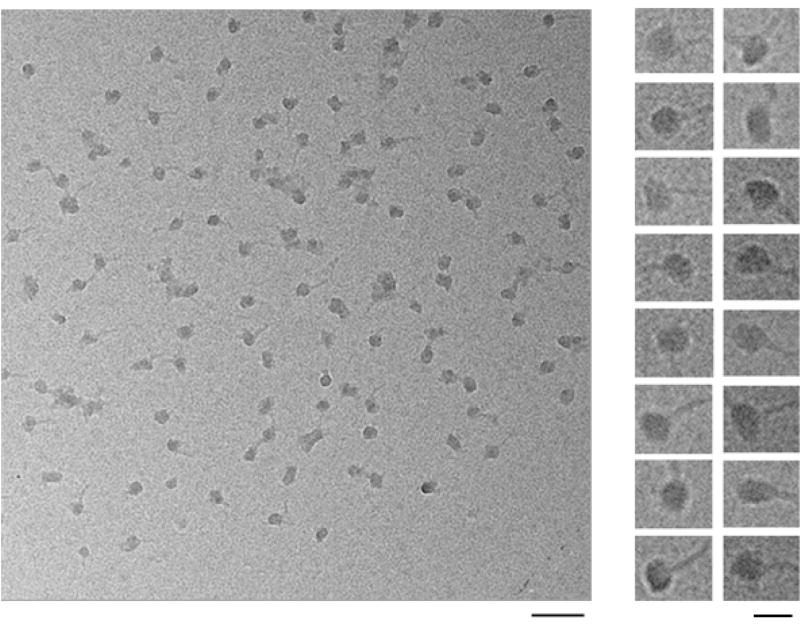
Fig. 7. Cryo-images of native spliceosomes.
An EM field (left panel) and a gallery (right panel) of ice-embedded native spliceosomes on continuous carbon film. An RNA tail, likely covered with proteins, can be visualized emanating from each complex, with an appearance of a head and a tail. The left-hand scale bar represents 100 nm; the right-hand scale bar represents 25 nm. Adapted from Ref [37].
Northern blot analysis of the U snRNA composition of the native spliceosomes and supraspliceosomes revealed a peak containing the five spliceosomal U snRNAs in the 200S region of the supraspliceosomal gradient, and a peak at the 60–70S region of native spliceosomes [37]. Thus, the native spliceosome contains all stable RNA elements known to be required for splicing. We have further demonstrated that both supraspliceosomes and native spliceosomes are active in splicing. These studies confirmed that each of the four substructures of the supraspliceosomes is equivalent to the native spliceosome, and the four native spliceosomes are interconnected via the pre-mRNA within the supraspliceosome [29, 37].
3.3. Structure of the native spliceosome by cryo-EM single particle technique
The isolation of native spliceosomes and their relative stability enabled us to perform 3D cryo-EM structural analysis by the single particle technique [29]. A 3D structure was reconstructed from 9297 raw single-particle images by first calculating an initial model using the common-lines approach [67]. This initial model was then refined by using cycles of multi-reference alignment to reprojections of the model. Due to complete random orientation in the vitreous ice, representative angular views of native spliceosomes were equally populated. After refinement, the Fourier shell correlation function [68] was used to calculate the resolution of the 3D reconstruction giving values of 20 Å for the 3σ threshold criteria, and 22 Å for the 0.5 threshold criteria.
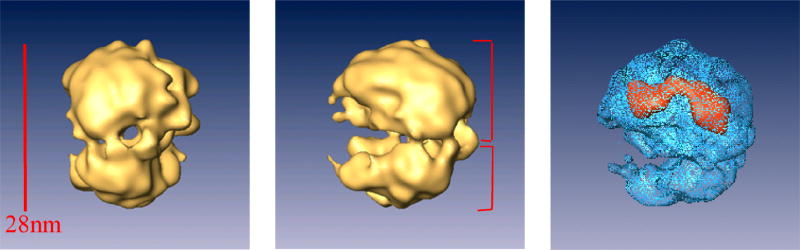
As shown in Fig. 8, the structure revealed new information about the organization of the native spliceosome depicting it as an elongated globular particle made up of two distinct subunits. This finding is consistent with our STEM mass measurements, which revealed two major, equally populated, distinct groups of small particles with masses of 1.5 MDa and 3.1 MDa, which together add up to close to the 4.8 MDa mass of the native spliceosome [30]. The two subunits are interconnected to each other leaving a tunnel in between, which is large enough to allow the pre-mRNA to pass through. The other side of the native spliceosome exposes a cavity that could provide a place to transiently store the labile pre-mRNA, and protect it from non-specific degradation (Fig. 8).
Fig. 8. Structure of the native spliceosome.
Different views of a surface representation of the native spliceosome reconstructed at 20 Å resolution from cryo-images [29]. The structure was contoured to a STEM-measured mass of 4.8 MDa [30]. Right panel: High-threshold rendering (blue surface) shows the high-density mass region, which represents the stable RNAs within the surface representation of the 3D reconstruction of the native spliceosome. The large subunit of the native spliceosome is thus a suitable candidate to harbor the five-spliceosomal U snRNPs. Adapted from Ref [29].
The two subunits vary also with respect to the distribution of high densities within the native spliceosome. Because RNA is denser than protein, the localization of regions of high density can provide some information about internal organization of RNA and protein components. We have shown that the large subunit is a suitable candidate to accommodate the five spliceosomal U snRNP, as the high density regions were found within the large subunit (Fig. 8), and the mass and volume of the large subunit are similar to the estimated mass and volume of the five-spliceosomal U snRNPs [29].
3.4. A unique spatial arrangement of the U snRNPs within the native spliceosome emerges from in-silico studies
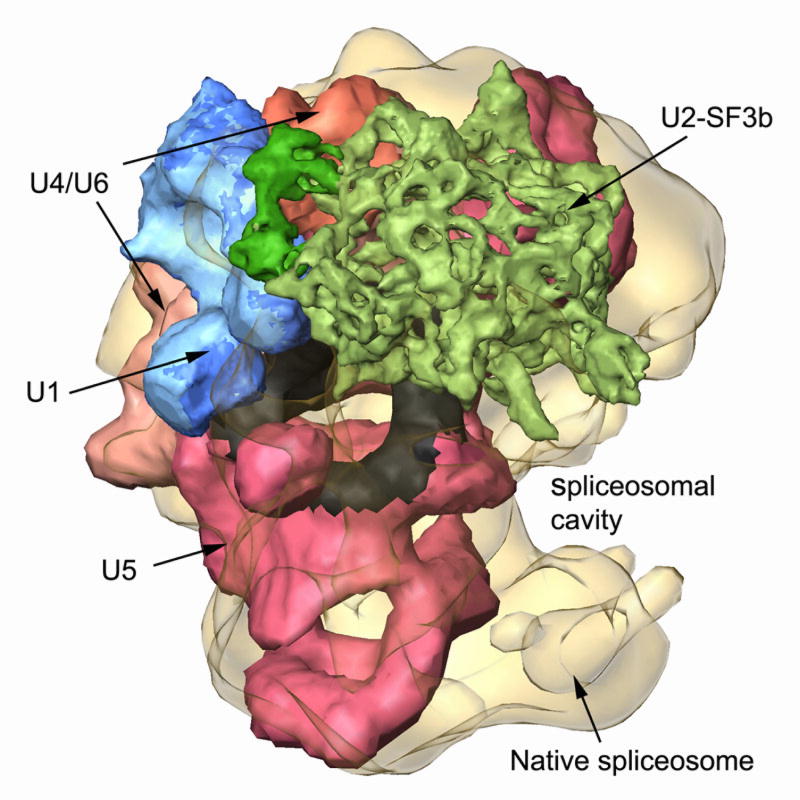
We further used computational tools to localize known structures of the spliceosomal U snRNPs within the native spliceosome [69]. We designed a new computational procedure to efficiently fit thousands of conformers into the native spliceosome envelope. Despite the low-resolution limitations, we obtained only one model that complies with the available biochemical data. Our model localizes the five-spliceosomal U snRNPs mostly within the large subunit of the native spliceosome, requiring only minor conformation changes (Fig. 9). The remaining free volume presumably accommodates additional spliceosomal components. The ample free volume may indicate that structural modulations of the U snRNPs can be tolerated while keeping the integrity of the spliceosome assembly. The constituents of the active core of the native spliceosome are juxtaposed, forming a continuous surface deep within the large spliceosomal cavity, which provides a sheltered environment for the splicing reaction [69]. Though the above unique spatial arrangement of the U snRNPs within the native spliceosome is limited by the 20 Å resolution of the structure of the native spliceosome, the latter arrangement of the constituents of the active core of the spliceosome is consistent with the recent high-resolution structural studies of spliceosome subcomplexes [11–17, 19].
Fig. 9. A unique spatial arrangement of the U snRNPs within the native spliceosome emerges from in-silico studies.
[69]. The native spliceosome is transparent; U4/U6.U5 tri-snRNP is colored by functional regions, with U5 snRNP in pink and the region attributed to loop I in black [83]; U4/U6 in beige-orange; U2 snRNP subcomponent SF3b is in green; and U1 snRNP is blue. Adapted from Ref [69].
4. Pol II transcripts at different stages of splicing are assembled in supraspliceosomes
4.1. Affinity purification of specific Pol II transcripts at defined functional states
The structural analysis performed thus far was of the general population of nuclear pre-mRNAs, each harboring a different number of introns and is likely to represent a different step of the splicing process. For better characterization of the splicing machine, we further prepared specific complexes assembled on specific pre-mRNAs, trapped at specific functional states (pre-mRNA or splicing intermediates), The isolation is based on preparation of stable cell lines transcribing specific tagged transcripts, and the isolation of specific functional states is based on mutations of genes encoding specific pre-mRNAs [38].
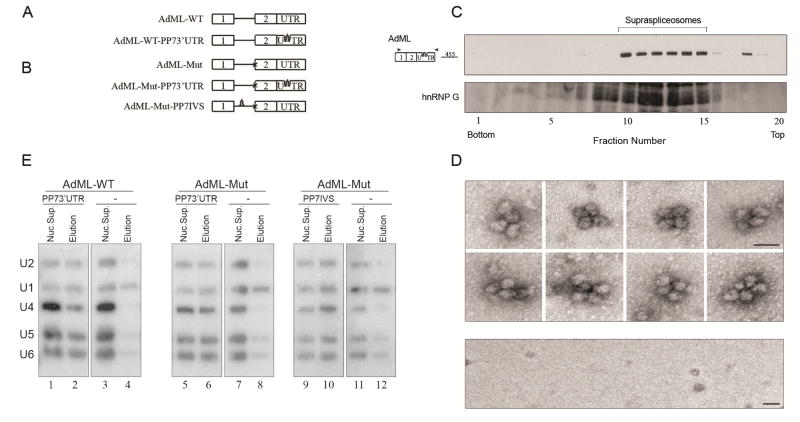
The Pseudomonas aeruginosa phage 7 (PP7) RNA binding site/recombinant PP7 coat protein (PP7CP) system [70], was used to affinity purify PP7-tagged splicing complexes [38]. A series of stable cell lines was prepared (Fig. 10A,B), each expressing a PP7-tagged wild type (WT), or mutant (Mut) AdML, having a mutation at the 3’SS that inhibits the second step of splicing, and having the PP7-tag either at the 3’ UTR or at the intron. Cell lines lacking the PP7 tag served as controls. Nuclear supernatants enriched for supraspliceosomes were prepared from each of the above cell lines (as described in section 2.1), and each was subjected to affinity purification using the PP7CP system. Specific affinity purification of the AdML assembled complexes from each of the cell lines expressing PP7-tagged AdML was achieved, and revealed that they were assembled in supraspliceosomes, as determined by density centrifugation and EM visualization (Fig. 10C,D) [38]. AdML-WT supraspliceosomes were found assembled on mature RNA, and AdML Mut supraspliceosomes were found assembled on pre-mRNA [38].
Fig. 10. The five-spliceosomal U snRNAs are associated with in vivo assembled supraspliceosomes at all splicing stages.
(A, B) Schemes of AdML minigenes, each expressed from a stable cell line from which specific supraspliceosomes at defined splicing stages were affinity purified using the PP7/PP7CP system [70]. Stable cell lines expressing the respective AdML transcripts lacking tag were used as controls. Open boxes represent exons, lines represent introns, and stem-loops represent the PP7 tag inserted either at the 3’UTR or the intron. (A) Constructs expressing AdML-WT. (B) constructs expressing AdML-Mut, having a mutated 3' splice site, designated “x”. (C, D) The affinity purified splicing complexes of AdML-WT-PP73’UTR are assembled in vivo on supraspliceosomes. (C) Upper panel: Affinity purified splicing complexes assembled on the AdML-WT-PP73’UTR transcript, were fractionated in 10–45% glycerol gradients [37] and RNA extracted from each fraction was analyzed by RT-PCR (the size of the PCR product (nt) is given). Lower panel: Western blot analysis of nuclear supernatants enriched for supraspliceosomes prepared from HeLa cells and fractionated in 10–45% glycerol gradients using antibodies against hnRNP G, which is associated with supraspliceosomes [39]. (D) Upper panel: Gallery of affinity purified splicing complexes assembled on AdML-WT-PP73’UTR transcript, visualized by EM, after negative staining with 1% uranyl acetate. Lower panel: Control, EM of eluted material from the control sample (AdML-WT transcript without the PP7-tag). The bar represents 50 nm. (E) Northern blot analysis with probes directed against the five-spliceosomal U snRNAs was performed on RNA extracted from nuclear supernatants (Nuc. Sup.), and from affinity purified supraspliceosomes (Elution), prepared from HeLa cell-lines expressing the AdML constructs either with the PP7 tag or without it (−), as indicated. AdML-WT supraspliceosomes were found assembled on mature RNA, and AdML Mut supraspliceosomes were found assembled on pre-mRNA [38]. Left, the AdML-WT-PP73' UTR supraspliceosomes; middle, the AdML-Mut-PP73'UTR supraspliceosomes; Right, AdML-Mut-PP7IVS supraspliceosomes. The identity of the U snRNA probes is given on the left. Adapted from Ref [38].
4.2. The endogenous spliceosome is assembled with all five-spliceosomal U snRNPs during all steps of the splicing reaction
To determine the U snRNP composition of the affinity purified specific supraspliceosomes, Northern blot analysis were performed and revealed that all five spliceosomal U snRNAs are associated with each of the above affinity-purified AdML supraspliceosomes: AdML-WT-PP73’ UTR supraspliceosomes that are assembled on mature mRNA (Fig. 10E, lane 2); as well as on AdML-Mut-PP73’UTR and AdML-Mut-PP7IVS supraspliceosomes that are assembled on AdML pre-mRNA (Fig. 10E, Lanes 6 and 10, respectively) [38]. Very low levels of U snRNAs were found in the control samples that lack the PP7 tag (lanes 4, 8, and 12), with some non-specific binding of U1 snRNA, known to be in excess in the cell nucleus [71]. It can therefore be concluded that supraspliceosomes isolated from mammalian cell nuclei have all five spliceosomal U snRNAs associated with them at all splicing stages. These findings highlight the important role of large, pre-formed, complexes in pre-mRNA splicing in vivo.
The apparent discrepancy between a stepwise assembly pathway of the spliceosome in vitro and the occurrence of a pre-formed splicing complex in vivo, has been explained by a “holospliceosome” model, in which the sequential complexes represent ordered modulations within the in vivo assembled spliceosome without the loss of components [4]. It has also been pointed out that such intermediate states in spliceosome assembly in vitro may not occur in vivo [72, 73].
4.3. Protein composition of affinity purified specific supraspliceosomes
Mass spectrometry analysis of the affinity purified supraspliceosomes revealed an average of 275 protein groups, higher amount than the 177 protein groups detected previously for the analysis of a general population of supraspliceosomes purified from HeLa cells [74]. This difference likely reflects the difference in purification methods and instrumentation used. Core components of the supraspliceosome were detected both in the AdML-WT and AdML-Mut assembled supraspliceosomes. These include all five spliceosomal U snRNPs, in agreement with the presence of the five U snRNAs at both populations [38]; the hPRP19/CDC5L complex, which was shown to be necessary for both splicing stages [75], and was also identified by the proteomic analysis of the general supraspliceosome population [74]; several splicing factors and other proteins involved in RNA metabolism such as the RNA binding proteins SR and hnRNP families [38]. The SR proteins, which play a role in alternative and constitutive RNA splicing [76], were previously shown to be part of the supraspliceosome [28, 39, 40, 52].
Another group of proteins detected in both supraspliceosome populations (assembled on AdML-WT or AdML-Mut transcripts) were proteins responsible for pre-mRNA processing, such as the cleavage and polyadenylation specificity factor (CPSF) that is responsible for 3’ end processing [77]; CBPs, ADAR editing enzymes; and the ALY/REF proteins which play a role in RNA export from the nucleus [76], consistent with previous finding of 5’-end and 3’-end processing components [35] and ADAR enzymes [43, 78] in supraspliceosomes.
The proteomic analyses of supraspliceosomes assembled on AdML-WT and AdML-Mut transcripts strengthen the above conclusion that the supraspliceosome is the pre-mRNA processing machine that is pre-assembled throughout all splicing stages, as the core components of the spliceosome can be seen in all these stages. The analyses also show the dynamic nature of the supraspliceosome during the splicing stages, as different hnRNP isoforms were found in supraspliceosomes from the different steps of splicing [38].
5. Structure and function of the endogenous spliceosome
5.1 The pre-mRNA is required for the reconstitution of supraspliceosomes from native spliceosomes
We have shown that oligonucleotide-directed RNase H digestion of the pre-mRNA within supraspliceosomes gave rise to native spliceosomes, yet, native spliceosomes could not be reconstituted into supraspliceosomes just by the addition of magnesium cations [29]. These experiments demonstrated that the pre-mRNA connects the native spliceosomes within the supraspliceosome, and highlighted the essential role of the pre-mRNA for the assembly of the supraspliceosome. We confirmed the conclusion that the pre-mRNA is required for supraspliceosome assembly, by showing that supraspliceosomes can be reconstituted by back-addition of pre-mRNA. Incubation of purified native spliceosomes with in vitro transcribed pre-mRNAs having an intron flanked by two exons, followed by TEM visualization of the negatively stained complexes revealed that native spliceosomes were thus reconstituted into tetrameric supraspliceosomes [37] (see also Section 5.2). This transition occurred with an efficiency of ~ 40–60%, as determined for 2185 particles. The reconstituted supraspliceosomes, obtained from each of three tested pre-mRNAs, are composed of four native spliceosomes and are similar to supraspliceosomes isolated from living cells with respect to their morphology and dimensions. Control experiments showed that no supraspliceosomes were reconstituted upon addition of double or single stranded DNA encoding for the respective pre-mRNAs. These experiments highlighted the essential role of the pre-mRNA for the integrity of the supraspliceosome [37].
5.2 A supraspliceosome is assembled on a single pre-mRNA
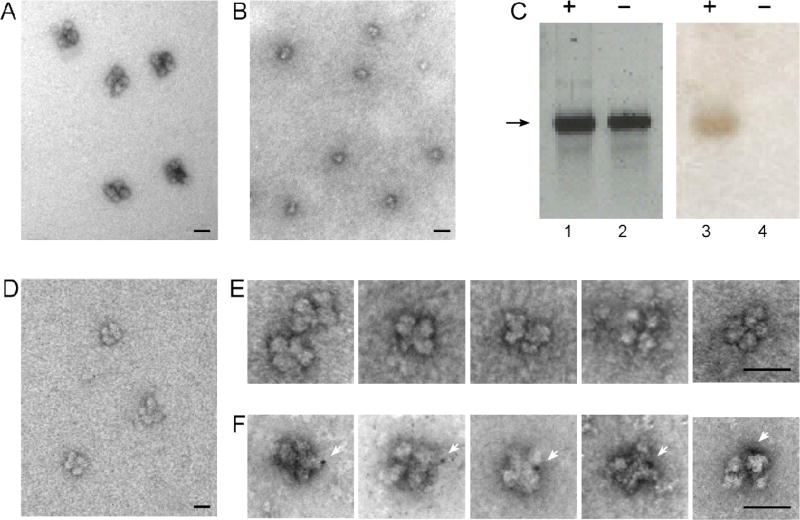
A prerequisite for efficient and precise alternative splicing and pre-mRNA processing within the supraspliceosome is that each complex should process a single RNA transcript at a time, and this hypothesis has been implied in the supraspliceosome model [6, 31, 32, 37]. To test this hypothesis, we used our above procedure for reconstituting tetrameric supraspliceosomes from monomeric native spliceosomes [37], this time, with an exogenously added gold-tagged synthetic pre-mRNA [40]. Tagging of the RNA with gold nanoclusters enhances visualization by EM, as exemplified by gold-tagged synthetic mRNAs, labeled at multiple sites, and visualized by EM and AFM [79]. For this experiment (Fig. 11), native spliceosomes were prepared from supraspliceosomes by specific cleavage of the endogenous pre-mRNAs as described [29]. Next, an in vitro transcribed human β-globin pre-mRNA was ligated to a thiolated 12-nt oligodeoxynucleotide, the RNA-oligo-SH was next tagged at its 3’-end with a single gold nanoparticle of 1.4 nm in diameter (Nanogold by Nanoprobes), and the gold-tagged and control untagged RNAs were analyzed by gel electrophoresis (Fig. 11C). Specific staining of the gold by silver enhancement, confirmed the tagging of the β-globin pre-mRNA with Nanogold [40]. Purified native spliceosomes were then incubated with the in vitro transcribed β-globin pre-mRNA (tagged or not-tagged with Nanogold). EM visualization (Fig. 11F) revealed that supraspliceosomes reconstituted with Nanogold-tagged pre-mRNA have only one Nanogold cluster per reconstituted supraspliceosome [40]. Of 97 reconstituted supraspliceosomes with Nanogold tagged pre-mRNA, 75 particles (77%) showed one Nanogold particle per supraspliceosomes, the remaining were not tagged with gold. These experiments confirm that each supraspliceosome is assembled on one pre-mRNA [40].
Fig. 11. A supraspliceosomes is assembled on one pre-mRNA molecule (assembled from native spliceosomes and Nanogold-tagged pre-mRNA).
Native spliceosomes were prepared from supraspliceosomes by specific cleavage of the pre-mRNA as described [29], and supraspliceosomes were reconstituted by incubating native spliceosomes with β-globin pre-mRNA tagged or untagged with Nanogold as described [37]. (A, B, D) Visualization by EM of a field of supraspliceosomes (A); Native spliceosomes (B); and reconstituted supraspliceosomes (D), negatively stained with 1% uranyl acetate. (C) Gel electrophoresis (2% agarose) analysis of untagged (lanes 2 and 4) and Nanogold-tagged β-globin pre-mRNA (lanes 1 and 3), used for the reconstitution experiment. Left panel, staining with ethidium bromide; right panel, staining of the blotted gel by silver enhancement. (E, F) Galleries of images of reconstituted supraspliceosomes visualized by EM after staining with 1% uranyl acetate and gold enhancement. (E) Gallery of supraspliceosomes reconstituted from native spliceosomes and untagged β-globin pre-mRNA. (F) Gallery of supraspliceosomes reconstituted from native spliceosomes and Nanogold-tagged β-globin pre-mRNA. White arrowheads point to gold-enhanced Nanogold. Bar represents 50 nm. Adapted from Ref [40].
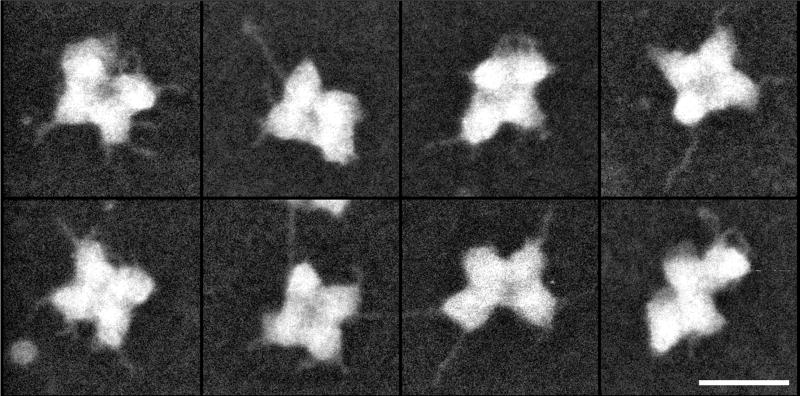
5.3. Arrangement of native spliceosomes within the supraspliceosome
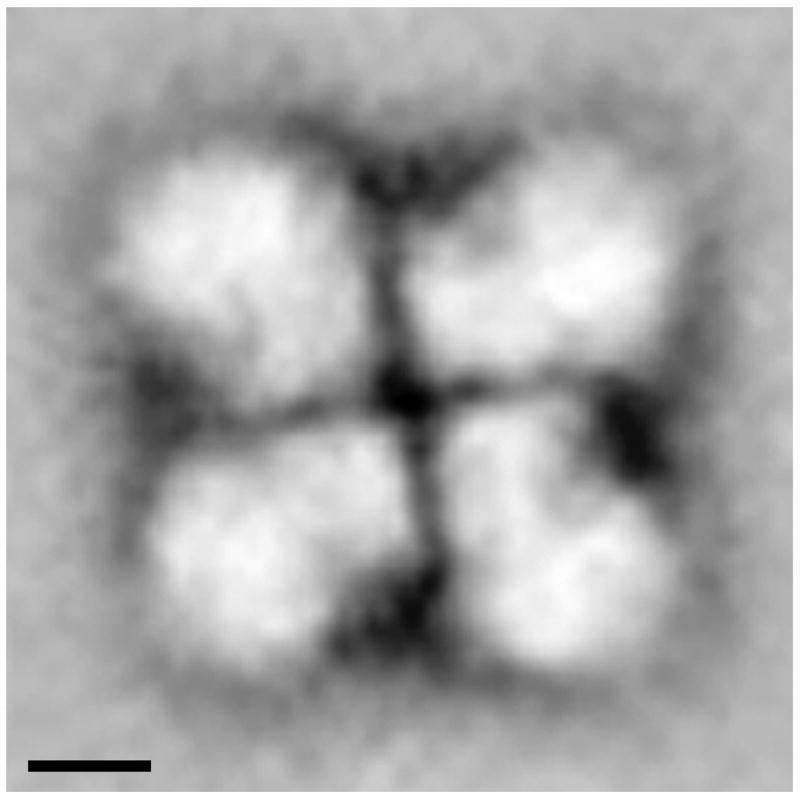
Structural analysis of native spliceosomes in the context of intact supraspliceosomes, using electron microscopy combined with image processing revealed good correlation between the structure of the isolated native spliceosome, solved by cryo-EM, and the native spliceosome within the intact supraspliceosome [80]. Furthermore, this study enabled us to study the arrangement of the native spliceosomes within the intact particle (Fig. 12), revealing that the four native spliceosomes are coordinated by interactions between their small subunits. The edges of the small subunits, which are in the center of the supraspliceosome, form a right angle and thus facilitate close contacts between the small subunits generating a four-fold pattern. This pattern is observed in individual and averaged images [80].
Fig. 12. The arrangement and orientation of native spliceosomes in the intact supraspliceosome.
Intact supraspliceosomes were classified by Correspondence Analysis and Hierarchical Ascendant Classification [80]. One of the classes is depicted showing the close contact between neighboring native spliceosomes in the center of the supraspliceosome, where the small subunits of the native spliceosomes are located. The contacts between the neighboring small subunits form a right-angled cross that reflects a four-fold arrangement. Scale bars represent 10 nm. Adapted from Ref [80].
5.4. The supraspliceosome model
The supraspliceosome is a stand-alone macromolecular machine (Fig. 13), composed of four native spliceosomes [29, 30, 37] that are connected by a single pre-mRNA molecule [40], capable of performing splicing of its pre-mRNA [37] – independent of its length or number of introns [6, 31]. The small subunit of each native spliceosome is placed at the center of the supraspliceosome [80], thus allowing communication between the native spliceosomes, which is a crucial element for regulated alternative splicing and for quality control of the resulting mRNAs. This setting places the large subunit of each native spliceosome, where catalysis by the U snRNPs presumably takes place, in the periphery of the supraspliceosome. This setting was supported by the in-silico study showing unique localization of the U snRNPs within the native spliceosome [69]. The supraspliceosome structure provides a platform to juxtapose exons about to be spliced, and each of the four native spliceosomes, resembling an in vitro assembled spliceosome, can splice the intron wound around it (Fig. 13). The supraspliceosome is a multiprocessor machine that can simultaneously splice four introns – not necessarily in a consecutive manner. This configuration enables examination, prior to introns excision, if correct splice junctions will be combined, and allows rearrangement of splice junction combinations to select the appropriate ones, thus ensuring the fidelity of splicing and alternative splicing.
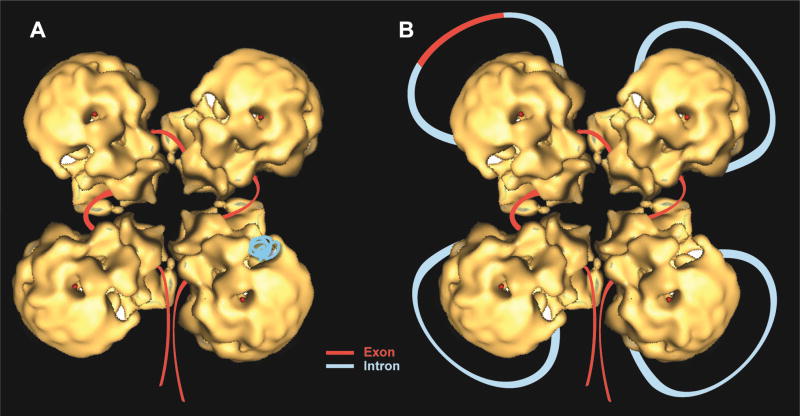
Fig. 13. A refined schematic model of the supraspliceosome.
Correctly orientated native spliceosome volume [37] was placed on a class average, and an idealized supraspliceosome particle was formed by rotating the native volume by 90°, 180° and 270° to generate a four-fold pattern. The rotated volumes fitted well into the parts of neighboring sub-complexes seen in the average image [80]. Schematic models of the supraspliceosome in which the pre-mRNA (introns in blue, exons in red) is connecting four native spliceosomes [29, 37, 80]. The supraspliceosome presents a platform onto which the exons can be aligned and splice junctions can be checked before splicing occurs. (A) The pre-mRNA that is not being processed is folded and protected within the cavities of the native spliceosome. (B) When a staining protocol that allows visualization of nucleic acids was used, RNA strands and loops were seen emanating from the supraspliceosomes [30]. Under these conditions, the RNA kept in the cavity likely unfolded and looped-out. In the looped-out scheme an alternative exon is depicted in the upper left corner.
The supraspliceosome model predicts that each transcript will be assembled in a tetrameric supraspliceosome. Splicing of a multi-intronic pre-mRNA can be facilitated by the translocation of the pre-mRNA through the complex in a ‘rolling model’ fashion. After processing of four introns the RNA roles in to place a new subset of introns for processing. At the other hand, transcripts with less than four introns are also assembled in supraspliceosomes [37, 38]. The interactions of the RNA with the native spliceosomes are presumably sufficient to hold the structure together.
The characterization of the supraspliceosome was performed on the nucleoplasmic complex. Thus, information on the initial stages of its assembly in vivo, whether it occurred in a step-wise manner, or it involved pre-assembled components is yet lacking. However, it is established, that unlike the in vitro spliceosome, all five-spliceosomal U snRNPs are associated with the nucleoplasmic supraspliceosome at all stages of splicing. The unique spatial arrangement of the U snRNPs within the native spliceosome that emerged from our in-silico studies [69], also revealed that there is ample free volume left within the native spliceosome after localization of the spliceosomal U snRNPs, indicating that structural modulations of the U snRNPs during the steps of the splicing reaction can be tolerated while keeping the integrity of the spliceosome assembly. This is consistent with the recent high-resolution structures of spliceosome complexes by cryo-EM revealing dynamic changes between the spliceosome complexes. Notably, movement of the pre-mRNA within the supraspliceosome, proposed in our model is consistent with movement of pre-mRNA elements observed between the different spliceosome subcomplexes [11–17, 19]. The supraspliceosome model differs from the model derived from in vitro studies that predicts the assembly of a spliceosome on each intron [81]. It is however consistent with the cotranscriptional splicing model and especially with the model of cotranscriptional commitment to splicing [23, 82].
The supraspliceosome provides a unique and general machine that encompasses the extensive network of interactions and offers coordination and regulation of the different splicing events that a multiintronic pre-mRNA has to undergo. It regulates alternative splicing [39, 40], and harbors a quality control mechanism (reviewed in Ref [31, 41]), as well as all the additional processing activities, such as 5’-end and 3’-end processing, and RNA editing [35, 43]. Furthermore, intronic pre-microRNAs were found in supraspliceosomes and their processing occurs there [44, 45]. Also, a sub-fraction of SNORDs assembled in non-canonical RNPs were found in supraspliceosomes likely playing a role in alternative splicing [46]. Thus, supraspliceosomes harbor components of all pre-mRNA processing activities, thus representing the nuclear pre-mRNA processing machine. Although the pattern and kinetics of the processing activities will likely be determined for each individual transcript, according to sequences of cis elements of that pre-mRNA, and the various factors that interact with these elements, the supraspliceosome is a general machine, capable of performing and regulating all the pre-mRNA processing activities that the pre-mRNA has to undergo before it can exit from the nucleus to the cytoplasm.
The recent advancements in cryo-EM, enabling structure determination at subnanometric resolution [7] have revolutionized the field of structural molecular biology, including the splicing field [8–19].. In view of this revolution in cryo-EM, further high-resolution imaging studies of the endogenous spliceosome are required. We Take advantage of the recent revolution in the field of structural analysis by cryo-EM, and of our advance in analyzing specific supraspliceosomes at defined functional states and alternatively spliced supraspliceosomes and native spliceosomes, for the cryo-EM structural analysis of supraspliceosomes and native spliceosomes using both cryo-ET and single particle technique as well as subtomograms averaging and in silico segmentations, in order to decipher the mechanism of regulation of alternative splicing, and how is the network of processing activities within the endogenous spliceosome coordinated.
Acknowledgments
We thank Aviva Petcho for excellent technical assistance and Minna Angenitski for help with the electron microscopy, and members of the Sperling labs for their contributions. This work was supported in part by grants from the US National Institutes of Heath [RO1 GM079549 to R.S. and J.S.], The USA-Israel Binational Science Foundation (BSF), the Israel science Foundation (ISF) and the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (to J.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
I declare no conflict of interests.
References
- 1.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3(7):a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papasaikas P, Valcarcel J. The Spliceosome: The Ultimate RNA Chaperone and Sculptor. Trends Biochem Sci. 2016;41(1):33–45. doi: 10.1016/j.tibs.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–60. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 5.Luhrmann R, Stark H. Structural mapping of spliceosomes by electron microscopy. Current Opinion in Structural Biology. 2009;19(1):96–102. doi: 10.1016/j.sbi.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Sperling J, Azubel M, Sperling R. Structure and Function of the Pre-mRNA Splicing Machine. Structure. 2008;16(11):1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Bai XC, McMullen G, Scheres HW. How cryo-EM is revolutionizing structural biology. Trends Bioche, Sciences. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science (New York, N.Y. 2016;351(6272):466–75. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature. 2016;530(7590):298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Luhrmann R, Stark H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science (New York, N.Y. 2016;351(6280):1416–20. doi: 10.1126/science.aad2085. [DOI] [PubMed] [Google Scholar]
- 11.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science (New York, N.Y. 2016;353(6302):904–11. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 12.Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, Stark H, Luhrmann R. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science (New York, N.Y. 2016;353(6306):1399–1405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 13.Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science (New York, N.Y. 2016;353(6302):895–904. doi: 10.1126/science.aag2235. [DOI] [PubMed] [Google Scholar]
- 14.Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537(7619):197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast step II catalytically activated spliceosome. Science (New York, N.Y. 2016 doi: 10.1126/science.aak9979. [DOI] [PubMed] [Google Scholar]
- 16.Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, Nagai K. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542(7641):377–380. doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science (New York, N.Y. 2015;349(6253):1182–91. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523(7558):47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science (New York, N.Y. 2015;349(6253):1191–8. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 20.Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Function of alternative splicing. Gene. 2013;514(1):1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akerman M, Fregoso OI, Das S, Ruse C, Jensen MA, Pappin DJ, Zhang MQ, Krainer AR. Differential connectivity of splicing activators and repressors to the human spliceosome. Genome biology. 2015;16:119. doi: 10.1186/s13059-015-0682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–98. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 24.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends in Molecular Medicine. 2012;18(8):472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Current opinion in cell biology. 2012;24(3):323–32. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212(1):13–27. doi: 10.1083/jcb.201510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sperling R, Sperling J, Levine AD, Spann P, Stark GR, Kornberg RD. Abundant nuclear ribonucleoprotein form of CAD RNA. Mol. Cell Biol. 1985;5:569–575. doi: 10.1128/mcb.5.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miriami E, Angenitzki M, Sperling R, Sperling J. Magnesium cations are required for the association of U small nuclear ribonucleoproteins and SR proteins with pre-mRNA in 200 S large nuclear ribonucleoprotein particles. J. Mol.Biol. 1995;246(2):254–263. doi: 10.1006/jmbi.1994.0081. [DOI] [PubMed] [Google Scholar]
- 29.Azubel M, Wolf SG, Sperling J, Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol. Cell. 2004;15(5):833–9. doi: 10.1016/j.molcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 30.Müller S, Wolpensinger B, Angenitzki M, Engel A, Sperling J, Sperling R. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J. Mol. Biol. 1998;283(2):383–394. doi: 10.1006/jmbi.1998.2078. [DOI] [PubMed] [Google Scholar]
- 31.Shefer K, Sperling J, Sperling R. The supraspliceosome-a multi-task-machine for regulated pre-mRNA processing in the cell nucleus. Computational and Structural Biotechnology Journal. 2014;11:113–122. doi: 10.1016/j.csbj.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperling R. The nuts and bolts of the endogenous spliceosome. Wiley interdisciplinary reviews RNA. 2017;8(1) doi: 10.1002/wrna.1377. [DOI] [PubMed] [Google Scholar]
- 33.Spann P, Feinerman M, Sperling J, Sperling R. Isolation and visualization of large compact ribonucleoprotein particles of specific nuclear RNAs. Proc. Natl. Acad. Sci. USA. 1989;86:466–470. doi: 10.1073/pnas.86.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sperling R, Koster AJ, Melamed-Bessudo C, Rubinstein A, Angenitzki M, Berkovitch-Yellin Z, Sperling J. Three-dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J. Mol. Biol. 1997;267:570–583. doi: 10.1006/jmbi.1997.0898. [DOI] [PubMed] [Google Scholar]
- 35.Raitskin O, Angenitzki M, Sperling J, Sperling R. Large nuclear RNP particles-the nuclear pre-mRNA processing machine. J. Struct. Biol. 2002;140(1):123–30. doi: 10.1016/s1047-8477(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 36.Medalia O, Typke D, Hegerl R, Angenitzki M, Sperling J, Sperling R. Cryoelectron microscopy and cryoelectron tomography of the nuclear pre-mRNA processing machine. J. Struct. Biol. 2002;138(1–2):74–84. doi: 10.1016/s1047-8477(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 37.Azubel M, Habib N, Sperling J, Sperling R. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J. Mol. Biol. 2006;356:955–966. doi: 10.1016/j.jmb.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 38.Kotzer-Nevo H, de Lima Alves F, Rappsilber J, Sperling J, Sperling R. Supraspliceosomes at Defined Functional States Present portray the Pre-Assembled Nature of the pre-mRNA Processing Machine in the Cell Nucleus. Int. J. Mol. Sci. 2014;15:11637–11664. doi: 10.3390/ijms150711637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, Sperling J, Sperling R, Stamm S. Heterogeneous Nuclear Ribonucleoprotein G Regulates Splice Site Selection by Binding to CC(A/C)-rich Regions in Pre-mRNA. J. Biol. Chem. 2009;284(21):14303–15. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sebbag-Sznajder N, Raitskin O, Angenitzki M, Sato TA, Sperling J, Sperling R. Regulation of alternative splicing within the supraspliceosome. J. Struct. Biol. 2012;177(1):152–9. doi: 10.1016/j.jsb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperling J, Sperling R. Nuclear surveillance of RNA polymerase II transcripts. RNA Biol. 2008;5(4):220–224. doi: 10.4161/rna.7162. [DOI] [PubMed] [Google Scholar]
- 42.Kamhi E, Raitskin O, Sperling R, Sperling J. A potential role for initiator-tRNA in pre-mRNA splicing regulation. Proc. Natl. Acad. Sci. USA. 2010;107(25):11319–24. doi: 10.1073/pnas.0911561107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc. Natl. Acad. Sci. USA. 2001;98(12):6571–6. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agranat-Tamir L, Shomron N, Sperling J, Sperling R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucl Acids Res. 2014;42:4640–51. doi: 10.1093/nar/gkt1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Falaleeva M, Agranat-Tamur L, Pages AP, Eyras E, Sperling EJ, Sperling R, Stamm S. The 5’ untranslated region of the serotonin receptor 2C pre-mRNA generates miRNAs and is expressed in non-neuronal cells. Exp Brain Res. 2013;230(4):387–394. doi: 10.1007/s00221-013-3458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falaleeva M, Pages A, Matsuzek Z, Hidmi S, Agranat-Tamir L, Korotkov K, Nevo Y, Eyras E, Sperling R, Stamm S. dual function of C/d box snoRNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci U S A. 2016;113:E1625–34. doi: 10.1073/pnas.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassarman DA, Steitz JA. A base-pairing interaction between U2 and U6 small nuclear RNAs occurs in >150S complexes in HeLa cell extracts: Implications for the spliceosome assembly pathway. Proc. Natl. Acad. Sci. USA. 1993;90:7139–7143. doi: 10.1073/pnas.90.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iborra FJ, Jackson DA, Cook PR. The path of transcripts from extra-nucleolar synthetic sites to nuclear pores: transcripts in transit are concentrated in discrete structures containing SR proteins. J. Cell Science. 1998;111:2269–2282. doi: 10.1242/jcs.111.15.2269. [DOI] [PubMed] [Google Scholar]
- 49.Padgett RA, Wahl GM, Coleman PF, Stark GR. N-(Phosphonacetyl)-L-aspartateresistant hamster cells overaccumulate a single mRNA coding for the multifunctional protein that catalyzes the first steps of UMP synthesis. J. Biol. Chem. 1979;254(3):974–80. [PubMed] [Google Scholar]
- 50.Wahl GM, Padgett RA, Stark GR. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. The Journal of biological chemistry. 1979;254(17):8679–89. [PubMed] [Google Scholar]
- 51.Sperling R, Sperling J. The lnRNP particle - A naturally assembled complex of pre-mRNA and splicing factors. In: Schenkel J, editor. RNP Particles, Splicing and Autoimmune Diseases. Springer: 1998. pp. 29–47. [Google Scholar]
- 52.Yitzhaki S, Miriami E, Sperling J, Sperling R. Phosphorylated Ser/Arg-rich proteins: Limiting factors in the assembly of 200S large nuclear ribonucleoprotein particles. Proc. Natl. Acad. Sci. USA. 1996;93:8830–8835. doi: 10.1073/pnas.93.17.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sperling R, Sperling J. Large nuclear ribonucleoprotein particles of specific RNA polymerase II transcripts. In: Strauss PR, Wilson SH, editors. The Eukaryotic Nucleus, Molecular Biochemistry and Macromolecular Assemblies. Telford Press; Caldwell, NJ: 1990. pp. 453–476. [Google Scholar]
- 54.Markus MA, Heinrich B, Raitskin O, Adams DJ, Mangs H, Goy C, Ladomery M, Sperling R, Stamm S, Morris BJ. WT1 interacts with the splicing protein RBM4 and regulates its ability to modulate alternative splicing in vivo. Exp. Cell Res. 2006;312(17):3379–88. doi: 10.1016/j.yexcr.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y-HJ, Markus AM, Mangs HA, Raitskin O, Sperling R, Morris BJ. ZRANB2 localizes to supraspliceosomes and influences the alternative splicing of multiple genes in the transcriptome. Molecular biology reports. 2013;40(9):5381–5395. doi: 10.1007/s11033-013-2637-9. [DOI] [PubMed] [Google Scholar]
- 56.Medalia O, Koster AJ, Tocilj A, Angenitzki M, Sperling J, Berkovitch YZ, Sperling R. Automated electron tomography of large nuclear RNP (lnRNP) particles- the naturally assembled complexes of precursor messenger RNA and splicing factors. J. Struct. Biol. 1997;120(3):228–236. doi: 10.1006/jsbi.1997.3926. [DOI] [PubMed] [Google Scholar]
- 57.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Quart. Rev. Biophys. 1988;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 58.Uzgiris EE, Kornberg RD. Two-dimensional crystallization technique for imaging macromolecules, with application to antigen--antibody--complement complexes. Nature. 1983;301(5896):125–9. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- 59.Kornberg RD, Darst SA. Two-dimensional crystals of proteins on lipid layers. Curr. Opin. Struct. Biol. 1991;1:642–646. doi: 10.1016/0959-440x(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 60.Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science (New York, N.Y. 2002;298(5596):1209–13. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 61.Schiske P. Zur Frage der Bildrekonstruktion durch Fokusreihen (On the question of image "restoration" from focal series); Proc. 4th Eur. Reg. Conf. on Electron Microscopy, Rome; 1968. pp. 145–146. [Google Scholar]
- 62.Typke D, Hegerl K, Kleinz J. Image restoration for biological objects using external TEM control and electronic image recording. Ultramicroscopy. 1992;46:157–173. [Google Scholar]
- 63.Tanford C. Physical Chemistry of Macromolecules. John Wiley & Sons; New York: 1961. [Google Scholar]
- 64.Dubochet J, Ducommun M, Zollinger M, Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J. Ultrastruct. Res. 1971;35:147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- 65.Miriami E, Motro U, Sperling J, Sperling R. Conservation of an open-reading frame as an element affecting 5' splice site selection. J. Struct. Biol. 2002;140(1):116–22. doi: 10.1016/s1047-8477(02)00539-7. [DOI] [PubMed] [Google Scholar]
- 66.Nevo Y, Kamhi E, Jacob-Hirsch J, Amariglio N, Rechavi G, Sperling J, Sperling R. Genome-wide activation of latent donor splice sites in stress and disease. Nucleic Acids Res. 2012;40(21):10980–94. doi: 10.1093/nar/gks834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Penczek PA, Zhu J, Frank J. A common-lines based method for determining orientations for N > 3 particle projections simultaneously. Ultramicroscopy. 1996;63(3–4):205–18. doi: 10.1016/0304-3991(96)00037-x. [DOI] [PubMed] [Google Scholar]
- 68.Van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21(2):111–23. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 69.Frankenstein Z, Sperling J, Sperling R, Eisenstein M. A unique spatial arrangement of the snRNPs within the native spliceosome emerges from In silico studies. Structure. 2012;20:1097–1106. doi: 10.1016/j.str.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA (New York, N.Y. 2007;13(6):868–80. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baserga SJ, Steitz JA. The diverse world of amsll ribonucleoproteins. In: Gesteland RF, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1993. pp. 359–381. [Google Scholar]
- 72.Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9(1):31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 73.Nilsen TW. The spliceosome: no assembly required? Mol. Cell. 2002;9(1):8–9. doi: 10.1016/s1097-2765(02)00430-6. [DOI] [PubMed] [Google Scholar]
- 74.Chen YI, Moore RE, Ge HY, Young MK, Lee TD, Stevens SW. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hogg R, McGrail JC, O'Keefe RT. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem Soc Trans. 2010;38(4):1110–5. doi: 10.1042/BST0381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taniguchi I, Ohno M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Molecular and cellular biology. 2008;28(2):601–8. doi: 10.1128/MCB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiang K, Tong L, Manley JL. Delineating the structural blueprint of the pre-mRNA 3'-end processing machinery. Molecular and cellular biology. 2014;34(11):1894–910. doi: 10.1128/MCB.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agranat L, Raitskin O, Sperling J, Sperling R. The editing enzyme ADAR1 and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proc. Natl. Acad. Sci. USA. 2008;105(13):5028–33. doi: 10.1073/pnas.0710576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Medalia O, Heim M, Guckenberger R, Sperling R, Sperling J. Gold-Tagged RNA-A Probe for Macromolecular Assemblies. J. Struct. Biol. 1999;127(2):113–119. doi: 10.1006/jsbi.1999.4134. [DOI] [PubMed] [Google Scholar]
- 80.Cohen-Krausz S, Sperling R, Sperling J. Exploring the architecture of the intact supraspliceosome using electron microscopy. J. Mol. Biol. 2007;368(2):319–27. doi: 10.1016/j.jmb.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 81.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92(3):315–26. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 82.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15(3):163–75. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Luhrmann R, Stark H. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol. Cell. 2006;24(2):267–78. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]