Abstract
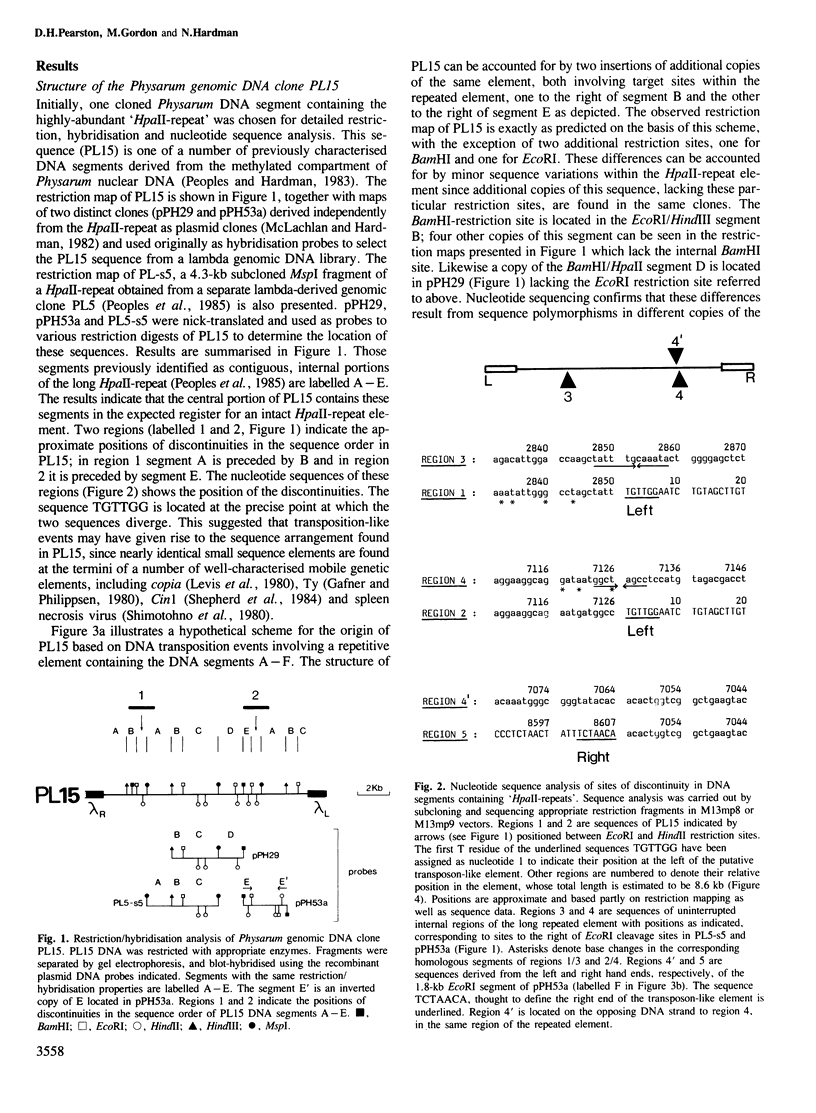
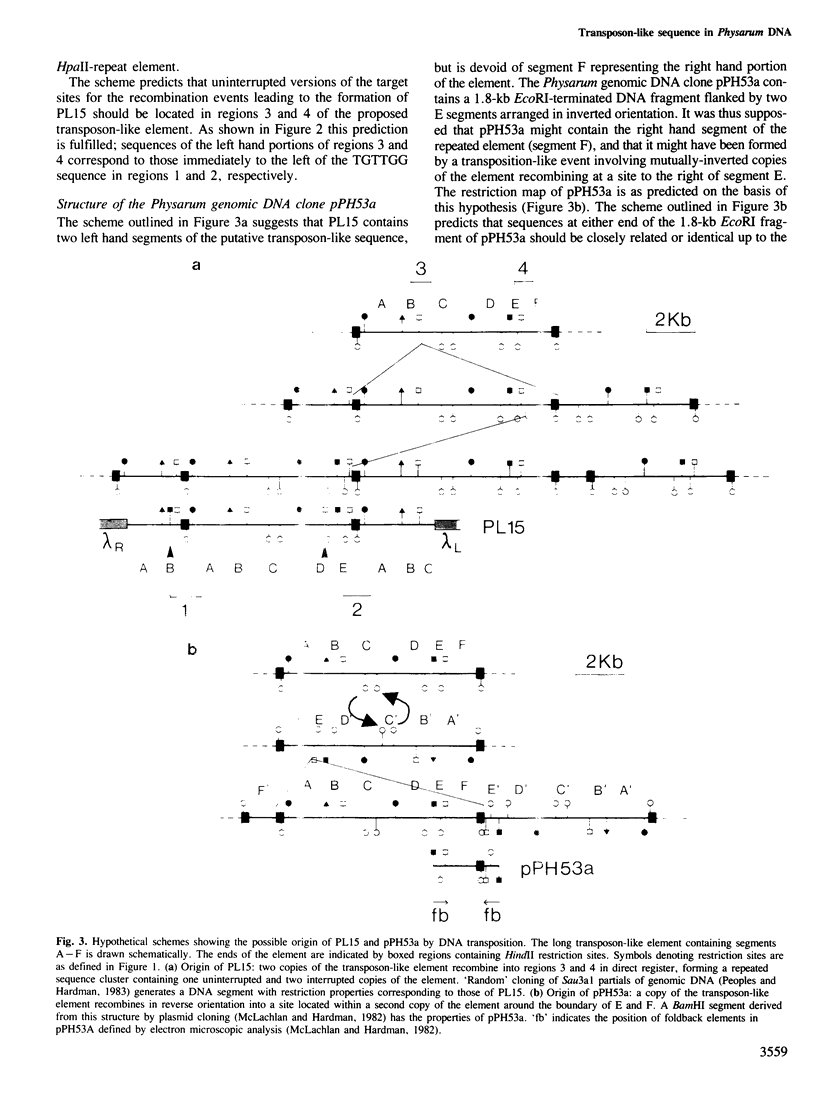
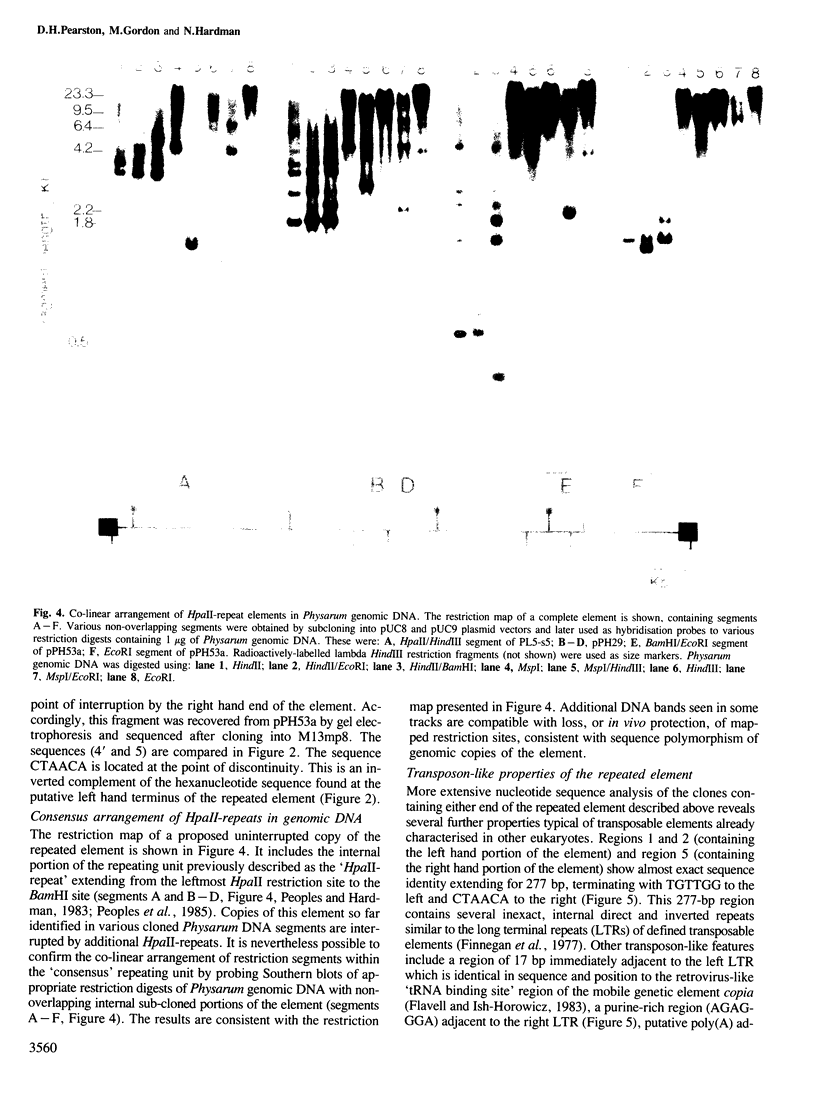
A family of long, highly-repetitive sequences, referred to previously as `HpaII-repeats', dominates the genome of the eukaryotic slime mould Physarum polycephalum. These sequences are found exclusively in scrambled clusters. They account for about one-half of the total complement of repetitive DNA in Physarum, and represent the major sequence component found in hypermethylated, 20-50 kb segments of Physarum genomic DNA that fail to be cleaved using the restriction endonuclease HpaII. The structure of this abundant repetitive element was investigated by analysing cloned segments derived from the hypermethylated genomic DNA compartment. We show that the `HpaII-repeat' forms part of a larger repetitive DNA structure, ∼8.6 kb in length, with several structural features in common with recognised eukaryotic transposable genetic elements. Scrambled clusters of the sequence probably arise as a result of transposition-like events, during which the element preferentially recombines in either orientation with target sites located in other copies of the same repeated sequence. The target sites for transposition/recombination are not related in sequence but in all cases studied they are potentially capable of promoting the formation of small `cruciforms' or `Z-DNA' structures which might be recognised during the recombination process.
Keywords: Physarum, repetitive DNA, transposon
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Cappello J., Cohen S. M., Lodish H. F. Dictyostelium transposable element DIRS-1 preferentially inserts into DIRS-1 sequences. Mol Cell Biol. 1984 Oct;4(10):2207–2213. doi: 10.1128/mcb.4.10.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke D. J., Dee J. Methods for the isolation and analysis of plasmodial mutants in Physarum polycephalum. Genet Res. 1974 Oct;24(2):175–187. doi: 10.1017/s0016672300015202. [DOI] [PubMed] [Google Scholar]
- D'Eustachio P., Ruddle F. H. Somatic cell genetics and gene families. Science. 1983 May 27;220(4600):919–924. doi: 10.1126/science.6573776. [DOI] [PubMed] [Google Scholar]
- Di Nocera P. P., Digan M. E., Dawid I. B. A family of oligo-adenylate-terminated transposable sequences in Drosophila melanogaster. J Mol Biol. 1983 Aug 25;168(4):715–727. doi: 10.1016/s0022-2836(83)80071-0. [DOI] [PubMed] [Google Scholar]
- Dowsett A. P., Young M. W. Differing levels of dispersed repetitive DNA among closely related species of Drosophila. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4570–4574. doi: 10.1073/pnas.79.15.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring H. P., Tillmann E., Starlinger P. DNA sequence of the maize transposable element Dissociation. Nature. 1984 Jan 12;307(5947):127–130. doi: 10.1038/307127a0. [DOI] [PubMed] [Google Scholar]
- Eden F. C., Musti A. M., Sobieski D. A. Clusters of repeated sequences of chicken DNA are extensively methylated but contain specific undermethylated regions. J Mol Biol. 1981 May 15;148(2):129–151. doi: 10.1016/0022-2836(81)90509-x. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. The origin of extrachromosomal circular copia elements. Cell. 1983 Sep;34(2):415–419. doi: 10.1016/0092-8674(83)90375-6. [DOI] [PubMed] [Google Scholar]
- Gafner J., Philippsen P. The yeast transposon Ty1 generates duplications of target DNA on insertion. Nature. 1980 Jul 24;286(5771):414–418. doi: 10.1038/286414a0. [DOI] [PubMed] [Google Scholar]
- Gerrie L. M., Humphreys J., Peoples O. P., Hardman N. Sequence organisation in nuclear DNA from Physarum polycephalum. Arrangement of highly-repeated sequences. Biochim Biophys Acta. 1983 Nov 17;741(2):214–223. doi: 10.1016/0167-4781(83)90061-1. [DOI] [PubMed] [Google Scholar]
- Hardman N., Bell A. J., McLachlan A. Organisation of inverted repeat sequences in hamster cell nuclear DNA. Biochim Biophys Acta. 1979 Oct 25;564(3):372–389. doi: 10.1016/0005-2787(79)90029-7. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L., Brown A. J., McLachlan A. Distribution of inverted repeat sequences in nuclear DNA from Physarum polycephalum. Eur J Biochem. 1979 Feb 15;94(1):179–187. doi: 10.1111/j.1432-1033.1979.tb12884.x. [DOI] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. Sequence organization in Dictyostelium: unique structure at the 5'-ends of protein coding genes. Nucleic Acids Res. 1983 Jan 25;11(2):541–552. doi: 10.1093/nar/11.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- McLachlan A., Hardman N. Analysis of foldback sequences and repetitive sequences in cloned segments of nuclear DNA from Physarum polycephalum. Biochim Biophys Acta. 1982 Apr 26;697(1):89–100. doi: 10.1016/0167-4781(82)90049-5. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Lafer E. M., Peck L. J., Wang J. C., Stollar B. D., Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982 Dec;31(2 Pt 1):309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- Peoples O. P., Hardman N. An abundant family of methylated repetitive sequences dominates the genome of Physarum polycephalum. Nucleic Acids Res. 1983 Nov 25;11(22):7777–7788. doi: 10.1093/nar/11.22.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples O. P., Whittaker P. A., Pearston D., Hardman N. Structural organization of a hypermethylated nuclear DNA component in Physarum polycephalum. J Gen Microbiol. 1985 May;131(5):1157–1165. doi: 10.1099/00221287-131-5-1157. [DOI] [PubMed] [Google Scholar]
- Saigo K., Kugimiya W., Matsuo Y., Inouye S., Yoshioka K., Yuki S. Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature. 1984 Dec 13;312(5995):659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd N. S., Schwarz-Sommer Z., Blumberg vel Spalve J., Gupta M., Wienand U., Saedler H. Similarity of the Cin1 repetitive family of Zea mays to eukaryotic transposable elements. Nature. 1984 Jan 12;307(5947):185–187. doi: 10.1038/307185a0. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980 Oct;21(3):599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Tabata S., Pachl C. The clustered and scrambled arrangement of moderately repetitive elements in Drosophila DNA. Cell. 1979 Dec;18(4):1231–1246. doi: 10.1016/0092-8674(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Whittaker P. A., Hardman N. Methylation of nuclear DNA in Physarum polycephalum. Biochem J. 1980 Dec 1;191(3):859–862. doi: 10.1042/bj1910859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]





