To the Editor
An estimated 30% to 70% of children with asthma outgrow their asthma symptoms, indicating that there is considerable variability in the possible disease course of asthma. Asthma is characterized by an oversensitivity of the airways to external stimuli. This airway hyperresponsiveness (AHR), when measured by challenge with methacholine inhalation, is a cardinal feature of asthma, and worse AHR correlates with worse asthma. AHR can be used to confirm a diagnosis of asthma, and the remission of AHR is an indicator of improved prognosis and reduced symptoms. Genetic variants influencing AHR have been identified, and variant analysis shows that AHR has significant genetic heritability. We here extend these results through an investigation of microRNAs (miRNAs, miRs): small noncoding single-stranded RNA molecules that regulate gene expression by degrading mRNA transcripts and/or inhibiting the translation of target genes and their ability to predict longitudinal AHR remission.
As the primary response variable, we used remission of AHR, measured through the methacholine challenge (provocative concentration required to achieve a 20% reduction in FEV1, hereafter PC20). We examined expression levels of circulating miRNAs from the serum of 160 non-Hispanic white children with asthma arbitrarily selected from the Childhood Asthma Management Program (CAMP) cohort, a clinical trial of mild-to-moderate persistent childhood asthma; subjects enrolled were 5 to 12 years old and displayed AHR at baseline, as evidenced by a PC20 value of 12.5 mg/mL or less.1 Baseline demographic, pulmonary function testing, and methacholine challenge data were used in our investigations, in addition to at least annual PC20 measurements through up to 16 years of follow-up. We have previously shown that these 160 subjects are similar to the remainder of the CAMP cohort.2 Serum microRNA was previously assayed for 738 unique human microRNAs using TaqMan OpenArray microRNA quantitative PCR primers (Life Technologies Megaplex RT Primers, Human Pool Set v3.0; Omaha, Neb). MicroRNA extraction, preparation, post-PCR data processing, and quality control have been previously described and the data accessible on the National Center for Biotechnology Information Gene Expression Omnibus under accession GSE74770,2 although we use here a detection threshold of 75% of the 160 subjects plus 13 replicates (173 total) as a minimum criteria for including miRNAs in our analysis.
We considered subjects with PC20 value of 37.5 mg/mL or more to display no AHR. We found that the greatest number of subjects fit this description at 14 years (168 months) after randomization (see Fig E1 in this article’s Online Repository at www.jacionline.org); we henceforth considered all subjects at or above 37.5 mg/mL PC20 at 14 years to be in AHR remission. Cohort characteristics at baseline are presented in Table I. We found that sex, baseline PC20, FEV 1/forced vital capacity, and bronchodilator response were significantly different between those CAMP subjects with AHR remission and those without, and included these baseline clinical measurements in our predictive modeling process.
TABLE I.
Cohort characteristics, stratified by AHR remission phenotype
| Characteristic | No remission (n = 128) | AHR remission (n = 32) | P value |
|---|---|---|---|
| Baseline age (y), mean ± SD | 8.88 ± 2.15 | 8.62 ± 2.02 | .53 |
| Sex: male, % | 49.20 | 75.00 | .0086 |
| Baseline height (cm), mean ± SD | 133.25 ± 13.85 | 130.70 ± 12.78 | .35 |
| Baseline PC20 (log mg/mL), mean ± SD | 1.48 ± 1.83 | 3.82 ± 3.29 | 2.4 × 10−7 |
| Baseline FEV1 (L), mean ± SD | 1.65 ± 0.48 | 1.62 ± 0.38 | .77 |
| Baseline FEV1 (percent predicated),mean ± SD | 93.70 ± 14.92 | 96.97 ± 13.80 | .26 |
| Baseline FEV1/FVC, mean ± SD | 78.03 ± 8.93 | 81.50 ± 7.25 | .044 |
| Baseline bronchodilator response (%), mean ± SD | 12.85 ± 11.00 | 7.28 ± 7.75 | .0077 |
| Baseline vitamin D insufficiency, count (%) | 31 (24.22) | 4 (12.50) | .15 |
| Oral steroid bursts required through trial, mean ± SD | 2.90 ± 3.53 | 2.72 ± 3.68 | .8 |
| Reduced growth longitudinal lung function pattern, n (%) | 57 (44.53) | 15 (46.88) | .81 |
| Early decline longitudinal lung function pattern, n (%) | 54 (42.19) | 14 (43.75) | .87 |
| Steroid responsiveness endophenotype, mean ± SD | 0.64 ± 1.11 | 0.48 ± 0.91 | .45 |
Reduced growth and early decline longitudinal lung function patterns as described in McGeachie et al.8 Steroid responsiveness endophenotype as described in Clemmer et al.9 P values computed using the chi-square test or the regression test.
FVC, Forced vital capacity; PC20, provocative concentration of methacholine required to effect a 20% decrease in FEV1.
We were able to learn a Bayesian network (BN) of miRNA within those with asthma predictive of AHR at 14 years in the CAMP cohort, using the CGBayesNets software package.3 We obtained an area under the receiver operating characteristic curve convex hull (AUC)4 of 86.5%, which was indicative of strong predictive performance (Fig 1, B), for example, achieving a sensitivity of 84% with a specificity of 70%. Furthermore, permutation testing showed that the risk of overfitting was low; performance of the 86.5% AUC was determined to be significantly better than might be obtained by chance (permutation test, P = .006; see Fig E2 in this article’s Online Repository at www.jacionline.org). This network contained 12 variables predictive of 14-year AHR remission: miR-106a-5p, miR-126-5p, miR-1290, miR-146b-5p, miR-17-5p, miR-185-5p, miR-199a-3p, miR-19b-3p, miR-25-3p, miR-30a-5p, miR-328-3p, and baseline PC20. By far the strongest statistical association within the network was between AHR and miR-30a-5p, with a Bayes factor5 (a Bayesian log-likelihood ratio) of 42.9 (Fig 1, arrow thickness), indicating that a dependence between the 2 is e42.9 times (4.3 × 1018) more likely than independence; mean Bayes factor for edges within Fig 1 is 14.8 ± 12.8.
FIG 1.
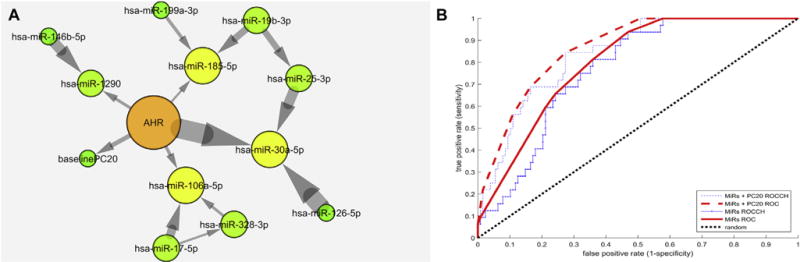
A, Markov neighborhood of conditional Gaussian BN predictive of AHR remission at 14 years. Included are 11 miRs and baseline PC20. Arrows indicate statistical dependence of the target node on the source node. Arrow thickness indicates strength of connection in log Bayes factor. Node color and size indicate the number of connections. See this article’s Methods section in the Online Repository at www.jacionline.org and McGeachie et al3 for additional details. B, Receiver operating characteristic curve and convex hull (ROCCH) for prediction of 14-year AHR remission using Bayesian in Fig 1, A. The convex hull of raw receiver operating characteristic curves represents achievable tradeoffs between false positives and false negatives using the classifiers. Area under the convex hull of the curve (AUC) is 86.5% for the Bayesian model including baseline PC20 and 80.0% for the Bayesian model of just miRs.
Logistic regression analysis did not reveal any miRs significantly associated with 14-year AHR remission (see Fig E3 in this article’s Online Repository at www.jacionline.org). This is indicative of the nonlinear combination of miRs in our BN; microRNA acting in networks is consistent with the way many genes are targeted by multiple miRs, each of which may have an additive or even negative effect on expression. These effects are sometimes masked and missed by single-miR analysis.6 Similarly, no miRs were significantly associated with sex in a logistic regression test, both before and after adjusting for baseline PC20. At least 6 of the 11 miRs included in our predictive model have previously been associated with asthma or pulmonary conditions: mirR-106a, mirR-126, mirR-19, mirR-30, mirR-146, and mirR-25.
Our BN included the baseline PC20 value in the prediction of AHR remission at 14 years after baseline, achieving an AUC of 86.5%. We also tested the predictive ability of the model based only on microRNAs, removing the baseline PC20 and recomputing parameters of a BN based on the remaining 11 miRs. This model achieved an AUC of 80.0%, suggesting good prediction, but which was significantly different from the full model’s 86.5% (P = .034). The microRNA-only BN was not strongly correlated with baseline PC20 values (r2 = 0.26), showing that the miRs provide additional, orthogonal information about future AHR remission.
We examined the expression and function of miR-30a-5p in airway smooth muscle (ASM) cells in vitro. Analysis of miR-deep sequencing data7 showed that miR-30a is among the most abundant miRs expressed in the human primary ASM cells. Because other miR-30 family members (miR-30d, miR-30e) are also highly expressed in ASM cells, we transfected mimics of these miRs in ASM cells and examined their effects on ASM growth and hypertrophy in vitro. There was no significant effect by any of the miRs on ASM growth (data not shown). However, miR-30a, miR-30d, and miR-30e all significantly increased the size of ASM cells (P < .01; see Fig E5 in this article’s Online Repository at www.jacionline.org), indicating potential roles for miR-30 family members including miR-30a in ASM hypertrophy and AHR in those with asthma.
Although the present model should be validated in other longitudinal asthma cohorts, this work demonstrates that micro-RNAs have the potential to be powerful prognostic biomarkers for future AHR remission and asthma prognosis. Additional longitudinal profiling of microRNAs alongside longitudinal clinical observation will be key in moving these models forward.
METHODS
Childhood Asthma Management Program
We used subjects from the CAMP cohort. CAMP was a clinical trial of mild-to-moderate persistent childhood asthma; subjects enrolled were 5 to 12 years old and displayed AHR at baseline, as evidenced by a PC20 level of 12.5 mg/mL or less.E1 Baseline demographic, pulmonary function testing, and methacholine challenge data were used. CAMP subjects were followed for additional observation after completing the trial through CAMP Continuation Studies (CAMPCS)E2 and CAMPCS2 and CAMPCS3, for a total of up to 18 years of observation. Subjects remaining in the studies went through methacholine challenges annually. Informed consent was obtained from all CAMP, CAMPCS, CAMPCS2, and CAMPCS3 subjects or guardians at the start of each study. The CAMP miRNA study has been approved by the Brigham and Women’s Hospital institutional review board.
Airway hyperresponsiveness
The primary phenotype was remission of AHR, a cardinal feature of asthma. This was measured through the methacholine challenge (PC20), when an interpolated value of 37.5 mg/mol was considered to be remission or absence of AHR. Across the period of observation through CAMP and CAMP continuations, subjects performed the methacholine challenge at least annually; at each challenge time point, we evaluated the number of subjects meeting the 37.5 mg/mL limit, and chose the time point with the maximum number of subjects at or above this level as our primary outcome.
MicroRNA assay
Serum microRNA was previously assayed for 738 unique human micro-RNAs using TaqMan OpenArray microRNA quantitative PCR primers (Life Technologies Megaplex RT Primers, Human Pool Set v3.0). MicroRNA extraction, preparation, post-PCR data processing, and quality control have been previously described and the data accessible on the NCBI Gene Expression Omnibus under accession GSE74770E3. Initial quality control was performed per manufacturer protocol using predefined thresholds for amplification scores (>1.24) and Cq (>0.80) CIs. MicroRNAs were annotated using miRBase release 21 (www.mirbase.org).E4 To assess technical replication, analysis of biological replicates was performed in approximately 10% of the cohort (13 subjects) and overall demonstrated high miRNA-miRNA correlations (rank correlations >0.90 for replicate samples). Quantile normalization was performed on the detected miRNAs samplewise to the mean of the data matrix using MatLab (MathWorks Inc, Natick, Mass). An miRNA was included in the analysis if expression was detected in at least 75% of 173 subjects (including 13 replicates). Based on this criterion, a total of 99 miRs were included in the analysis. Data were further normalized and centered for BN analysis, which used conditional Gaussian BNs and performs better on normal-distributed data.
BN prediction
We used CGBayesNets to learn a BN predictive of AHR following our previous methodology.E5 For predictive network discovery, we included 99 miRNAs that were identified in at least 75% of the 173 samples, along with known clinical indicators for AHR and asthma remission: sex, baseline PC20, baseline FEV1/forced vital capacity, baseline bronchodilator response. Using the CGBayesNets package, we used the K2 backtracking search procedure to learn a BN predictive of AHR remission. This iteratively identified the most likely connection between pairs of variables, where likelihood is measured by the posterior probability of the data given the model and using a Gamma prior over parameters of Gaussian variables. Hyperparameters for these were chosen to implement a stringent complexity penalty during network structure learning (hyperparameter nu = 10, minimum log Bayes factor = 3).E6 The K2-algorithm continued adding edges to the network until no further edges increased the posterior likelihood above threshold. Predictive accuracy was measured by AUC. Permutation testing to rule out overfitting was performed following the same protocols, except with randomized shuffling of phenotype labels.
Transfection, proliferation, and cell size measurement in human airway smooth muscle (HASM) cells
HASM cells were transfected with 10 nM of either scramble control (AllStars Negative Control siRNA, Qiagen) or miR mimic (Qiagen, Venlo, The Netherlands) using RNAiMax (ThermoFisher Scientific, Waltham, Mass) according to the manufacturer’s protocol. Seventy-two hours after transfection, cells were trypsinized and then measured for both cell number and cell size using the Moxi Z Cell Analyzer (Orflo Technologies, Ketchum, Idaho). Cell growth was presented as the percentage of cell number relative to scramble control. Average cell diameter (μm) was compared in mimic-transfected versus scramble-transfected HASM cells. Data (mean ± SE) were obtained from 3 independent experiments, using the same cell line. MicroRNAs in ASM cells were sequenced by small RNA-seq, and was previously described.E7
Supplementary Material
Acknowledgments
This work is supported by the National Institutes of Health (NIH) (grant nos. R01 HL127332, R01 HL129935, and U01 HL65899). M.J.M. is supported by a grant from the Parker B. Francis Foundation. J.S.D. is supported by the NIH (grant no. T32 HL007427). A.T.K. is supported by the NIH (grant no. K25 HL091124). A.D. is supported by grant no. K01 HL130629. J.E.S. is supported by grant no. R00-HL109162. M.S. and Q.L. are supported by grant no. R01 HL114769. S.T.W. is supported by grant nos. P01 HL132825 and R37 HL066289.
Footnotes
Disclosure of potential conflict of interest: M. J. McGeachie’s institution has received the Parker B. Francis Grant for this work. J. S. Davis’s institute received a grant from the National Institutes of Health for this work and grants from the National Institutes of Health for other works. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 2.Kho AT, Sharma S, Davis JS, Spina J, Howard D, McEnroy K, et al. Circulating microRNAs: association with lung function in asthma. PLoS One. 2016;11:e0157998. doi: 10.1371/journal.pone.0157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGeachie MJ, Chang HH, Weiss ST. CGBayesNets: conditional Gaussian Bayesian network learning and inference with mixed discrete and continuous data. PLoS Comput Biol. 2014;10:e1003676. doi: 10.1371/journal.pcbi.1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Provost F, Fawcett T. Robust classification for imprecise environments. Mach Learn. 2001;44:203–31. [Google Scholar]
- 5.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–95. [Google Scholar]
- 6.Garcia-Garcia F, Panadero J, Dopazo J, Montaner D. Integrated gene set analysis for microRNA studies. Bioinformatics. 2016;32:2809–16. doi: 10.1093/bioinformatics/btw334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu R, Pan W, Fedulov AV, Jester W, Jones MR, Weiss ST, et al. MicroRNA-10a controls airway smooth muscle cell proliferation via direct targeting of the PI3 kinase pathway. FASEB J. 2014;28:2347–57. doi: 10.1096/fj.13-247247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGeachie MJ, Yates KP, Zhou X, Guo F, Sternberg AL, Van Natta ML, et al. Patterns of growth and decline in lung function in persistent childhood asthma. N Engl J Med. 2016;374:1842–52. doi: 10.1056/NEJMoa1513737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemmer GL, Wu AC, Rosner B, McGeachie MJ, Litonjua AA, Tantisira KG, et al. Measuring the corticosteroid responsiveness endophenotype in asthmatic patients. J Allergy Clin Immunol. 2015;136:274–81.e8. doi: 10.1016/j.jaci.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


