To the editor
Each of the major haplotypes of the HBB gene cluster in sickle cell anemia are associated with different fetal hemoglobin (HbF) levels. Four of these HBB haplotypes originated in Africa (Benin, Central African Republic, Cameroon, and Senegal) while the Arab-Indian haplotype arose independently in the Arabian Peninsula or in India. Although HbF is the major modulator of disease severity, the genetic elements that underlie the association of HbF and HBB haplotypes are not fully understood1.
Saudi sickle cell patients from the Southwestern Province, whose HBB gene cluster is of African origin, have HbF levels of about 10%; African origin patients with the Benin haplotype have HbF levels of about 6%. Saudi patients have a genetic population structure similar to other Arabs, which does not resemble African-origin patients2. We hypothesized that while Saudi and African American Benin haplotype homozygotes have similar HBB clusters, there might be common variants in the Saudi Benin patients that are associated with their increased HbF; conversely, African American Benin patients might have common variants that are associated with reduced HbF relative to Saudi patients.
To study the genetic differences between Saudi and African American Benin haplotype patients that might be associated with HbF, we imputed genome-wide association study (GWAS) data from the Cooperative Study of Sickle Cell Disease (CSSCD) and patients from the Southwestern Province of Saudi Arabia to 1000 Genomes Phase 3 v5 reference panel. Pre-imputation quality control was performed using PLINK3, and imputation was carried out through the Michigan Imputation server and phased imputed data was obtained using Eagle4. We classified haplotypes using haplotypeClassifier available from https://github.com/eshaikho/haplotypeClassifier. Only homozygous Benin haplotype cases were selected for downstream analysis. We then removed related samples and outliers (based on their genetic markers) in the first 20 principal components (PCs) using King and EIGENSOFT, respectively5,6. There were 293 African American patients not taking hydroxyurea, 153 males, and 140 females, aged between 2 and 68 years with an average HbF of 6.38%, and 63 Saudi Benin haplotype patients 27 of them are taking hydroxyurea, 36 males and 27 females, aged between 4 and 43 years, with an average HbF of 10.38%. A linear model adjusted for age and sex to predict the effect of PCs HbF level showed insignificance of the first 20 components indicating absence of population substructure within each population; the first five PCs were used in the final GWAS analysis to account for any potential bias due to these components. Log10 HbF levels were employed as a quantitative phenotype to find the most significant SNPs in each cohort. Efficient and Parallelizable Association Container Toolbox (EPACTS; https://github.com/statgen/EPACTS) adjusted for sex and the first five PCs were used for the final analysis. To avoid false associations due to small sample size we only considered SNPs with minor allele frequency (MAF) greater than 0.05. Including age as covariate doesn’t improve the goodness of the model fit, thus it was excluded from the final model. Both cohorts were analyzed separately, and subjected to the same analytical methods except for hydroxyurea adjustment in Saudi Benin patients.
In African American Benin haplotype patients, only rs1427407 in BCL11A met GWAS significance levels for association with HbF (Figure 1). Six intronic SNPs in BCL11A had marginal genome-wide significance; 3 intronic SNPS in LARGE1, NEDD9 and PAK2 also showed marginal GWAS significance with p-values between 8.97E-07 and 2.61E-07 (Table 1). In Saudi Benin cases, there were no associations with HbF meeting GWAS significance levels; however the small sample size reduced the statistical power of the study to detect an association. The allele frequencies for the top 10 associated SNPs in African American patients were similar to that in Saudi patients except for rs6706648 and rs7606173 where the MAFs were 0.4 and 0.44 in African American and 0.11 and 0.25 in Saudi cases. To examine the effect of rs1427407, rs6706648, and rs7606173 we examined the distribution of HbF levels by the genotypes of these SNPs. We took advantage of the phased imputed data to examine the three SNPs haplotype effect on HbF. Homozygosity for a TCG haplotype of rs1427407, rs6706648 and rs7606173, respectively, was associated with 10 % HbF in African American patients. This haplotype was found in 29% of Saudi and 24% of African American Benin haplotype patients. Homozygosity for the T allele rs1427407 was always associated with homozygosity for the C allele of rs6706648 and G allele of rs7606173. The homozygosity for a GTC haplotype of rs1427407, rs6706648 and rs7606173, respectively, was associated with 4.5 HbF in African American Benin. GTC haplotype has frequency of 0.40 in African American Benin, and 0.11 in Saudi Benin patients.
Figure 1.
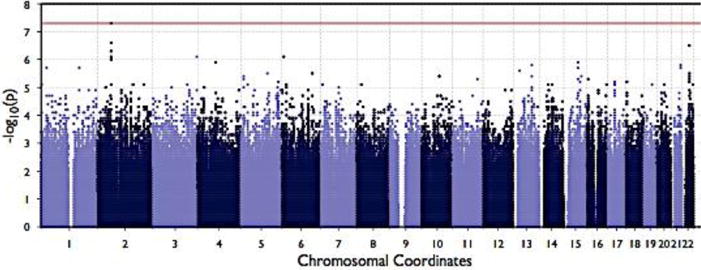
Manhattan plot for African American Benin haplotype and HbF. There is a clear signal at chromosome 2 which corresponds to BCL11A SNPs.
Table 1.
Top associations with HbF in African Americans homozygous for the Benin haplotype with MAF = [0.05,0.5]. NS denotes number of samples and BETA represents the effect of a SNP.
| CHR | BEGIN | END | RSID | MARKER_ID | NS | MAF | PVALUE | BETA |
|---|---|---|---|---|---|---|---|---|
| 2 | 60718043 | 60718043 | rs1427407 | 2:60718043_T/G_Intron:BCL11A | 293 | 0.25 | 5.44E-08 | −0.20 |
| 2 | 60722040 | 60722040 | rs6706648 | 2:60722040_C/T_Intron:BCL11A | 293 | 0.40 | 2.61E-07 | −0.16 |
| 22 | 33862330 | 33862330 | rs557939075 | 22:33862330_G/GT_Insertion:LARGE | 293 | 0.12 | 3.05E-07 | −0.25 |
| 2 | 60725451 | 60725451 | rs7606173 | 2:60725451_G/C_Intron:AC009970.1|BCL11A | 293 | 0.44 | 5.13E-07 | −0.16 |
| 2 | 60724086 | 60724086 | rs1896295 | 2:60724086_T/C_Intron:AC009970.1|BCL11A | 293 | 0.27 | 7.52E-07 | −0.18 |
| 2 | 60724087 | 60724087 | rs1896296 | 2:60724087_G/T_Intron:AC009970.1|BCL11A | 293 | 0.27 | 7.52E-07 | −0.18 |
| 6 | 11287332 | 11287332 | rs4713339 | 6:11287332_C/T_Intron:NEDD9 | 293 | 0.23 | 7.93E-07 | −0.18 |
| 3 | 196544117 | 196544117 | rs13080125 | 3:196544117_T/C_Intron:PAK2 | 293 | 0.35 | 8.29E-07 | 0.16 |
| 2 | 60719970 | 60719970 | rs766432 | 2:60719970_C/A_Intron:BCL11A | 293 | 0.27 | 8.97E-07 | −0.18 |
| 2 | 60720951 | 60720951 | rs4671393 | 2:60720951_A/G_Intron:BCL11A | 293 | 0.27 | 8.97E-07 | −0.18 |
However, even with these differences in the frequencies of TCG and GTC haplotypes, BCL11A variants does not explain the difference in HbF level seen between Saudi Benin and African America Benin.
It is known that 3-base pair deletion in the HBS1L-MYB intergenic polymorphisms (HMIP) region has effect in HbF. However, examining the allele frequencies of rs9399137 which is in high LD with 3bp deletion showed that the frequencies are very low in Saudi Benin as well as in African American Benin (MAF is 0.037 & 0.047, respectively). This result would indicate that the 3-base pair deletion does not explain the difference between Saudi Benin and African American Benin.
The difference in HbF between Saudi Benin and African Americans Benin may due to one or more variants in Saudi Benin that couldn’t be detected due to the small sample size of Saudi Benin. Availability of whole genome sequence may help in identifying variants that specific or rare in both populations.
Acknowledgments
Funded in part by R01 HL 068970, RC2 HL 101212, R01 87681 (MHS), T32 HL007501 (EMS), from the NIH Bethesda, MD.
Footnotes
Conflict-of-interest disclosure: The authors declare no competing interests.
Contribution: PS, JJF, MHS supervised the research and edited the paper, AA provided important samples and edited the paper, EMS wrote the paper and performed the analysis.
References
- 1.Steinberg MH, Forget BG, Higgs DR, Weatherall DJ. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. 2. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- 2.Alsultan A, Solovieff N, Aleem A, et al. Fetal hemoglobin in sickle cell anemia: Saudi patients from the Southwestern province have similar HBB haplotypes but higher HbF levels than African Americans. American journal of hematology. 2011;86(7):612–614. doi: 10.1002/ajh.22032. [DOI] [PubMed] [Google Scholar]
- 3.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loh P-R, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nature genetics. 2016;48(7):811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patterson N, Price AL, Reich D. Population Structure and Eigenanalysis. PLOS Genetics. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]


