Abstract
Objective
Our aim was to evaluate the bacterial profiles of young monkeys as they were weaned into peer groups with a particular focus on Prevotella, an important taxon in both human and nonhuman primates. The weaning of infants and increased social contact with peers is a developmental stage that is likely to affect the gut microbiome.
Methods
Gut bacteria were assessed in 63 rhesus monkeys living in social groups comprised of 4–7 individuals. Two groups were assessed prospectively on Day 1 and 2 weeks after rehousing away from the mother and group formation. Ten additional groups were assessed at 2 weeks after group establishment. Fecal genomic DNA was extracted and 16S ribosomal RNA sequenced by Illumina MiSeq (5 social groups) and 454-amplicon pyrosequencing (7 social groups).
Results
Combining weaned infants into small social groups led to a microbial convergence by 2 weeks (p<.001). Diversity analyses indicated more similar community structure within peer groups than across groups (p<.01). Prevotella was the predominant taxon and its abundance differed markedly across individuals. Indices of richness, microbial profiles and less abundant taxa were all associated with the Prevotella levels. Functional KEGG analyses suggested corresponding shifts in metabolic pathways.
Conclusions
The formation of small groups of young rhesus monkeys was associated with significant shifts in the gut microbiota. The profiles were closely associated with the abundance of Prevotella, a predominant taxon in the rhesus monkey gut. Changes in the structure of the gut microbiome are likely to induce differences in metabolic and physiologic functioning.
Keywords: monkey, microbiome, Prevotella, infant, weaning, diversity
INTRODUCTION
Recent research has highlighted the significance of the gut microbiome for promoting the health and normal development of young infants, not only for digestive and immune function, but also via effects of the enteric nervous system and gut-associated lymphoid tissue on the brain (1–7). Other studies have demonstrated that the mode of delivery, vaginal versus caesarian section, as well as parental decisions regarding breast- or formula-feeding, can influence the establishment the infant’s gut microbiota, affecting both the diversity and richness of the microbial community (4–6). In addition, there appears to be another important developmental transition that coincides with weaning. Social contact with peers and cohabitation may further influence the microbiome composition of the developing individual (7–9). Our study focused on this second maturational stage in young monkeys as they were transferred away from their mothers into small social groups with peers. Typically, in a natural troop, this progressive movement away from the mother is gradual and takes place over several months, but in captive settings, such as a laboratory, it is common to re-house an entire group of weanlings on the same day.
This social transition in young monkeys results in a period of behavioral agitation and activates stress physiology for several days followed by recovery as they become familiar with the new environment and with peers (10–12). Using traditional microbiological culture methods, it was shown that there are alterations in the infant monkeys’ gut microbiota, which were maximal at 3 days after weaning, followed by what appeared to be a return to the original set points after 1–2 weeks (13). However, those analyses were done with selective media to culture specific taxa, including Lactobacilli and Bifidobacteria. Thus, we were interested in employing more current 16S rRNA gene sequencing and phylogenetic analyses to evaluate the microbial community. An initial experiment prospectively compared the microbiota as monkeys were rehoused away from the mother to their profiles 2 weeks later. Then, 2 more experiments evaluated how Prevotella, a prominent taxon in the adult monkey gut, was associated with the microbial profiles that emerged in peer groups.
Although Prevotella spp are commonly found to be a dominant component of the gut microbiota in nonhuman primates (14,15), they appear to be a major factor only in certain human populations (16,17). Differences in the abundance of Prevotella in humans reflect a strong influence of diet. For example, studies comparing urban and rural populations reported that Prevotella were more common when children or adults consumed a high fiber-based diet (6,18,19). In keeping with this perspective, experimental diet manipulations showed that increased fiber intake favors Prevotella (20,21) and that glucoregulatory function in both humans and mice is improved by fiber intake in a Prevotella-dependent manner (22). In addition, Prevotella also appears to correlate with other microbial differences between lean and obese adults, associated with both the overall bacterial composition and digestive efficiency (23,24). In contrast to these seemingly positive effects, some clinical studies concluded that elevations in Prevotella may be a signature feature of the dysbiosis seen in autoimmune conditions, such as rheumatoid arthritis (25–27).
When comparing across species, it should not be assumed that Prevotella would necessarily be indicative of better or worse health in nonhuman primates. A longitudinal evaluation of infant monkeys that were either breast-fed or raised on formula by humans found that Prevotella were more abundant in the mother-reared infants (28). Both more Prevotella and Campylobacter were associated with healthier concentrations of arachidonic acid in the infants’ stool, which appeared to contribute to a beneficial increase in the number of regulatory T cells (Th17) in circulation. These microbial differences in the mother-reared monkeys persisted after the infants were rehoused at 6 months of age, which is the developmental stage we evaluated. Our analyses focused on individual differences in Prevotella abundance as a potential biomarker of the bacterial community structure and the predicted impact on metabolic processes in the infant’s gut. Because we examined 12 different groups, we also looked for signs of microbial convergence among members of each group.
METHODS AND MATERIALS
Subjects
Sixty-three mother-reared infant rhesus monkeys (Macaca mulatta) were assessed in small social groups. Both the mothers and infants were part of a long-established breeding colony, which was housed indoors under controlled environmental conditions. Infants were 7–8 months of age when specimens were obtained during the first 2 weeks after being simultaneously rehoused into groups of 4–7 peers. Only healthy infants were selected, excluding any with diarrheic symptoms or loose stool prior to or during the assessments. Infant monkeys of this age are already eating the solid food diet provided at our facility.
Mothers and peer groups were all fed a standardized diet of laboratory monkey chow (PMI Nutrition International, Richmond, IN), supplemented with fresh fruits and vegetables. Water was available ad libitum. Ambient temperature was controlled at a mean 21 ºC, and the light dark schedule was 16:8 with lights on at 0600. Specimens were collected between 0930–1130. During the maternal phase, the infants lived in different cages, either with just the mother or with another dyad. Peer groups were housed in larger pens or runs.
Ethics Statement
Husbandry and experimental procedures were approved by the Animal Care and Use Committee at the University of Wisconsin-Madison.
Specimen Collection
To analyze each animal’s microbiota, rectal swabs were obtained using a BBL CultureSwab™ Collection & Transport System swab/tube (Becton Dickinson, Cockeysville, MD, USA) and stored at −60 ºC. until analyzed. For the first study, 11 infants were assessed as they were rehoused into 2 peer groups. Rectal swabs were obtained on the first day as well as at 2 weeks after group formation. The second study was comprised of two datasets. The first dataset combined the 2-week samples from the 2 peer groups in the first study with rectal swabs obtained from 3 additional groups that had also been established for 2 weeks (19 additional animals). We employed the Illumina MiSeq method to analyze the samples from Study 1 as well as the additional samples for Study 2. In addition, a second set of samples was incorporated into Study 2 analyses as a replication. This second set consisted of rectal swabs from 7 other peer groups, comprised of 33 monkeys in total, which were also collected at the 2-week time point. This comparison enabled us to determine if similar conclusions would be reached about the infants’ microbiota using the earlier methodology of 454-amplicon pyrosequencing.
Isolation of Genetic Material
After thawing 41 rectal swabs for the Illumina MiSeq analysis, total DNA was isolated using the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA), according to manufacturer’s protocol. The purified genomic DNA extracts were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA), and stored at −20 °C in 10mM Tris buffer until the DNA was sequenced.
For the 16S rRNA gene amplicon 454 pyrosequencing, specimens were analyzed similar to previously described methods (29). In brief, fecal material from the swabs was re-suspended in RLT buffer (Qiagen, Valencia, CA) (with β-mercaptoethanol). Bacterial lyses were generated in a Qiagen TissueLyser (Qiagen, Valencia, CA), and the DNA recovery protocol followed instructions for the QIAamp DNA Mini Kit (Qiagen, Valencia, CA). The genomic extract was eluted and quantified using a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France).
16S Ribosomal RNA Gene Sequencing
PCR amplification of the V4 variable region of the 16S rRNA gene using V4 region specific primers (515F-806R) and amplicon sequencing were performed by Institute for Genomics & Systems Biology at the Argonne National Laboratory (Argonne, IL) on the Illumina MiSeq Platform per manufacturer’s guidelines.
For the 454 Amplicon pyrosequencing (bTEFAP), the methods of Dowd et al were utilized (30,31). The 16S rRNA gene V4 variable region PCR primers 515/806 (32) were employed with a HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA). Samples were sequenced on a Roche 454 FLX titanium sequencer utilizing reagents following the manufacturer’s guidelines.
Data Processing and Statistical Analyses
The sequences were analyzed using QIIME (Quantitative Insights into Microbial Ecology) (33). Sequences were de-multiplexed and quality filtered, per QIIME defaults (minimum sequence length: 200; maximum sequence length: 1000; minimum average qual score: 25; maximum ambiguous bases: 6; maximum length of homopolymer: 6; maximum number of primer mismatches: 0; maximum number of barcode errors: 1.5; and did not retain unassigned reads). Operational taxonomic units (OTUs) were assigned to the Greengenes (v. 2013_May) database (34–36) using PyNAST at 97% similarity through uclust and the closed reference OTU picking in QIIME (37). Because the number of genomic 16S copies varies across the bacterial taxa, not taking 16S copy number into account can lead to significant errors in reported relative abundances (38). Therefore, we normalized the OTU tables by dividing each OTU by the known, or predicted, 16S copy number through implementation in PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (39). Because of our interest in testing for differential abundance of specific taxa, in addition to differences in microbial community structure), we followed current recommendations against the rarefaction of 16S rRNA data, and instead accounted for unequal sampling by normalizing our data through the parametric method, metagenomeSeq, implemented in QIIME (40). One case that exhibited less than 1000 sequences per sample (703.0 sequences) was discarded prior to the diversity analyses or taxonomic assignments. Though rarefactions were not utilized for any analyses, rarefaction curves were generated and inspected before any subsequent analyses in order to determine whether sampling depth was sufficiently accurate to characterize the gut microbiota.
Further, we generated the Faith’s Whole Tree index of phylogenetic diversity (as well as other common indices: Shannon index, chao1 and Observed Species) to examine sample richness, or alpha diversity, and weighted UNIFRAC dissimilarity matrices to examine differences in microbial community structure (beta diversity) while accounting for both microbial abundances and phylogenetic relatedness, as well as unweighted UNIFRAC which only accounts for taxa presence/absence (41,42). These measures were generated using QIIME and principal coordinates (PCoA) obtained through EMPeror (43). PICRUSt was also used to predict microbial metagenomes from KEGG (Kyoto Encyclopedia of Genes and Genomes) Orthologs in order to assess potential shifts in the metabolic activity of the microbiota with respect to functional pathways (39). Although the nonhuman primate gut microbiota composition is relatively similar in composition to humans, the rhesus gut microbiome is not a commonly reported biosphere. Therefore, we used the weighted Nearest Sequenced Taxon Index (NSTI) to inspect the extent to which the bacteria in our samples were related to sequenced reference genomes as a quality control for PICRUSt (39).
Significance testing was carried out by various implementations in QIIME, and PAST (PAleontological STatistics) (44), including Analysis of Similarity for beta diversity indices (ANOSIM, if PERMDISP showed significant inequality of variances across comparison group variance, or PERMNOVA, if PERMDISP indicated equality of variance across groups), nonparametric t-tests (Kruskal-Wallis), Pearson’s correlations (Bootstrapped when appropriate), and graphing. All permutation-based comparisons were carried out with 999 permutations. Taxonomic analyses were restricted to abundant bacteria (abundant taxa were retained by filtering out bacteria that did not meet a minimum 1% composition in total observations across samples), presented and analyzed at the genus level only. When multiple comparisons were performed on taxonomic units, p-values were False Discovery Rate-adjusted (FDR; Benjamini-Hochberg FDR procedure).
RESULTS
Several quality control measures were generated. Alpha rarefaction curves (Figure S1, Supplemental Digital Content 1), NSTI scores and sample sequence counts all reached satisfactory quality to proceed with the subsequent analyses. NSTI scores for the longitudinal study, cross-sectional 454 and Illumina datasets were 0.11 (SD=0.02), 0.14 (SD=0.01) and 11.4 (SD=0.2), respectively. The average sequence counts per sample are presented in Table 1.
Table 1.
Descriptive statistics for the 2 studies, including N, and for quartiles of Prevotella abundance
| N
|
Sex
|
Age (months)
|
Days Since Weaning
|
Sequences Per Sample
|
Prevotella Abundance
|
|
|---|---|---|---|---|---|---|
| Weaning Study | ||||||
| Day 1 | 11 | 4 F, 7 M | 7.0 ± 0.3 | 1.0 ± 0.0 | 2914 ± 326 | 11.0% ± 1.6% |
| 2 Week | 11 | 4 F, 7 M | 7.5 ± 0.3 | 14.0 ± 0.0 | 2778 ± 238 | 8.4% ± 1.2% |
| Cross-Sectional Studies | ||||||
|
|
||||||
| 454-Amplicon Pyrosequencing | ||||||
| 1st Quartile | 8 | 6 F, 2 M | 8.0 ± 0.8 | 18.0 ± 4.3 | 2994 ± 970 | 9.1% ± 3.0% |
| 2nd Quartile | 8 | 5 F, 3 M | 7.4 ± 0.3 | 16.5 ± 3.5 | 2676 ± 1462 | 12.5% ± 0.5% |
| 3rd Quartile | 8 | 6 F, 2 M | 7.9 ± 0.8 | 15.9 ± 2.7 | 3589 ± 1312 | 14.0 ± 0.5 |
| 4th Quartile | 9 | 3 F, 6 M | 7.6 ± 0.4 | 17.4 ± 2.7 | 2707 ± 836 | 17.5% ± 2.1% |
| Illumina MiSeq | ||||||
| 1st Quartile | 7 | 4 F, 3 M | 8.1 ± 0.7 | 16.0 ± 2.0 | 2866 ± 143 | 6.3% ± 0.9% |
| 2nd Quartile | 7 | 3 F, 4 M | 7.6 ± 0.4 | 14.3 ± 2.2 | 2778 ± 248 | 8.3% ± 0.4 |
| 3rd Quartile | 8 | 5 F, 3 M | 8.3 ± 0.5 | 17.7 ± 1.5 | 2748 ± 207 | 9.3% ± 0.4 |
| 4th Quartile | 8 | 3 F, 5 M | 7.6 ± 0.6 | 17.4 ± 2.1 | 2383 ± 327 | 10.9% ± 1.3% |
Means and standard deviations
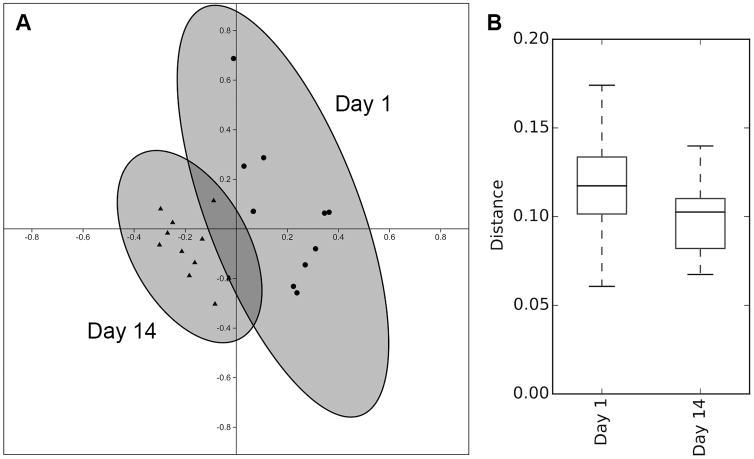
Weighted UNIFRAC distances were calculated for the 11 monkeys sampled on the day of group formation and at 2 weeks. Two-axis PCoA coordinates were generated to summarize dissimilarities in microbial profiles, and revealed that the two sets of 11 samples clustered separately (Figure 1A). ANOSIM analysis indicated a significant difference in community structure over time from the day of formation to when the group was more established at 2 weeks (R=0.603, p<.001). Unweighted UNIFRAC analyses yielded comparable results (R=0.395, p<0.001). Analyses of beta diversity provided further evidence for a convergence in microbial profiles (Figure 1B). Unpaired t-test comparisons of mean dissimilarity distances among gut microbiota profiles indicated that sample-to-sample distances decreased by 2 weeks in the group-living monkeys compared to when the infants had first come from different cages with just their mothers (t=4.28, p<.001). In spite of this convergence in microbial community structure, there was not a net loss of richness or bacterial diversity between Day 1 and two weeks (t=0.39, p=0.740). Taxonomic analyses indicated shifts in bacterial composition at the genus level. Prevotella significantly decreased by two weeks (t=4.45, p=.002), although it continued to be the predominant taxon at both time points. Paralleling the lower Prevotella abundance, increments were observed in the proportions of other common genera, evident in those more prevalent than 1% (Table 2).
Figure 1.
Analyses of beta diversity based on weighted UNIFRAC. (A) There was a significant shift in the microbial community structure from the day of rehousing away from the mother to 2 weeks after peer group formation. The relative distance between samples illustrates the magnitude of dissimilarity in the profiles. (B) At 2 weeks, the dissimilarity in microbial community structure was significantly lower than on the first day, indicating a convergence in gut microbiota profiles.
Table 2.
Microbiota composition ranked by abundance at the genus level for Study 1
| Day 1 | 2 Week | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Rank | Genus | Average Abundance | S.D. | Rank | Genus | Average Abundance | S.D. |
|
|
|
|
|
|
|
|
|
| 1 | Prevotella* | 11.0% ± 1.6% | 1 | Prevotella * | 8.3% ± 1.2% | ||
| 2 | Blautia * | 5.0% ± 0.7% | 2 | Lactobacillus * | 5.8% ± 1.5% | ||
| 3 | Lactobacillus * | 3.8% ± 1.5% | 3 | Blautia * | 4.1% ± 0.7% | ||
| 4 | Ruminococcus * | 2.9% ± 0.6% | 4 | Ruminococcus * | 3.8% ± 0.7% | ||
| 5 | Faecalibacterium | 2.5% ± 0.7% | 5 | Faecalibacterium | 2.1% ± 0.4% | ||
| 6 | Coprococcus | 2.3% ± 0.4% | 6 | Coprococcus | 2.0% ± 0.3% | ||
| 7 | Roseburia | 2.2% ± 0.6% | 7 | Roseburia | 1.7% ± 0.4% | ||
| 8 | Oscillospira | 1.2% ± 0.2% | 8 | Streptococcus | 1.4% ± 0.7% | ||
| 9 | Streptococcus | 1.2% ± 0.6% | 9 | Oscillospira | 1.3% ± 0.2% | ||
| 10 | Dorea | 1.1% ± 0.2% | 10 | Clostridium * | 1.3% ± 0.2% | ||
| 11 | Clostridium * | 1.0% ± 0.2% | 11 | Dorea | 1.2% ± 0.2% | ||
| Unclassified genera | 48.2% | 12 | Treponema | 1.0% ± 0.4% | |||
| Genera below 1% | 17.6% | Unclassified genera | 50.4% | ||||
| Genera below 1% | 15.6% | ||||||
Indicates statistically significant differences; Prevotella (t=−4.45, p=.002); Blautia (t=−3.02, p=.018); Lactobacillus (t=3.17, p=.015); Ruminococcus (t=3.52, p=.009)); and Clostridium (t=2.76, p=.028); P-values were FDA-adjusted by Benjamini-Hochberg method.
PIRCRUSt analysis referencing KEGG Orthologs database predicted potential shifts in functional pathways based on the estimated genomic content represented by the microbiota present in each monkey. The predictions suggested shifts in many metabolic pathways, including those related to genetic and environmental information processing (i.e., gene regulation, signaling and membrane transport) and cellular processes (i.e., cell growth and death, transport and catabolism), which would be expected under new environmental pressures and with a reshaping of the microbial community (Table 3). The KEGG predictions also indicated potential increases in carbohydrate and lipid metabolism, xenobiotic biodegradation and metabolism, as well as biochemical pathways implicated in infectious disease in humans. Functional pathway predictions further suggested that the microbiota changes associated with the social group transition would lead to a decrease in processes related to energy metabolism, glycan biosynthesis, metabolism of cofactors and vitamins, terpenoid and polyketides, as well as affect the regulation of host digestive and immune processes (Table 3). More detailed summaries of these pathway analyses are presented in Table S1, Supplemental Digital Content 2.
Table 3.
KEGG Ortholog predictions based on changes in microbiota profiles
| T-Test Statistic | FDR Adjusted P-Value | Day 1 | Week 2 | Relative Change | |
|---|---|---|---|---|---|
|
|
|
|
|
|
|
| Cellular Processes | |||||
| Transport and Catabolism | −8.72 | <.001 | 0.28% | 0.23% | −17.84% |
| Unclassified | −5.79 | <.001 | 3.98% | 3.82% | −4.06% |
| Environmental Information Processing | |||||
| Membrane Transport | 8.72 | <.001 | 9.88% | 11.68% | 18.30% |
| Genetic Information Processing | |||||
| Folding, Sorting and Degradation | −6.21 | <.001 | 2.76% | 2.51% | −8.92% |
| Transcription | 4.27 | .001 | 2.51% | 2.80% | 11.50% |
| Translation | −2.62 | .035 | 6.60% | 6.46% | −2.09% |
| Unclassified | −2.72 | .032 | 2.68% | 2.63% | −1.84% |
| Metabolism | |||||
| Carbohydrate Metabolism | 5.70 | <.001 | 9.68% | 10.45% | 7.96% |
| Energy Metabolism | −6.50 | <.001 | 6.30% | 5.87% | −6.80% |
| Glycan Biosynthesis and Metabolism | −7.84 | <.001 | 2.93% | 2.36% | −19.45% |
| Lipid Metabolism | 4.62 | .001 | 2.58% | 2.70% | 4.80% |
| Metabolism of Cofactors and Vitamins | −9.63 | <.001 | 4.63% | 4.25% | −8.19% |
| Metabolism of Other Amino Acids | −3.69 | .004 | 1.53% | 1.48% | −2.90% |
| Metabolism of Terpenoids and Polyketides | −3.73 | .004 | 1.81% | 1.74% | −4.30% |
| Xenobiotics Biodegradation and Metabolism | 7.15 | <.001 | 1.45% | 1.66% | 14.43% |
| Unclassified | 2.59 | .036 | 2.38% | 2.42% | 1.61% |
| Organismal Systems | |||||
| Digestive System | −3.16 | .013 | 0.09% | 0.06% | −26.45% |
| Immune System | −8.47 | <.001 | 0.10% | 0.09% | −15.16% |
FDR-adjustment: Benjamini-Hochberg method; Nonsignificant KEGG pathways were omitted from this table.
Assessment of 5 social groups at the 2-week time point using Illumina MiSeq (by considering 3 additional groups) and 7 other peer groups using 454 pyrosequencing (also swabbed at 2 weeks after group formation) yielded similar results on the abundance of Prevotella relative to other abundant taxa (Table 4). There was significant variation in Prevotella across individuals. Prevotella abundance ranged between 5.1 – 13.6% in the MiSeq data, and between 3.0 – 21.6% in the 454 data (Table 1). Two weeks after rehousing, the Illumina MiSeq method detected a total of 400 distinct bacteria at the genus level, though many were classifiable only at higher taxonomic levels. Eighteen genera passed the 1% abundance threshold for our taxonomic comparisons. The 454 pyrosequencing method detected 173 bacteria, 16 of which passed the 1% abundance threshold.
Table 4.
Microbiota composition ranked by abundance at genus level for Study 2
| 454 Pyrosequencing | Illumina MiSeq | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Rank | Genus | Average Abundance | S.D. | Rank | Genus | Average Abundance | S.D. |
|
|
|
|
|
|
|
||
| 1 | Prevotella | 13.2% ± 3.9% | 1 | Prevotella | 8.8% ± 1.9% | ||
| 2 | Lactobacillus | 8.2% ± 3.1% | 2 | Lactobacillus | 5.4% ± 1.5% | ||
| 3 | Faecalibacterium | 4.3% ± 1.5% | 3 | Blautia | 4.1% ± 0.8% | ||
| 4 | Blautia | 4.2% ± 1.5% | 4 | Ruminococcus | 3.3% ± 0.8% | ||
| 5 | Coprococcus | 2.3% ± 0.8% | 5 | Faecalibacterium | 2.2% ± 0.5% | ||
| 6 | Roseburia | 2.2% ± 1.2% | 6 | Coprococcus | 2.1% ± 0.3% | ||
| 7 | Ruminococcus | 1.7% ± 0.8% | 7 | Roseburia | 1.7% ± 0.5% | ||
| 8 | Oscillospira | 1.5% ± 0.5% | 8 | Streptococcus | 1.5% ± 0.7% | ||
| 9 | Dorea | 1.1% ± 0.5% | 9 | Oscillospira | 1.3% ± 0.3% | ||
| 10 | Bacteroides | 1.0% ± 0.4% | 10 | Clostridium | 1.2% ± 0.2% | ||
| Unclassified genera | 46.5% | 11 | Dorea | 1.2% ± 0.2% | |||
| Genera below 1% | 13.9% | Treponema | 1.0% ± 0.3% | ||||
| Unclassified genera | 49.6% | ||||||
| Genera below 1% | 16.5% | ||||||
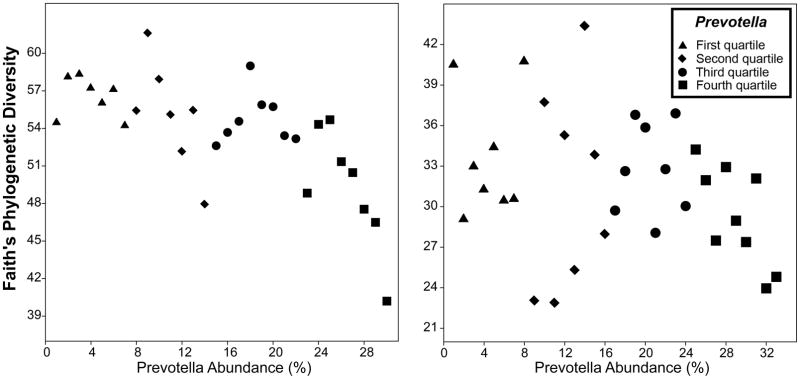
Analyses of alpha diversity, using both MiSeq and pyrosequencing methods, revealed lower phylogenetic richness (Faith’s Phylogenetic Diversity index) with increasing Prevotella abundance (Figure 2A and 2B) (r=−.349, p=.047, and r=−.733, p<.001, respectively). Other indices of richness that do not account for genetic relatedness were also examined, yielding similar results. Using the pyrosequencing method, Chao1, Observed Species and Shannon indices also indicated a negative trend, but they did not reach statistical significance (p=0.239, p=0.236 and p=0.272, respectively). However, the Illumina MiSeq method detected strong and significant negative correlations between Prevotella and these same indices (r=−0.518, r= −0.635 and r= −0.671 and p=0.003 p<0.001 and p<0.001, respectively). The abundance of other common genera (more than 1%) was also significantly correlated with Prevotella, although these relationships again were stronger when quantified by MiSeq (Table S2, SDC 3). Bivariate correlations across all abundant taxa are presented in Figures S2 and S3 (SDC 1).
Figure 2.
Examination of sample richness in young rhesus monkeys at 2 weeks after formation of small social groups indicating that high Prevotella abundance was associated with lower phylogenetic diversity. This relationship was found both with the Illumina MiSeq (A) and 454-pyrosequencing (B) platforms.
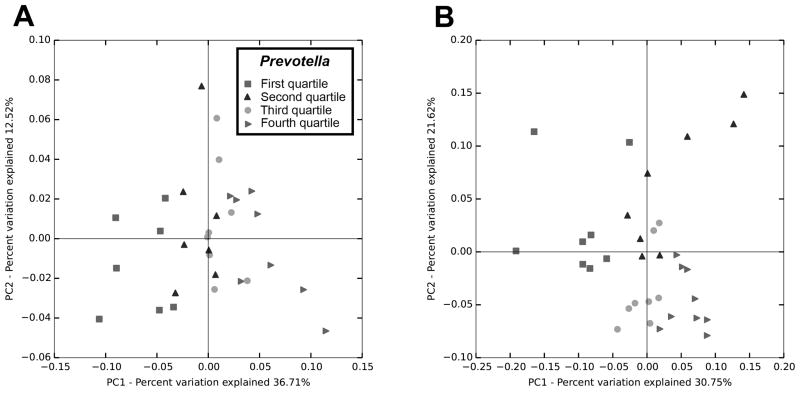
To analyze beta diversity, the monkeys were categorized into standardized quartiles according to Prevotella levels within each dataset. Beta diversity indices for both pyrosequencing and MiSeq methods were sufficiently sensitive to show a main effect of Prevotella classification on the compositional structure of the microbiota community across the quartiles (Figure 3). Analyses of pyrosequencing data using Weighted and unweighted UNIFRAC showed significant differences in beta diversity (PERMANOVA, pseudo-F=5.42, p<0.001; PERMDISP, p=0.063) and (PERMANOVA, pseudo-F=1.545, p<0.001; PERMDISP, p=0.486), respectively; as did the analyses of the MiSeq data, using both weighted and unweighted UNIFRAC (PERMANOVA, pseudo-F=4.66, p<0.001; PERMDISP, p=0.77) and (ANOSIM, R=0.141, p=0.002; PERMDISP, p=0.027), respectively.
Figure 3.
PCoA of weighted UNIFRAC measures of beta diversity. Analyses of dissimilarity indicated differences in microbiota profiles across quartiles of Prevotella abundance when sequenced either by Illumina MiSeq (A) or 454-pyrosequencing (B).
Individual differences in Prevotella abundance and associated microbial taxa were predictive of major metabolic differences, whether assessed at the broad functional level (Table 5) or with respect to specific pathways (Table S1). KEGG Orthologs derived predictions suggested significant differences in cellular growth and death processes as well as in the sensing, processing and regulation of genetic and environmental information. Our analyses of KEGG predictions suggested that shifts in Prevotella abundance are accompanied by genetic and physiological processes associated with the reshaping of the microbiome. While our assessment did not allow us to determine whether Prevotella can exert a direct influence on the metabolic potential of the host, the abundance of Prevotella was strongly associated with predicted metabolic shifts within the microbiota.
Table 5.
KEGG pathways that were correlated with increased Prevotella abundance
| 454 Pyrosequencing | Illumina MiSeq | |||
|---|---|---|---|---|
|
|
|
|||
| Pearson’s Correlation Coefficient | FDR Adjusted P-Value | Pearson’s Correlation Coefficient | FDR Adjusted P-Value | |
|
|
|
|
|
|
| Cellular Processes | ||||
| Cell Growth and Death | 0.51 | <.001 | 0.54 | <.001 |
| Cell Motility | −0.21 | >.99 | −0.41 | .041 |
| Transport and Catabolism | 0.51 | <.001 | 0.59 | <.001 |
| Unclassified | 0.60 | <.001 | 0.71 | <.001 |
| Environmental Information Processing | ||||
| Membrane Transport | −0.80 | <.001 | −0.89 | <.001 |
| Signal Transduction | −0.59 | <.001 | −0.77 | <.001 |
| Signaling Molecules and Interaction | 0.01 | >.99 | 0.45 | <.001 |
| Genetic Information Processing | ||||
| Folding, Sorting and Degradation | 0.86 | <.001 | 0.84 | <.001 |
| Replication and Repair | 0.31 | >.99 | 0.65 | <.001 |
| Transcription | −0.64 | <.001 | −0.65 | <.001 |
| Translation | 0.14 | >.99 | 0.56 | <.001 |
| Metabolism | ||||
| Biosynthesis of Other Secondary Metabolites | 0.67 | <.001 | 0.39 | .015 |
| Carbohydrate Metabolism | −0.53 | <.001 | −0.50 | .015 |
| Energy Metabolism | 0.67 | <.001 | 0.80 | <.001 |
| Enzyme Families | 0.56 | <.001 | 0.65 | <.001 |
| Glycan Biosynthesis and Metabolism | 0.86 | <.001 | 0.91 | <.001 |
| Lipid Metabolism | −0.55 | <.001 | −0.79 | <.001 |
| Metabolism of Cofactors and Vitamins | 0.71 | <.001 | 0.80 | <.001 |
| Metabolism of Other Amino Acids | 0.78 | <.001 | 0.88 | <.001 |
| Metabolism of Terpenoids and Polyketides | 0.79 | <.001 | 0.94 | <.001 |
| Nucleotide Metabolism | 0.40 | >.99 | 0.79 | <.001 |
| Xenobiotics Biodegradation and Metabolism | −0.65 | <.001 | −0.50 | <.001 |
| Unclassified | −0.23 | >.99 | −0.75 | <.001 |
| Organismal Systems | ||||
| Digestive System | 0.88 | <.001 | 0.90 | <.001 |
| Endocrine System | 0.38 | .010 | 0.04 | >.99 |
| Excretory System | −0.21 | >.99 | −0.81 | <.001 |
| Immune System | 0.47 | .010 | 0.68 | <.001 |
| Nervous System | 0.59 | <.001 | 0.21 | >.99 |
FDR-adjustment: Benjamini-Hochberg method; Nonsignificant KEGG pathways were omitted from this table.
According to our KEGG predictions, glycan biosynthesis and metabolism (which includes the biosynthesis of lipopolysaccharides) would likely increase. Shifts in Prevotella abundance and associated taxa would also affect the metabolism of cofactors and vitamins, influencing the efficiency of energy utilization. Individual differences in Prevotella also correlated significantly with pathways affecting carbohydrate, mineral and protein digestion and absorption, which would potentially influence energetic harvesting efficiency. Conversely, a downregulation of carbohydrate and lipid metabolism was also predicted by high levels of Prevotella. Prevotella abundance was further associated with multiple pathways involved in the metabolism of terpenoids and polyketides, some known to moderate the effects of phytochemicals and responses to pharmaceutical drugs. Overall, the predicted shift in metabolic potential of the gut microbiota was similar for both the pyrosequencing and MiSeq data. In addition, the differences in metabolic pathways observed for monkeys with low Prevotella were similar to the predicted changes observed when comparing monkeys in Study 1 from Day 1 to 2 weeks as Prevotella abundance decreased after group formation.
Notwithstanding the heterogeneity of the gut bacterial composition across animals, examination of PCoA plots of beta diversity indicated a degree of convergence (Figure 1) and clustering of gut microbiota profiles among the members of a peer group (Figure S4, SDC 1). Statistically significant differences in beta diversity were found in spite of variability among abundant taxa and overlap across some groups,. Indices of dissimilarity were significantly greater across groups than within groups in the MiSeq data (PERMANOVA, pseudo-F=2.16, p=.004, PERMDISP, p=.712) and (PERMANOVA, pseudo-F=1.34, p=.001, PERMDISP, p=.217), weighted and unweighted UNIFRAC respectively, as well as in the pyrosequencing data (ANOSIM, R=.269, p=.002, PERMDISP, p=0.003) and (PERMANOVA, pseudo-F=1.42, p=.001, PERMDISP, p=.288), weighted and unweighted UNIFRAC respectively.
DISCUSSION
Our analyses confirm a previous paper reporting that the transition of a young monkey from its mother to living with other weaned monkeys can affect gut bacteria (13). That finding was based on traditional culturing methods and documented transient changes in gram-negative bacteria and select taxa, including Lactobacilli and Bifidobacteria. Using more current methods, we were able to evaluate the bacterial community structure, which evinced significant change, but in other ways remained fairly stable after the new peer groups were established. This interpretation concurs with a different study of mother-reared and human-reared infant monkeys, which concluded that lasting differences in the gut microbiota were induced by early rearing conditions, but that the microbiome remained constant at 9 and 12 months of age, after all of the monkeys had been rehoused at 6 months of age (28).
When separated from the mother at this age, infant rhesus monkeys are already feeding on solid food. Rhesus mothers wean their infants from breastmilk at about four months after birth, at which point infants already rely mostly on solid foods (45). Thus it is likely that the transient changes observed in the gut microbiota were induced by the psychological disturbance associated with the relocation away from the mother and the contact with new conspecifics, including exposure to their fecal matter. Both transient and persistent changes in gut bacteria and function have been observed following acute and chronic psychological stress (29,46). Prior research also indicated that Prevotella can be specifically responsive to acute psychological stressors, even in the absence of significant shifts in diversity or structure (47). Although our methods did not allow us to establish the directionality of effects, the association between Prevotella abundance and the community structure and predicted metabolic function are consistent with other studies indicating that Prevotella can affect the overall composition and stability of the gut microbiota in humans (6,16,17,48–50). However, we also cannot rule out the possibility that the observed effects were, at least in part, driven by taxa that covary with Prevotella, even when present at much lower abundances. It is known that Prevotella can establish symbiotic relationships with other commensal bacteria in other body compartments (51–53). Further research is needed to determine the role of Prevotella in constricting and modulating other taxa.
Reference to the KEGGs orthologs, and the genes associated with the represented microbial taxa, generated predictions about the likely metabolic potential of these microbial profiles. The predicted metabolic shift after group formation makes intuitive sense with respect to the energetic and metabolic challenges faced by young monkeys during this transition. Changes associated with group formation were also consistent with the prediction from cross-sectional evaluation of individual monkeys based just on Prevotella abundance. Furthermore, the predictions concur with how Prevotella spp and Prevotella-associated taxa have been linked to caloric harvesting (18,54–56) and its modulatory effects on host immune responses (54–59). Future research will now have to determine if these metabolic predictions hold up with an evaluation of systemic metabolism in monkeys that vary in Prevotella abundance.
Notwithstanding our assessment of many monkeys, and employing two different platforms for the bacterial sequencing and identification, these analyses have several limitations. The bacterial DNA was isolated from rectal swabs and thus reflects the microbiota present in fecal matter. Fortunately, research by other investigators who had access to gut tissue and lumenal content from monkeys already documented that the relative prevalence of the primary taxa in stool is reflective of their numbers in the distal gut (15). A second limitation was our focus on one time point after group formation. Additional microbial changes may occur over time as the groups became more established. However, previous studies on weaned monkeys indicated that the major stress-related changes subside by one week (13). Specimen collection at one time point enabled us to maximize the number of social groups that could be assessed, evaluating all monkeys at the same age and stage.
In sum, this research characterized the microbial community structure of young rhesus monkeys, and identified the major taxa to be similar to reports on adult rhesus and cynomolgus monkeys (14,15). It likely also reflects the consumption of solid foods, which typically begins by 3 months of age in monkeys as the mother naturally weans her infant. Thus, the microbial community structure is different than the one seen in very young infants still actively breastfeeding (28). Our analyses also highlight the importance of Prevotella as an important feature of the bacterial community structure. Given its abundance, Prevotella is likely to contribute not only to energetic harvesting and the adaptation to various diets, but also to the neurochemical signaling to the gut and nervous system.
In spite of extensive variability in the bacterial community structure, analyses of beta diversity indicated that members of a peer group tended to be more similar among themselves than between groups. The clustering of gut microbiota profiles by peer group is consistent with our first experiment showing that microbiotic dissimilarity within a group decreased by two weeks after group formation. Microbial convergence within a group would also be in keeping with reports of similar microbial profiles among members of the same household, for farm animals in a shared pen, and even between children and their pet companion animals (57–61). Cohousing and cohabitation effects on the microbiota have also been noted in other rodent and primate studies (7–9). To directly test this question, our current research is determining if developing monkeys will continue to maintain stable differences in the prevalence of specific taxa, which may offer protective or metabolic advantages.
Supplementary Material
Acknowledgments
This research was supported by grants from the NIMH (MH104198) and NICHD (HD080201). All authors participated in the data collection and reviewed the manuscript. No conflicts of interest to report.
Acronyms
- rRNA
ribosomal RNA
- OTUs
Operational taxonomic units
- PCoA
principal coordinate analyses
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- ANOSIM
Analysis of Similarity
- FDR
False Discovery Rate
- NSTI
Nearest Sequenced Taxon Index
References
- 1.Borre YE, O’Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. Trends Mol Med. 9. Vol. 20. Elsevier Ltd; 2014. Jun 20, Microbiota and neurodevelopmental windows: implications for brain disorders; pp. 509–18. [DOI] [PubMed] [Google Scholar]
- 2.Yang I, Corwin EJ, Brennan PA, Jordan S, Murphy JR, Dunlop A. The Infant Microbiome. Nurs Res. 2016;65(1):76–88. doi: 10.1097/NNR.0000000000000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly D, King T, Aminov R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res. 2007;622:58–69. doi: 10.1016/j.mrfmmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Kashtanova DA, Popenko AS, Tkacheva ON, Tyakht AV, Alexeev DG, Boytsov SA. Nutrition. Elsevier Inc; 2015. Association Between the Gut Microbiota and Diet: Fetal Life, Early Childhood, and Further Life; pp. 1–8. [DOI] [PubMed] [Google Scholar]
- 5.Meropol SB, Edwards A. Development of the infant intestinal microbiome: A bird’s eye view of a complex process. Birth Defects Res Part C - Embryo Today Rev. 2015 doi: 10.1002/bdrc.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012 Jun 14;486(7402):222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tung J, Barreiro LB, Burns MB, Grenier JC, Lynch J, Grieneisen LE, Altmann J, Alberts SC, Blekhman R, Archie EA. Social networks predict gut microbiome composition in wild baboons. Elife. 2015;2015(4):e05224. doi: 10.7554/eLife.05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knight D, Clemente JC, Nakielny S, Gordon JI, Fierer N, Knight R. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2013(2):1–22. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, Ochman H. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. PNAS. 2012;109(32):13034–9. doi: 10.1073/pnas.1110994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coe CL, Erickson CM. Stress decreases lymphocyte cytolytic activity in the young monkey even after blockade of steroid and opiate hormone receptors. Dev Psychobiol. 1997;30(1):1–10. [PubMed] [Google Scholar]
- 11.Rilling JK, Winslow JT, O’Brien D, Gutman DA, Hoffman JM, Kilts CD. Neural correlates of maternal separation in rhesus monkeys. Biol Psychiatry. 2001;49(2):146–57. doi: 10.1016/s0006-3223(00)00977-x. [DOI] [PubMed] [Google Scholar]
- 12.Capitanio JP, Mendoza SP, Cole SW. Brain Behav Immun. 1. Vol. 25. Elsevier Inc; 2011. Jan, Nervous temperament in infant monkeys is associated with reduced sensitivity of leukocytes to cortisol’s influence on trafficking; pp. 151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol. 1999;35(2):146–55. [PubMed] [Google Scholar]
- 14.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Harris RA, Frias AE, Grove KL, Aagaard KM. Nat Commun. May. Vol. 5. Nature Publishing Group; 2014. Jan, High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model; p. 3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, Miler GM, Rowlett JK, Gevers D, Huttenhower C, Morgan XC. Cell Host Microbe. 3. Vol. 17. Elsevier Inc; 2015. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque; pp. 385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorvitovskaia A, Holmes SP, Huse SM. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome Microbiome. 2016;4(1):15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011 May 12;473(7346):174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, Vipperla K, Naidoo V, Mtshali L, Tims S, Puylaert PG, DeLany J, Krasinskas A, Benefiel AC, Kaseb HO, Newton K, Nicholson JK, de Vos WM, Gaskins HR, Zoetendal EG. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6(May 2014):6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014 Jan 23;505(7484):559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haro C, Montes-Borrego M, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Tinahones FJ, Landa BB, López-Miranda J, Camargo A, Pérez-Jiménez F. Two Healthy Diets Modulate Gut Microbial Community Improving Insulin Sensitivity in a Human Obese Population. J Clin Endocrinol Metab. 2016;101(1):233–42. doi: 10.1210/jc.2015-3351. [DOI] [PubMed] [Google Scholar]
- 22.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2014:971–82. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Kong LC, Holmes BA, Cotillard A, Habi-Rachedi F, Brazeilles R, Gougis S, Gausserès N, Cani PD, Fellahi S, Bastard JP, Kennedy SP, Doré J, Ehrlich SD, Zucker JD, Rizkalla SW, Clément K. Dietary patterns differently associate with inflammation and gut microbiota in overweight and obese subjects. PLoS One. 2014 Jan;9(10):e109434. doi: 10.1371/journal.pone.0109434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radilla-Vázquez RB, Parra-Rojas I, Martínez-Hernández NE, Márquez-Sandoval YF, Illades-Aguiar B, Castro-Alarcón N. Gut Microbiota and Metabolic Endotoxemia in Young Obese Mexican Subjects. Obes Facts. 2016;9(1):1–11. doi: 10.1159/000442479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandhya P, Danda D, Sharma D, Scaria V. Does the buck stop with the bugs?: an overview of microbial dysbiosis in rheumatoid arthritis. 2016:8–20. doi: 10.1111/1756-185X.12728. [DOI] [PubMed] [Google Scholar]
- 26.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno J. Prevotella copri and the microbial pathogenesis of rheumatoid arthritis. Reumatol Clin. 2015;11(2):61–3. doi: 10.1016/j.reuma.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Ardeshir A, Narayan NR, Méndez-Lagares G, Lu D, Rauch M, Huang Y, Van Rompay KK, Lynch SV, Hartigan-O’Connor DJ. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. 2014;6(252):252ra120. doi: 10.1126/scitranslmed.3008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Brain Behav Immun. 3. Vol. 25. Elsevier Inc; 2011. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation; pp. 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3(10) doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Lauber CL, Walters WA, Berg-lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. 2010 doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, Mcdonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat Publ Gr Nature Publishing Group. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R, Ley RE. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J Nature Publishing Group. 2012;6(1):94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J Nature Publishing Group. 2012;6(3):610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, Knight R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kembel SW, Wu M, Eisen JA, Green JL. Incorporating 16S Gene Copy Number Information Improves Estimates of Microbial Diversity and Abundance. 2012;8(10):16–8. doi: 10.1371/journal.pcbi.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Nat Biotechnol. 9. Vol. 31. Nature Publishing Group; 2013. Sep, Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences; pp. 814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mcmurdie PJ, Holmes S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. 2014;10(4) doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME J Nature Publishing Group. 2011;5(2):169–72. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2(1):16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammer Ø, Harper DAT, Ryan PD. Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9–18. [Google Scholar]
- 45.Fooden J. Systematic review of the rhesus macaque, Macaca mulatta (Zimmermann, 1780) Fieldiana Zool. 2000;96:1–180. [Google Scholar]
- 46.Bharwani A, Mian MF, Foster JA, Surette MG, Bienenstock J, Forsythe P. Psychoneuroendocrinology Structural & functional consequences of chronic psychosocial stress on the microbiome & host. Elsevier Ltd. 2016;63:217–27. doi: 10.1016/j.psyneuen.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Maslanik T, Tannura K, Mahaffey L, Loughridge AB, Benninson L, Ursell L, Greenwood BN, Knight R, Fleshner M. Commensal Bacteria and MAMPs Are Necessary for Stress-Induced Increases in IL-β and IL-18 but Not IL-6, IL-10 or MCP-1. PLoS One. 2012;7(12):1–10. doi: 10.1371/journal.pone.0050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the Prevotella-to-Bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl Environ Microbiol. 2014;80(3):1142–9. doi: 10.1128/AEM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dlugosz A, Winckler B, Lundin E, Zakikhany K, Sandström G, Ye W, Engstrand L, Lindberg G. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Sci Rep. 2015;5:8508. doi: 10.1038/srep08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2015:1–6. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pybus V, Onderdonk AB. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J Infect Dis. 1997;175:406–13. doi: 10.1093/infdis/175.2.406. [DOI] [PubMed] [Google Scholar]
- 52.Pybus V, Onderdonk AB. A commensal symbiosis between Prevotella bivia and Peptostreptococcus anaerobius involves amino acids: Potential significance to the pathogenesis of bacterial vaginosis. FEMS Immunol Med Microbiol. 1998;22(4):317–27. doi: 10.1111/j.1574-695X.1998.tb01221.x. [DOI] [PubMed] [Google Scholar]
- 53.Kolenbrander PE, Andersen RN, David S, Egland PG, Foster JS, Palmer RJ., Jr Communication among Oral Bacteria Communication among Oral Bacteria. Microbiol Mol Microbiol Rev. 2002;66(3):486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doerflinger SY, Throop AL, Herbst-Kralovetz MM. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J Infect Dis. 2014;209(12):1989–99. doi: 10.1093/infdis/jiu004. [DOI] [PubMed] [Google Scholar]
- 55.Larsen JM, Musavian HS, Butt TM, Ingvorsen C, Thysen AH, Brix S. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology. 2015;144(2):333–42. doi: 10.1111/imm.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Ramakrishnan A, Fletcher S, Prochownik EV, Genetics M. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2015;2(2):983–94. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi E-Y, Jin J-Y, Choi J-I, Choi IS, Kim S-J. DHA suppresses Prevotella intermedia lipopolysaccharide-induced production of proinflammatory mediators in murine macrophages. Br J Nutr. 2013;111(7):1221–30. doi: 10.1017/S0007114513003681. [DOI] [PubMed] [Google Scholar]
- 58.Tokuda M, Sakuta T, Fushuku a, Torii M, Nagaoka S. Regulation of interleukin-6 expression in human dental pulp cell cultures stimulated with Prevotella intermedia lipopolysaccharide. J Endod. 2001;27(4):273–7. doi: 10.1097/00004770-200104000-00008. [DOI] [PubMed] [Google Scholar]
- 59.Centanni M, Turroni S, Consolandi C, Rampelli S, Peano C, Severgnini M, Biagi E, Caredda G, De Bellis G, Brigidi P, Candela M. The enterocyte-associated intestinal microbiota of breast-fed infants and adults responds differently to a TNF-α-mediated pro-inflammatory stimulus. PLoS One. 2013;8(11):1–11. doi: 10.1371/journal.pone.0081762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics. 2006;118(2):511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 61.Konya T, Koster B, Maughan H, Escobar M, Azad MB, Guttman DS, Sears MR, Becker AB, Brook JR, Takaro TK, Kozyrskyj AL, Scott JA CHILD Study Investigators. Associations between bacterial communities of house dust and infant gut. Environ Res Elsevier. 2014;131:25–30. doi: 10.1016/j.envres.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Ludvigsen J, Svihus B, Rudi K. Rearing Room Affects the Non-dominant Chicken Cecum Microbiota, While Diet Affects the Dominant Microbiota. Front Vet Sci. 2016;3(February):1–7. doi: 10.3389/fvets.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.