Abstract
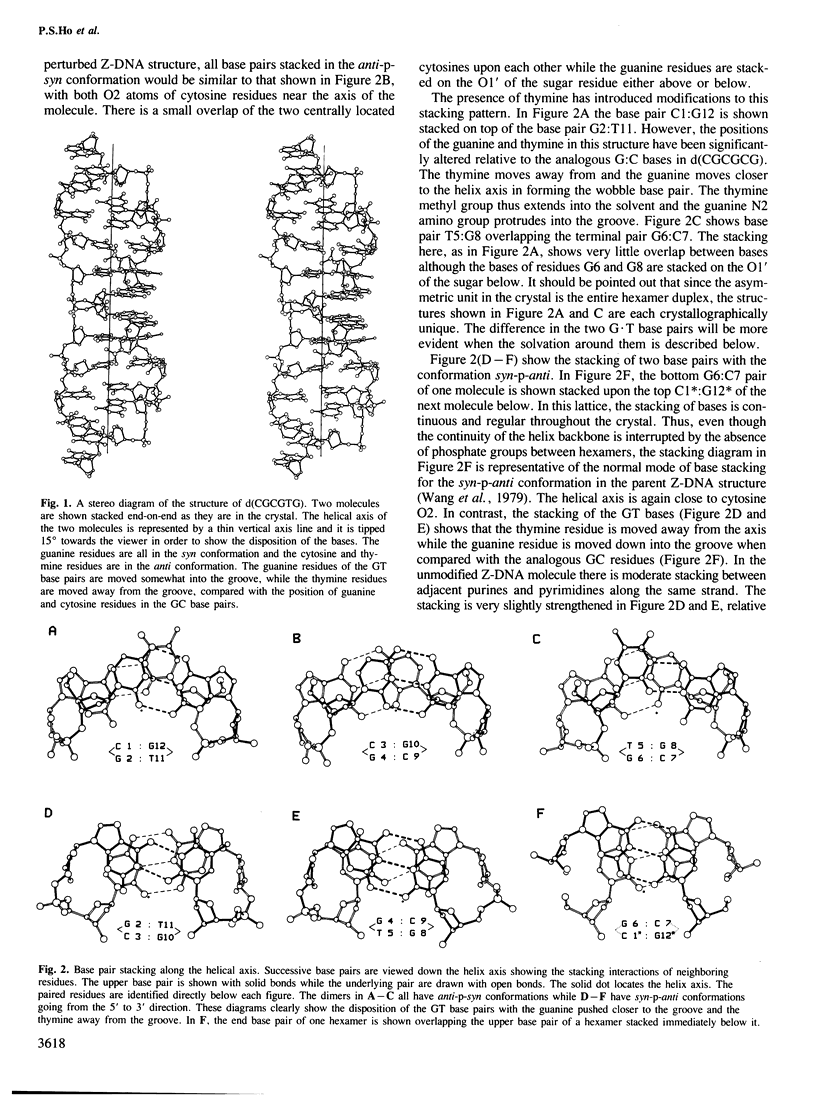
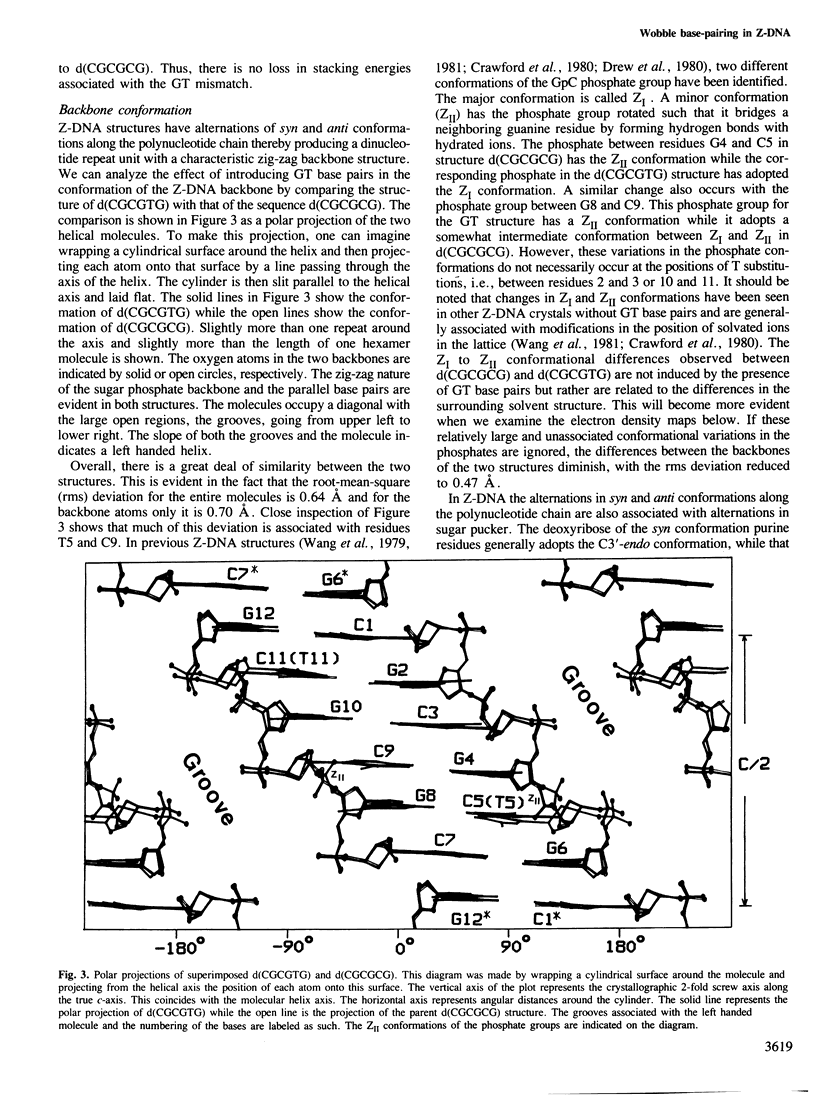
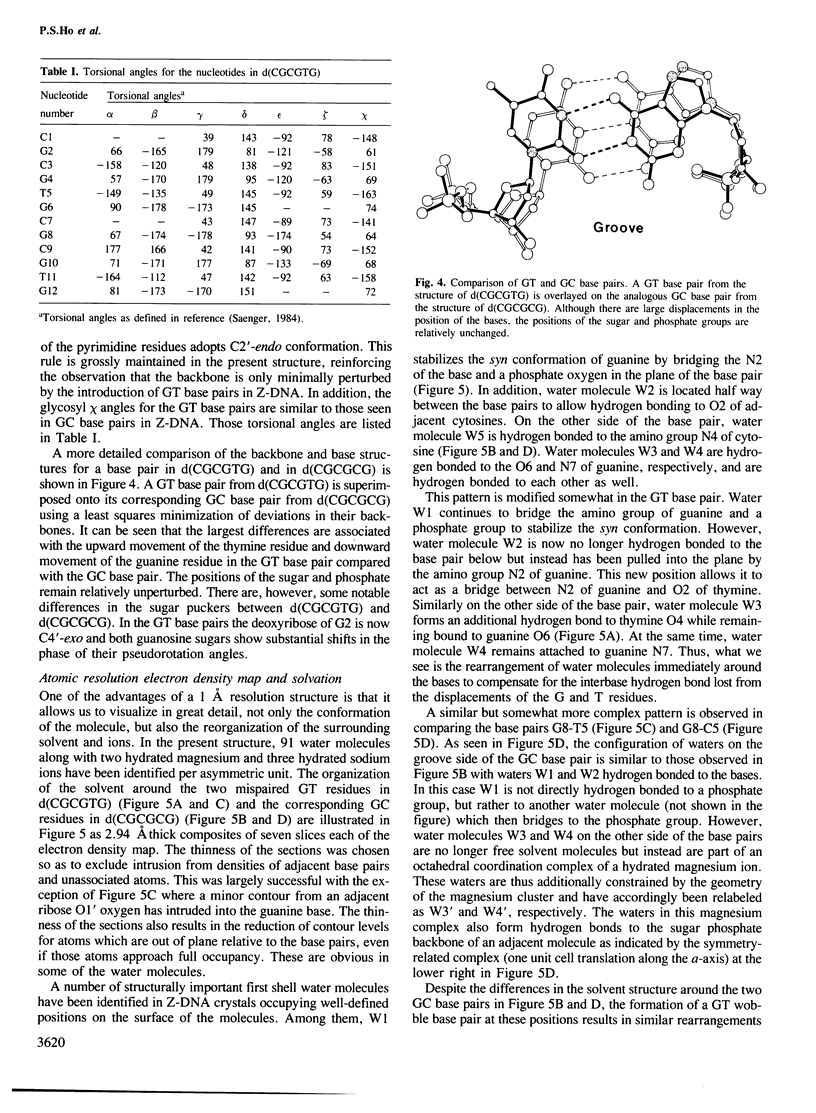
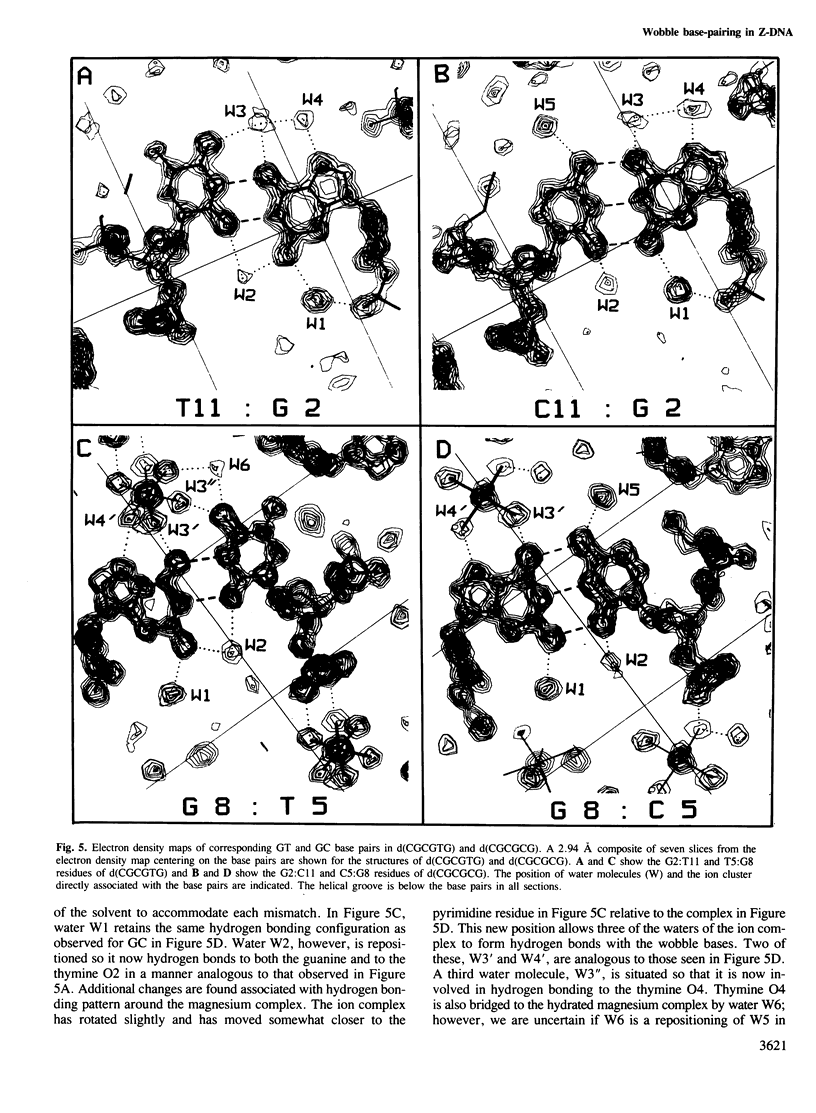
The DNA oligomer d(CGCGTG) crystallizes as a Z-DNA double helix containing two guanine-thymine base pair mismatches of the wobble type. The crystal diffracts to 1 A resolution and the structure has been solved and refined. At this resolution, a large amount of information is revealed about the organization of the water molecules in the lattice generally and more specifically around the wobble base pairs. By comparing this structure with the analogous high resolution structure of d(CGCGCG) we can visualize the structural changes as well as the reorganization of the solvent molecules associated with wobble base pairing. There is only a small distortion of the Z-DNA backbone resulting from introduction of the GT mismatched base pairs. The water molecules cluster around the wobble base pair taking up all of the hydrogen bonding capabilities of the bases due to wobble pairing. These bridging water molecules serve to stabilize the base-base interaction and, thus, may be generally important for base mispairing either in DNA or in RNA molecules.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown T., Kennard O., Kneale G., Rabinovich D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature. 1985 Jun 13;315(6020):604–606. doi: 10.1038/315604a0. [DOI] [PubMed] [Google Scholar]
- Crawford J. L., Kolpak F. J., Wang A. H., Quigley G. J., van Boom J. H., van der Marel G., Rich A. The tetramer d(CpGpCpG) crystallizes as a left-handed double helix. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4016–4020. doi: 10.1073/pnas.77.7.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Feigon J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Z-DNA forms without an alternating purine-pyrimidine sequence in solution. Science. 1985 Oct 4;230(4721):82–84. doi: 10.1126/science.4035359. [DOI] [PubMed] [Google Scholar]
- Gessner R. V., Quigley G. J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Structural basis for stabilization of Z-DNA by cobalt hexaammine and magnesium cations. Biochemistry. 1985 Jan 15;24(2):237–240. doi: 10.1021/bi00323a001. [DOI] [PubMed] [Google Scholar]
- Irikura K. K., Tidor B., Brooks B. R., Karplus M. Transition from B to Z DNA: contribution of internal fluctuations to the configurational entropy difference. Science. 1985 Aug 9;229(4713):571–572. doi: 10.1126/science.3839596. [DOI] [PubMed] [Google Scholar]
- Mandal C., Kallenbach N. R., Englander S. W. Base-pair opening and closing reactions in the double helix. A stopped-flow hydrogen exchange study in poly(rA).poly(rU). J Mol Biol. 1979 Dec 5;135(2):391–411. doi: 10.1016/0022-2836(79)90443-1. [DOI] [PubMed] [Google Scholar]
- Mirau P. A., Kearns D. R. Effect of environment, conformation, sequence and base substituents on the imino proton exchange rates in guanine and inosine-containing DNA, RNA, and DNA-RNA duplexes. J Mol Biol. 1984 Aug 5;177(2):207–227. doi: 10.1016/0022-2836(84)90453-4. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Leng M. Salt-dependent dynamic structure of poly(dG-dC) x poly(dG-dC). Nature. 1980 Nov 27;288(5789):413–414. doi: 10.1038/288413a0. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Gessner R. V., van der Marel G. A., van Boom J. H., Rich A. Crystal structure of Z-DNA without an alternating purine-pyrimidine sequence. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3611–3615. doi: 10.1073/pnas.82.11.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG). Cell. 1984 May;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang A. J., Quigley G. J., Kolpak F. J., van der Marel G., van Boom J. H., Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981 Jan 9;211(4478):171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]


