Abstract
There is limited data on methicillin-resistant Staphylococcus aureus (MRSA) carriage in dental clinics. 1300 specimens from patients, health personnel, and environmental surfaces of a dental clinic in Egypt were tested for MRSA. Antibiotic susceptibility, biofilm formation, Staphylococcal protein A (spa) typing, SCCmec typing, and PCR-based assays were used to detect mecA, mecC, vanA, Panton-Valentine Leukocidin toxin (PVL), and toxic shock syndrome toxin-1 (tst) genes. Among 34 mecA-positive MRSA isolates, five (14.7%) were PVL-positive, seventeen (50%) were tst-positive, ten (29.4%) were vanA-positive, while none harboured mecC. MRSA hand carriage rates in patients, nurses, and dentists were 9.8%, 6.6%, and 5%. The respective nasal colonization rates were 11.1%, 6.7%, and 9.7%. 1.3% of the environmental isolates were MRSA-positive. Strong and moderate biofilm-forming isolates represented 23.5% and 29.4% of MRSA isolates. 24 MRSA isolates (70.6%) were multi-resistant and 18 (52.9%) harboured SCCmec IV. Among eight spa types, t223 (26.5%), t267 (23.5%), and t14339 (23.5%) were predominant. We noted an alarming genetic relatedness between 7 (20.6%) MRSA isolates and the epidemic EMRSA-15 clone, as well as a combined occurrence of tst and PVL in 3 (8.8%) isolates. Results suggest high MRSA pathogenicity in dental wards highlighting the need for more efficient surveillance/infection control strategies.
Introduction
Staphylococcus aureus is an infectious human pathogen that can survive on inanimate environmental surfaces1. It can colonize skin, mucous membranes, and the anterior nares in about 30% of healthy individuals2, 3. Methicillin-resistant Staphylococcus aureus (MRSA) is associated with substantial morbidity and mortality in many regions of the world2. MRSA strains that can spread rapidly among patients are known as epidemic MRSA (EMRSA) strains4. At least 17 different EMRSA clones have been identified5. One of these clones, EMRSA-15, is of global health concern, because it is highly transmissible, with capability of spreading between different continents, which explains its dissemination from the UK (where it was first reported) to several other parts of the world6.
MRSA infections, especially its biofilm-forming variants, are often difficult to treat for a variety of reasons. Firstly, these infections are usually attributed to multiple virulence determinants, including the lukF/S-PV genes encoding the Panton-Valentine leukocidin (PVL) toxin and the tst gene encoding the toxic shock syndrome toxin-1 (TSST-1)3. Secondly, infections with biofilm-forming strains of MRSA are usually persistent and respond poorly to conventional antibiotic therapy7. Thirdly, MRSA strains possess high levels of resistance to multiple antibiotics as a result of both intrinsic and acquired mechanisms8, such as the mecA- or mecC-mediated methicillin resistance9, 10, and vanA-mediated vancomycin resistance11. It has to be highlighted that while mecA gene is located on a mobile staphylococcal cassette chromosome (SCC) element known as SCCmec, twelve different types of SCCmec (I to XII) have been defined to date, five of which (I to V) are globally distributed12–16.
MRSA can be health-care-associated MRSA (HA-MRSA) or community-associated MRSA (CA-MRSA)17. HA-MRSA infections are more common in individuals with predisposing risk factors, such as hospitalization or invasive medical procedure18. Many CA-MRSA infections still arise in individuals not exposed to these risk factors18. CA-MRSA strains tend to be susceptible to many non-β-lactam antibiotics, whereas HA-MRSA strains are normally resistant to many antibiotic classes19. Despite efforts, CA-MRSA infections are on the rise worldwide20. In general, CA-MRSA strains are considered to be more virulent, transmissible, and persistent than their HA-MRSA counterparts21, 22. On the genetic level, there are remarkable differences between the two categories. HA-MRSA strains usually carry SCCmec types I, II, or III, whereas the SCCmec types IV or V together with the PVL gene are strongly associated with CA-MRSA strains22. Various molecular typing techniques have been developed for effective epidemiological surveillance and control of MRSA, the most common of which are SCCmec typing, multilocus sequence typing (MLST), Staphylococcus protein A (spa) typing, pulsed-field gel electrophoresis (PFGE) typing, and PVL typing. In this regard, studies have already shown the cost-effectiveness and the efficacy of spa, SCCmec, and PVL techniques compared to PFGE and MLST23, 24.
MRSA can be transmitted through a variety of ways in dental settings. These can include one or more of the following: (1) direct contact with blood or saliva (2) indirect contact with contaminated instruments or environmental surfaces; and (3) exposure to microbial aerosols released from the oral cavity25–27. Therefore, it is likely that dental clinic surfaces and dental health-care personnel (DHCPs) contribute to MRSA transmission to patients or other DHCPs28, 29.
Compared to the number of studies on MRSA isolates from hospitals30–33, less attention has been paid to MRSA isolated from dental care settings. More specifically, data related to the genetic diversity and virulence gene determinants of clones in dental clinics in the region, including Egypt, is scarce. Similarly, little is known about the carriage frequency, the biofilm-forming capacity, and the antimicrobial resistance profiles of MRSA isolated from these settings. Therefore, with a focus on MRSA isolates from dental care settings in Egypt, the objectives of the current study were to: (i) determine the prevalence of these isolates in different dental wards; (ii) assess their carriage rates in patients, nurses, dentists, and environmental surfaces; (iii) determine their genetic lineages using SCCmec and spa genotyping techniques; (iv) characterize their antimicrobial resistance profiles by disk diffusion or agar dilution techniques; (v) determine the presence or absence of five genes (mecA, mecC, vanA, tst, and PVL) implicated in antimicrobial resistance or virulence; and (vi) investigate the biofilm-forming abilities of the isolates.
It is anticipated that a better understanding of virulence gene profiling and molecular characterization of the clones circulating in both community and hospital settings will help us to develop more effective management plans and control strategies for MRSA infections.
Results
Prevalence of MRSA and other staphylococci
In this study, a total of 1300 swab specimens were collected from six different wards within a dental clinic in Egypt, including: 1030 (79.2%) specimens from environmental surfaces and 270 (20.8%) specimens from hands (n = 182) and anterior nares (n = 88) of both patients and DHCPs. These 1030 specimens from environmental surfaces fall into two categories, those from clinical-contact surfaces (n = 602) and those from housekeeping surfaces (n = 428) (Table 1).
Table 1.
Distribution of the recovered isolates by site of specimen.
| Specimen site | No. (%) of specimens or isolates | |||
|---|---|---|---|---|
| Specimens collected n = 1300 | Recovered isolates n = 863 | Isolates positive for S. aureu n = 112s | Isolates positive for MRSA n = 34 | |
| Personnel | ||||
| Hand (patients) | 61 (4.69) | 44 (5.10) | 17 (15.18) | 6 (17.65) |
| Nares (patients) | 27 (2.08) | 21 (2.43) | 12 (10.71) | 3 (8.82) |
| Hand (nurses) | 61 (4.69) | 49 (5.68) | 12 (10.71) | 4 (11.76) |
| Nares (nurses) | 30 (2.31) | 26 (3.01) | 11 (9.82) | 2 (5.88) |
| Hand (dentists) | 60 (4.62) | 54 (6.26) | 7 (6.25) | 3 (8.82) |
| Nares (dentists) | 31 (2.38) | 32 (3.71) | 10 (8.93) | 3 (8.82) |
| Total personnel specimens | 270 (20.77) | 226 (26.19) | 69 (61.60) | 21 (61.76) |
| Environmental surfaces | ||||
| Clinical-contact surfaces | ||||
| Dental light arm | 66 (5.08) | 10 (1.16) | 5 (4.46) | 1 (2.94) |
| Dentist’s chair | 128 (9.85) | 91 (10.54) | 9 (8.04) | 3 (8.82) |
| Dentist’s drill | 112 (8.62) | 56 (6.49) | 8 (7.14) | 2 (5.88) |
| Dentist’s tool rack | 97 (7.46) | 68 (7.88) | 2 (1.79) | — |
| Patient’s sink faucet | 116 (8.92) | 98 (11.36) | 7 (6.25) | 3 (8.82) |
| X-ray switch | 83 (6.38) | 60 (6.95) | 1 (0.89) | — |
| Housekeeping surfaces | ||||
| Dentist/nurse hand washing sink | 68 (5.23) | 14 (1.62) | 1 (0.89) | — |
| Disinfectant containers | 46 (3.54) | 31 (3.59) | 2 (1.79) | — |
| Door knobs | 93 (7.15) | 72 (8.34) | 3 (2.68) | 3 (8.82) |
| Floors | 75 (5.77) | 44 (5.10) | 2 (1.79) | 1 (2.94) |
| Light switches | 94 (7.23) | 54 (6.26) | 2 (1.79) | — |
| Nurses’ desks | 52 (4.0) | 39 (4.52) | 1 (0.89) | — |
| Total environmental surface specimens | 1030 (79.23) | 637 (73.81) | 43 (38.40) | 13 (38.24) |
A minus sign (−) denotes the absence of an attribute.
Based on biochemical properties, 112 isolates (8.6%) from the total specimens collected were S. aureus, and 290 isolates (22.3%) were coagulase-negative Staphylococcus (CoNS). From any specimen source, the CoNS isolates were more predominant than S. aureus counterparts. For example, the CoNS carriage rates in hand, nasal, and environmental specimens were 23.6% (43/182), 40.9% (36/88), and 20.5% (211/1030), respectively, while the respective rates for S. aureus were 19.8% (36/182), 37.5% (33/88), and 4.2% (43/1030).
The isolates recovered from housekeeping surfaces demonstrated a lower prevalence of S. aureus as compared to those recovered from clinical-contact surfaces; however, this difference was statistically non-significant [4.3% versus 8.4%; P = 0.053 by Fisher’s exact test]. For clinical contact surfaces, the dentists’ chairs had the highest prevalence of S. aureus (8%), followed by dentists’ drills (7.1%) and patients’ faucet sinks (6.2%). For housekeeping surfaces, door knobs had the highest prevalence of S. aureus (2.68%), followed by disinfectant containers, floors, and light switches, which had equal prevalence rates of 7.1% each.
Screening for methicillin-resistant isolates was performed by the disk diffusion method [using oxacillin (1 μg) and cefoxitin (30 µg] and was subsequently verified by PCR targeting the mecA and mecC genes. Among the identified S. aureus isolates, 21.4% (24/112) were resistant to both antibiotics (Table 2). On the other hand, four isolates (Table 2, IDs: 11, 15, 20 and 68) showed an oxacillin-sensitive/cefoxitin-resistant profile, while six isolates (Table 2, IDs: 2, 25, 35, 70, 71 and 93) were oxacillin-intermediate but cefoxitin-resistant. All 34 isolates were positive for MRSA as indicated by PCR, leading to a MRSA prevalence of 30.4% (34/112) among all recovered S. aureus isolates.
Table 2.
Sources and characteristics of individual MRSA isolates (n = 34) in this study.
| Isolate ID | Specimen Source | Dental clinic/ward | Phenotypic resistance profilea | Biofilm forming abilityb | Genotypic characteristicsc | ||||
|---|---|---|---|---|---|---|---|---|---|
| PVL | vanA | tst | spa typed | SCCmec type | |||||
| Isolates recovered from personnel (n = 21) | |||||||||
| 1a | Hand (patient) | Dental surgery | FOX-OX | Moderate | − | ND | + | t14339 | II |
| 1b | Hand (patient) | Dental surgery | FOX-OX-VAN* | Moderate | − | − | − | t14339 | V |
| 2 | Hand (patient) | Dental surgery | FOX-DO-CN-OX* | Weak | − | ND | − | t267 | I |
| 7 | Hand (patient) | Prosthetic dentistry | FOX-DO-CN-LZD-OX-VAN | Strong | − | + | + | t267 | IV |
| 15 | Hand (patient) | Operative dentistry | FOX-CD-E-CN-LZD-VAN | Moderate | − | + | − | t084 | IV |
| 71 | Hand (patient) | Prosthodontics | FOX-E-OX* | Non | − | ND | − | t223 | I |
| 56 | Nares (patient) | Dental surgery | FOX-C-DO-CN-OX | Moderate | − | ND | − | t267 | IV |
| 63 | Nares (patient) | Prosthetic dentistry | FOX-CIP-CD-CN-LZD-OX-VAN* | Non | − | − | + | t3689 | I |
| 93 | Nares (patient) | Prosthodontics | FOX-E-CN-LZD-OX*-VAN* | Strong | − | − | − | t084 | IV |
| 23 | Hand (nurse) | Dental surgery | FOX-C*-DO-E-CN-OX-VAN* | Weak | − | − | + | t267 | I |
| 112 | Hand (nurse) | Dental surgery | FOX-DO-E-CN-LZD-OX | Non | + | ND | + | t14339 | IV |
| 30 | Hand (nurse) | Prosthetic dentistry | FOX-DO-CN-OX | Weak | − | ND | + | t223 | I |
| 44 | Hand (nurse) | Operative dentistry | FOX-DO-CN-OX-VAN | Moderate | − | + | − | t1339 | IV |
| 58 | Nares (nurse) | Dental surgery | FOX-CIP-CD-E-OX-VAN* | Non | − | − | − | t223 | Non-typeablee |
| 59 | Nares (nurse) | Dental surgery | FOX-C-CIP-DO*-E-CN-OX-VAN* | Weak | − | − | + | t14339 | I |
| 111 | Hand (dentist) | Dental surgery | FOX-CD*-DO-CN-OX | Non | + | ND | + | t8506 | IV |
| 109 | Hand (dentist) | Periodontics | FOX-CD-DO-CN-OX-VAN | Non | + | + | − | t380 | IV |
| 95 | Hand (dentist) | Endodontics | FOX-DO-CN-OX | Non | − | ND | − | t267 | IV |
| 65 | Nares (dentist) | Prosthetic dentistry | FOX-CIP-CD-CN-LZD-OX-VAN* | Moderate | − | − | − | t3689 | IV |
| 68 | Nares (dentist) | Operative dentistry | FOX-C*-CD-DO-E-CN-LZD-VAN | Moderate | − | + | − | t380 | I |
| 70 | Nares (dentist) | Periodontics | FOX-C-CD-E-OX* | Non | − | ND | − | t223 | I |
| Isolates recovered from environmental surfaces (n = 13) | |||||||||
| 5 | Dental light arm | Dental surgery | FOX-CIP-E-CN- OX | Strong | − | ND | + | t223 | IV |
| 11 | Dentist’s chair | Dental surgery | FOX-CD-LZD-VAN | Strong | + | − | + | t14339 | IV |
| 13 | Dentist’s chair | Periodontics | FOX-C*-DO-E-CN-LZD-OX-VAN | Strong | − | + | + | t223 | IV |
| 18 | Dentist’s chair | Prosthetic dentistry | FOX-C*-CD-DO-OX-VAN | Strong | − | − | + | t223 | I |
| 35 | Dentist’s drill | Periodontics | FOX-C-DO-CN-OX*-VAN | Moderate | − | + | + | t084 | IV |
| 42 | Dentist’s drill | Operative dentistry | FOX-DO-E-CN-OX-VAN* | Strong | − | − | + | t267 | IV |
| 25 | Patient’s sink faucet | Prosthetic dentistry | FOX-C*-E-OX*-VAN* | Weak | − | − | + | t14339 | I |
| 26 | Patient’s sink faucet | Prosthetic dentistry | FOX-DO-CN-OX-VAN | Moderate | − | + | − | t223 | I |
| 50 | Patient’s sink faucet | Periodontics | FOX-C*-E-OX-VAN* | Weak | − | − | + | t14339 | I |
| 20 | Door knob | Periodontics | FOX-E-CN-VAN* | Moderate | − | − | + | t267 | IV |
| 33 | Door knob | Operative dentistry | FOX-C*-DO-E-CN-OX-VAN | Strong | − | + | − | t14339 | I |
| 103 | Door knob | Endodontics | FOX-C-DO-E-CN-OX | Non | + | ND | − | t223 | IV |
| 38 | Floor | Operative dentistry | FOX-C-DO-E-CN-OX-VAN* | Weak | − | − | − | t267 | IV |
PVL: The gene encoding the Panton–Valentine leucocidin toxin; vanA: the gene encoding an enzyme that causes a structural change in the terminal amino acid of the pentapeptide chain of peptidoglycan, thus conferring vancomycin resistance; tst: the gene encoding the toxic shock syndrome toxin; spa: staphylococcal protein A; SCCmec: staphylococcal cassette chromosome mec; a plus sign (+) denotes the presence of a gene; a minus sign (−) denotes the absence of a gene; ND: not determined.
aFOX: cefoxitin, C: chloramphenicol, CIP: ciprofloxacin, CD: clindamycin, DO: doxycycline, E: erythromycin, CN: gentamicin, LZD: linezolid, OX: oxacillin, and VAN: vancomycin
*Denotes intermediate resistance to the antibiotic, according to CLSI guidelines and breakpoints88.
bThe isolates were classified as biofilm non producers, weak, moderate, and strong biofilm producers based on previously published criteria34.
cAll the MRSA isolates investigated in the current study were mecA-positive and mecC-negative.
dThe spa types shown are based on the Ridom StaphType software.
eNon-typeable: An isolate that was positive only for the mecA gene, with no PCR product obtained, or not in agreement with the predicted band patterns of SCCmec types I-V by the multiplex PCR method used.
The prevalence rate of MRSA was 0.98% (2/205) in samples collected from the endodontic ward, 2.9% (6/205) in samples from the operative dentistry, 2.4% (6/255) in samples from the periodontics, 3.9% (7/180) in samples from the prosthetic dentistry, 1% (2/200) in samples from the prosthodontics, and 4.3% (11/255) in samples from the dental surgery ward (Tables 2 and 3). This prevalence difference was found to be statistically non-significant (χ2 = 8.394, df = 5, P = 0.136).
Table 3.
Ward distribution of the specimens collected in this study.
| Dental ward | No. (%) of specimens or isolates | |||||
|---|---|---|---|---|---|---|
| Personnel | Environmental surfaces | Total no. (Personnel + Environmental) | ||||
| Specimens collected n = 270 | Recovered isolates n = 226 | Specimens collected n = 1030 | Recovered isolates n = 637 | Specimens collected n = 1300 | Recovered isolates n = 863 | |
| Endodontics | 38 (14.02) | 32 (14.16) | 167 (16.21) | 126 (19.78) | 205 (15.77) | 158 (18.31) |
| Operative dentistry | 40 (14.81) | 26 (11.51) | 165 (16.02) | 104 (16.33) | 205 (15.77) | 130 (15.06) |
| Periodontics | 45 (16.7) | 28 (12.39) | 210 (20.39) | 103 (16.17) | 255 (19.62) | 131 (15.18) |
| Prosthetic dentistry | 49 (18.15) | 48 (21.24) | 131 (12.72) | 90 (14.13) | 180 (13.85) | 138 (15.99) |
| Prosthodontics | 48 (17.8) | 46 (20.35) | 152 (14.76) | 118 (18.52) | 200 (15.38) | 164 (19.00) |
| Dental surgery | 50 (18.52) | 46 (20.35) | 205 (19.90) | 96 (15.07) | 255 (19.61) | 142 (16.46) |
| Total | 270 (100) | 226 (100) | 1030 (100) | 637 (100) | 1300 (100) | 863 (100) |
As shown in Table 1, the highest hand carriage rate of MRSA was detected in patients (9.8%, 6/ 61), followed by nurses (6.6%, 4/61), and dentists (5%, 3/60); however, this difference was not statistically significant (χ2 = 2.006, df = 2, P = 0.3666). The highest MRSA nasal colonization rate was observed in patients (11.1%, 3/27), followed by dentists (9.7%, 3/31), and nurses (6.7%, 2/30) (Table 1). This difference was also non-significant (χ2 = 0.5883, df = 2, P = 0.7452).
The environmental surfaces in 5 (83.3%) out of the 6 wards under study were contaminated with MRSA (Table 2). Environmental surfaces within the prosthetic dentistry ward showed the highest prevalence (2.3%, 3/131) of MRSA, followed by those from periodontics (1.9%, 4/210), operative dentistry (1.8%, 3/165), dental surgery (0.98%, 2/205), endodontics (0.60%, 1/167), while those within the prosthodontic ward were MRSA-free. This difference turned to be statistically significant (χ2 = 6.42, df = 5, P = 0.2675). The highest prevalence of MRSA in the environmental surfaces was observed in door knobs (3.2%, 3/93) and dentists’ chairs (2.3%, 3/128), while the lowest prevalence was found in dental light arms (1.5%, 1/66) and floors (1.3%, 1/75).
Characterization of the MRSA isolates
Genetic groups based on spa typing and SCCmec typing
The spa typing analysis revealed 8 distinct spa types within the tested MRSA isolates. The spa type attribution of each isolate is reported in Table 2. The frequencies, geographical spread, and repeat successions for each identified spa type are reported in Table 4. The spa type including the largest number of isolates was t223 (n = 9, 26.5% of all tested MRSA isolates). This was immediately followed by t14339 and t267 (each of which contained 8 isolates, 23.5%). The other spa types were less frequent, including t084 (n = 3), t3689 (n = 2), t380 (n = 2), t8506 (n = 1), and t1339 (n = 1).
Table 4.
Frequencies, geographical spread, repeat successions, and predicted clonal complexes (CCs) of the spa types detected in this study.
| Cluster group and spa-CCsa | Ridom spa typeb | No. (%) of MRSA isolates | Geographical spread* | spa repeat succession | Predicted MLST CCd | |||
|---|---|---|---|---|---|---|---|---|
| Total specimens n = 34 | Personnel specimens n = 21 | Environmental surface specimens | Relative global frequency* | |||||
| Cluster 1 spa-CC223 | t223 | 9 (26.5%) | 4 (19%) | 5 (38.5%) | 0.42% | Austria, Belgium, Chile, Czech Republic, Denmark, Detmold, France, Germany, Iceland, Ireland, Israel, Italy, Jordan, Minden, Netherlands, New Zealand, Norway, Romania, Spain, Sweden, Switzerland, Syria, United Arab Emirates, United Kingdom, the Gaza Strip (Palestine)50, Kuwait61 | 26-23-13-23-05-17-25-17-25-16-28 | CC22f |
| t14339 | 8 (23.5%) | 4 (19%) | 4 (30.8%) | 0.00% | Ireland | 26-23-13-23-36-17-25-17-25-16-28 | CC22 | |
| t3689 | 2 (5.9%) | 2 (9.5%) | — | 0.00% | Denmark | 26-23-13-23-05-17-25-17-25-25–16-28 | CC22 | |
| t8506 | 1 (2.9%) | 1 (4.8%) | — | 0.00% | Saudi Arabia | 26-23-13-16-05-17-25-17-25-16-28 | CC22 | |
| Singletons | t267 | 8 (23.5%) | 5 (23.8%) | 3 (23%) | 0.30% | Argentina, Austria, Belgium, Cyprus, Denmark, France, Gabon, Germany, Iceland, Iran, Israel, Italy, Jordan, Lebanon, Netherlands, New Zealand, Norway, Saudi Arabia, South Africa, Spain, Sweden, Taiwan, United Arab Emirates, United Kingdom, United States | 07-23-12-21-17-34-34-34-33-34 | CC80f |
| t084 | 3 (8.9%) | 2 (9.5%) | 1 (7.7%) | 1.76% | Argentina, Australia, Austria, Belgium, China, Denmark, Finland, France, Gabon, Germany, Greece, Iceland, Indonesia, Iran, Italy, Jordan, Lebanon, Netherlands, New Zealand, Nigeria, Norway, Poland, Romania, South Africa, Spain, Sweden, Switzerland, Taiwan, Uganda, United Arab Emirates, United Kingdom, United States, Germany | 07-23-12-34-34-12-12-23-02-12-23 | CC15e | |
| t1339 | 1 (2.9%) | 1 (4.8%) | — | 0.01% | Austria, Denmark, Germany, Iceland, Norway, Sweden, United Arab Emirates | 07-12-21-17-13-13-34-13-34-33-34 | CC80g | |
| Excluded | t380 | 2 (5.9%) | 2 (9.5%) | — | 0.00% | Germany, Lebanon, Sweden, United Kingdom | 26-34-33-34 | NP |
*Based on data from the Ridom StaphType database (http://spa.ridom.de/frequencies.shtml), last accessed on 15 November 2016, unless otherwise stated. Countries from the Arab world are double-underlined.
a spa-CC: denotes spa clonal complex as determined by the Based Upon Repeat Pattern (BURP) algorithm in the Ridom StaphType software with a distance cost of ≤ 5; excluded: denotes entries excluded from BURP clustering because the spa repeat pattern comprised fewer than five repeats.
bNomenclature according to Harmsen et al.91; the spa types shown are based on the Ridom StaphType software.
cA minus sign (−) indicates the absence of the spa type.
dMLST CC: denotes predicted clonal complexes based on multi-locus sequence typing.
eAccording to the Ridom SpaServer (http://spaserver.ridom.de).
fBased on data from98.
gBased on data from99.
NP: not predictable (not yet assigned to CC).
Two of the most frequent spa types in our study (t223 and t267) were disseminated in different wards, since they were recovered from five out of the six tested wards (Table 5). Conversely, the spa type 14339 was mainly predominant in the dental surgery ward (62.5%, 5/ 8).
Table 5.
Characteristics and detailed distribution of the spa-CCs and spa-types detected in this study.
| Cluster group and spa-CCs | Ridom spa typea | No. of MRSA isolates | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Based on specimen source | Based on dental ward | Based on presence of antibiotic resistance or virulence genes b | Based on biofilm forming ability c | |||||||||||||||||||||||
| Personnel (n = 21) | Environmental surfaces (n = 13) | End-odon-tics (n = 2) | Oper-ative denti-stry (n = 6) | Periodontics (n = 6) | Prost-hetic dent-istry (n = 7) | Prostho-dontics (n = 2) | Dental surgery (n = 11) | PVL (n = 5) | vanA (n = 10) | tst (n = 17) | Non (n = 9) | Weak (n = 7) | Mode-rate (n = 10) | Strong (n = 8) | ||||||||||||
| Patients | Nurses | Dentists | Clinical-contact | Housekeeping | ||||||||||||||||||||||
| Hand (n = 6) | Nares (n = 3) | Hand (n = 4) | Nares (n = 2) | Hand (n = 3) | Nares (n = 3) | D light arm (n = 1) | D chair (n = 3) | D drill (n = 2) | P sink faucet (n = 3) | Door knobs (n = 3) | Floors (n = 1) | |||||||||||||||
| Cluster 1 spa-CC223 | t223 (n = 9) | 1 | 0 | 1 | 1 | — | 1 | 1 | 2 | — | 1 | 1 | — | 1 | — | 2 | 3 | 1 | 2 | 1 | 2 | 4 | 4 | 1 | 1 | 3 |
| t14339 (n = 8) | 2 | — | 1 | 1 | — | — | — | 1 | — | 2 | 1 | — | — | 1 | 1 | 1 | — | 5 | 2 | 2 | 6 | 1 | 3 | 2 | 2 | |
| t3689 (n = 2) | — | 1 | — | — | — | 1 | — | — | — | — | — | — | — | — | — | 2 | — | — | — | — | 1 | 1 | — | 1 | — | |
| t8506 (n = 1) | — | — | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — | 1 | 1 | — | 1 | 1 | — | — | — | |
| Singletons | t267 (n = 8) | 2 | 1 | 1 | — | 1 | — | — | — | 1 | — | 1 | 1 | 1 | 2 | 1 | 1 | — | 3 | — | 1 | 4 | 1 | 3 | 2 | 2 |
| t084 (n = 3) | 1 | 1 | — | — | — | — | — | — | 1 | — | — | — | — | 1 | 1 | — | 1 | — | — | 2 | 1 | — | — | 2 | 1 | |
| t1339 (n = 1) | — | — | 1 | — | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | — | 1 | — | — | — | 1 | — | |
| Excluded | t380 (n = 2) | — | — | — | — | 1 | 1 | — | — | — | — | — | — | — | 1 | 1 | — | — | — | 1 | 2 | — | 1 | — | 1 | — |
spa-CC: denotes spa clonal complex as determined by the Based Upon Repeat Pattern (BURP) algorithm in the Ridom StaphType software with a distance cost of ≤ 5; excluded: denotes entries excluded from BURP clustering because the spa repeat pattern comprised fewer than five repeats; D light arm: Dental light arm; D chair: Dentist’s chair; D drill: Dentist’s drill; P sink faucet: Patient’s sink faucet; PVL: The gene encoding the Panton–Valentine leucocidin toxin; vanA: the gene encoding an enzyme that causes a structural change in the terminal amino acid of the pentapeptide chain of peptidoglycan, thus conferring vancomycin resistance; tst: the gene encoding the toxic shock syndrome toxin; spa: staphylococcal protein A; a minus sign (−) denotes the absence of an attribute.
aNomenclature according to Harmsen et al.91; the spa types shown are based on the Ridom StaphType software.
bAll the MRSA isolates investigated in the current study were mecA-positive and mecC-negative.
CThe isolates were classified as biofilm non-producers, weak, moderate, and strong biofilm producers based on previously published criteria34.
Only four spa types (t223, t14339, t267, and t084) coexisted in both personnel and environmental surface specimens, with higher frequencies of t267 and t084 in personnel specimens, an equal distribution of t14339 in both specimen categories, and higher frequency of t223 in environmental surface specimens (Table 5). On the other hand, the other four spa types (t3689, t8506, t1339, and t380) were only detected among isolates from personnel (Table 5).
The BURP algorithm (cost ≤ 5) assigned the isolates into a single clonal complex, spa-CC223 (n = 20 isolates, 58.8% of all tested MRSA isolates), as well as 3 singletons (n = 12 isolates, 35.2%), while excluding 2 isolates (5.8%) from the clustering (Table 4). The spa types in the spa-CC223 were: t223 (9 strains out of 20, 45%), t14339 (8, 40%), t3689 (2, 10%), t8506 (1, 5%) as presented in Fig. 1 and Table 4. Table 5 lists the characteristics and detailed distribution of different spa-CCs and spa types.
Figure 1.
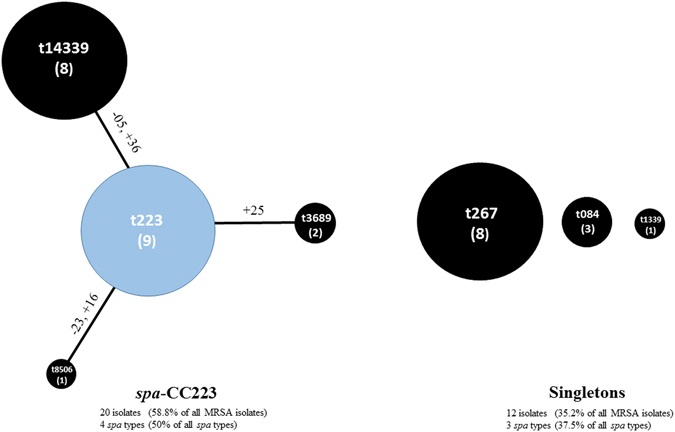
Population structure of the tested MRSA isolates (n = 34) based on BURP analysis. This analysis was performed using the Based Upon Repeat Pattern (BURP) algorithm of the Ridom StaphType software (Ridom GmbH, Würzburg, Germany) at a cost setting of ≤ 5 and excluding spa-types with 5 or fewer repeats. Each dot represents a different spa type, with the diameter of the dot being proportional to the quantity of the corresponding spa type. Clusters of linked spa types correspond to spa clonal complexes (spa-CCs). The predict founder of a cluster (which was used for defining the cluster) is shown in blue, while the others in black. Near the lines of connection, the mutations involved in the transition from a spa type to the next one are reported in detail. All DNA changes are meant to occur from the founder to the periphery. Legend: numbers along the lines refer to the repeat sequence involved in the mutation; +indicates the acquisition of a repeat sequence; - indicates the loss of a repeat sequence; within circles the numbers of the strains of each CC appear between brackets. In summary, the analysis identified a single clonal complex (spa-CC223) comprising spa types t223, t14339, t3689, and t8506; n = 20 isolates, and accounted for 58.8% of all tested MRSA isolates, as well as 3 singletons (t267, t084, and t1339; n = 12 isolates, 35.2%), while excluded 2 isolates (t380, 5.8%) from the clustering, as they consisted of four repeat units only.
For SCCmec typing, the multiplex PCR assay identified 18 out of 34 MRSA isolates (52.9%) with SCCmec type IV, 13 (38.2%) with SCCmec type I, 1 isolate (2.9%) with SCCmec type II, and 1 isolate (2.94) with SCCmec type V (Table 6). The SCCmec type III was not found in any isolate. One isolate (2.9%) could not be typed (Table 6 ).
Table 6.
Characteristics and detailed distribution of the SCCmec types detected in this study.
| SCCmec type | No. of MRSA isolates | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Based on specimen source | Based on dental ward | Based on presence of antibiotic resistance or virulence genes a | Based on biofilm orming ability b | ||||||||||||||||||||||
| Personnel (n = 21) | Environmental surfaces (n = 13) | Endodontics (n = 2) | Operative dentistry (n = 6) | Periodontics (n = 6) | Prosthetic dentistry (n = 7) | Prosthodontics (n = 2) | Dental surgery (n = 11) | PVL (n = 5) | vanA (n = 10) | tst (n = 17) | Non (n = 9) | Weak (n = 7) | Moderate (n = 10) | Strong(n = 8) | |||||||||||
| Patients | Nurses | Dentists | Clinical-contact | Housekeeping | |||||||||||||||||||||
| Hand (n = 6) | Nares (n = 3) | Hand (n = 4) | Nares (n = 2) | Hand (n = 3) | Nares (n = 3) | D. light arm (n = 1) | D. chair (n = 3) | D. drill (n = 2) | P. sink faucet (n = 3) | Door knobs (n = 3) | Floors (n = 1) | ||||||||||||||
| I ( n = 13) | 2 | 1 | 2 | 1 | — | 2 | — | 1 | — | 3 | 1 | — | — | 2 | 2 | 5 | 1 | 3 | — | 3 | 7 | 3 | 6 | 2 | 2 |
| II ( n = 1) | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 1 | — | — | 1 | — | — | 1 | — |
| III ( n = 0) | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| IV ( n = 18) | 2 | 2 | 2 | — | 3 | 1 | 1 | 2 | 2 | — | 2 | 1 | 2 | 4 | 4 | 2 | 1 | 5 | 5 | 7 | 9 | 5 | 1 | 6 | 6 |
| V ( n = 1) | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | — | 1 | — |
| Non-typeable ( n = 1) | — | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | — | — | — |
SCCmec: staphylococcal cassette chromosome mec; Non-typeable: an isolate with no PCR product obtained, or not in agreement with the predicted band patterns of SCCmec types I-V by the multiplex PCR method used; D light arm: Dental light arm; D chair: Dentist’s chair; D drill: Dentist’s drill; P sink faucet: Patient’s sink faucet; PVL: The gene encoding the Panton–Valentine leucocidin toxin; vanA: the gene encoding an enzyme that causes a structural change in the terminal amino acid of the pentapeptide chain of peptidoglycan, thus conferring vancomycin resistance; tst: the gene encoding the toxic shock syndrome toxin; spa: staphylococcal protein A; a minus sign (−) denotes the absence of an attribute.
aAll the MRSA isolates investigated in the current study were mecA-positive and mecC-negative.
bThe isolates were classified as biofilm non-producers, weak, moderate, and strong biofilm producers based on previously published criteria34.
Antimicrobial resistance profile
Antimicrobial susceptibility testing of the MRSA isolates revealed thirty one resistance profiles, in which nineteen and thirteen of these profiles were observed among the MRSA isolates recovered from personnel and environmental surfaces, respectively (Table 2).
The rates of full resistance among the 34 MRSA isolates tested in the current study were as follows: 100% (n = 34) for cefoxitin, 17.6% (n = 6) for chloramphenicol, 14.7% (n = 5) for ciprofloxacin, 26.5% (n = 9) for clindamycin, 55.8% (n = 19) for doxycycline, 52.9% (n = 18) for erythromycin, 73.5% (n = 25) for gentamycin, 26.4% (n = 9) for linezolid, 67.6% (n = 23) for oxacillin, and 29.4% (n = 11) for vancomycin. Conversely, all tested isolates were susceptible to cefaclor, ceftriaxone, imipenem, and neomycin.
Some isolates showed intermediate resistance to the tested antimicrobials, with a rate of 20.6% (n = 7) for chloramphenicol, 2.9% (n = 1) for doxycycline, 17.6% (n = 6) for oxacillin, and 35.3% (n = 12) for vancomycin. The overall rates of resistance (defined as the rate of intermediate resistance plus the rate of full resistance) to the previously mentioned antimicrobial agents were as follows: 38.2% (n = 13) for chloramphenicol, 58.8% (n = 20) for doxycycline, 85.3% (n = 29) for oxacillin, and 67.6% (n = 23) for vancomycin.
The majority of the tested MRSA isolates (n = 24, 70.6%) were multidrug resistant (non-susceptible to at least one agent in three or more of the tested antimicrobial classes, other than β-lactams). In this regard, the non-susceptibility rates, which include both intermediate and resistant isolates, to two, three, four, five, six, seven, and eight antimicrobials were 5.9%, 8.8%, 29.4%, 26.5%, 17.6, 5.9%, and 2.9%, respectively. There was no significant difference in the prevalence of multidrug resistance between the MRSA isolates recovered from environmental surfaces and those isolated from personnel (76.9% versus 66.67%, P = 0.704 by Fisher’s exact test).
The full resistance rates were generally higher for personnel isolates than for environmental surface isolates for all tested antimicrobial agents. For cefoxitin, no difference was found between the two specimen categories. For chloramphenicol, doxycycline, erythromycin, and vancomycin, full resistance rates for the environmental surface isolates were higher than their personnel counterparts.
A comparison of the occurrence of antimicrobial resistance among the investigated MRSA isolates in relation to different clonal lineages is presented in Table 7. A significantly higher frequency of ciprofloxacin resistance was recorded among isolates from spa type t3689 than other types (χ2 = 14.62, df = 7, P = 0.0412). Similarly, resistance to clindamycin occurred at significantly higher frequencies in MRSA with spa types t3689 or t380 as compared to other types (χ2 = 15.8, df = 7, P = 0.027). For the remaining antimicrobial agents, non-significant differences in resistance were observed between the spa types identified in the current study (Table 7).
Table 7.
Antimicrobial resistance rates among the investigated MRSA isolates in relation to different clonal lineages.
| Antimicrobial agent | No. (%) of MRSA isolates phenotypically resistant to | P value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| spa -CC223 | Singletons | Excluded | Total | |||||||
| t223 n = 9 | t14339 n = 8 | t3689 n = 2 | t8506 n = 1 | t267 n = 8 | t084 n = 3 | t1339 n = 1 | t380 n = 2 | n = 34 | ||
| Cefoxitin (FOX) | 8 (88.9%) | 8 (100%) | 2 (100%) | 1 (100%) | 8 (100%) | 3 (100%) | 1 (100%) | 1 (50%) | 32 (94.1%) | 0.259 |
| Chloramphenicol (C) | 2 (22.2%) | 1 (12.5%) | 0 | 0 (0%) | 2 (25%) | 1 (33.3%) | 0 | 0 | 6 (17.6%) | 0.937 |
| Ciprofloxacin (CIP) | 2 (22.2%) | 1 (12.5%) | 2 (100%) | 0 (0%) | 0 | 0 | 0 | 0 | 5 (14.7%) | 0.041 |
| Clindamycin (CD) | 3 (33.3%) | 1 (12.5%) | 2 (100%) | 0 (0%) | 0 | 1 (33.3%) | 0 | 2 (100%) | 9 (26.5%) | 0.027 |
| Doxycycline (DO) | 5 (55.6%) | 2 (25%) | 0 | 1 (100%) | 7 (87.5%) | 1 (33.3%) | 1 (100%) | 2 (100%) | 19 (55.9%) | 0.081 |
| Erythromycin (E) | 6 (66.7%) | 5 (62.5%) | 0 | 0 | 4 (50%) | 2 (66.7%) | 0 | 1 (50%) | 18 (52.9%) | 0.571 |
| Gentamicin (CN) | 5 (55.6%) | 3 (37.5%) | 2 (100%) | 1 (100%) | 8 (100%) | 3 (100%) | 1 (100%) | 2 (100%) | 25 (73.5%) | 0.073 |
| Linezolid (LZD) | 1 (11.1%) | 2 (25%) | 2 (100%) | 0 | 1 (12.5%) | 2 (66.7%) | 0 (0%) | 1 (50%) | 9 (26.5%) | 0.129 |
| Oxacillin (OX) | 7 (77.8%) | 6 (75%) | 2 (100%) | 1 (100%) | 6 (75%) | 0 | 1 (100%) | 1 (50%) | 24 (70.6%) | 0.209 |
| Vancomycin (VAN) | 3 (33.3%) | 2 (25%) | 0 | 0 | 1 (12.5%) | 2 (66.7%) | 1 (100%) | 2 (100%) | 11 (32.4%) | 0.140 |
Data indicate the number and percentage (%) of full resistance for each respective antimicrobial agent; isolates showing intermediate resistance to the tested antimicrobials are not included in the table.
All the tested isolates were susceptible to cefaclor, ceftriaxone, imipenem, and neomycin.
spa-CC: denotes spa clonal complex as determined by the Based Upon Repeat Pattern (BURP) algorithm in the Ridom StaphType software with a distance cost of ≤ 5; excluded: denotes entries excluded from BURP clustering because the spa repeat pattern comprised fewer than five repeats.
a P values calculated using chi-squared tests of 2 × 8 contingency tables. P values < 0.05 are highlighted in bold.
Prevalence of mecA, mecC, vanA, tst, and PVL-encoding genes
All the tested MRSA isolates possessed the mecA gene (n = 34, 100%), while mecC was not identified.
Ten (29.4%) of the tested MRSA isolates (distributed equally between environmental surfaces and personnel; n = 5 for each group) were positive for the vanA gene. Four (40%) of these isolates were associated with spa-CC223, four (40%) were associated with multiple spa types, and two (20%) had a spa type (t380) that was excluded from clustering (Tables 2 and 4). The vanA gene was detected only in isolates harbouring SCCmec type IV (n = 7, 70%) and SCCmec type I (n = 3, 30%) (Table 6).
Seventeen (50%) of the tested MRSA isolates were positive for the tst gene (distributed as follows: 9 from environmental surfaces, 6 from hand swabs, and 2 from nasal swabs). These isolates were predominantly associated with spa-CC223 corresponding to CC22 (n = 12, 70.6%), while the remaining tst-positive isolates were associated with spa type t267 corresponding to CC80 (n = 4, 23.5%) and spa type t084 corresponding to CC15 (n = 1, 5.9%) (Tables 2 and 4).
Five (14.7%) out of the 34 MRSA isolates contained the PVL gene, which seemed to be more associated with environmental surfaces than with personnel isolates (15.38% versus 14.29%), but this difference was non-significant (P > 0.9999 by Fisher’s exact test). While all of these PVL-positive MRSA isolates carried SCCmec type IV (Table 6), the majority (80%, 4/5) were associated with spa-CC223, while only one isolate (20%) had a spa type (t380) with non-predictable CC (Tables 2 and 4). Additionally, three of the PVL-positive strains were also positive for the tst gene, one of which also harboured the vanA gene (Table 2).
Biofilm formation
From the 34 MRSA isolates tested for biofilm formation, 8 (23.5%) isolates were classified as strong biofilm producers, 10 (29.4%) were moderate, 7 (20.6%) were weak, and 9 (26.5%) were non-biofilm producers (Fig. 2). This classification was based on the criteria established by Stepanovic and colleagues34.
Figure 2.
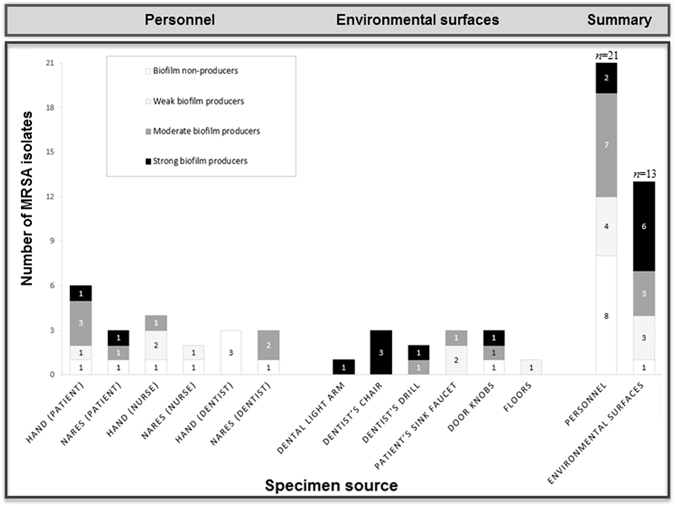
Biofilm-forming abilities of the tested MRSA isolates in relation to the specimen source.
All isolates from personnel were shown to be moderate, weak, or non-biofilm producers, except two isolates with strong biofilm-forming ability (Table 2, IDs: 7 and 93), which were derived from hand and nasal swabs of two different patients attending the prosthetic dentistry and prosthodontic wards, respectively. For isolates recovered from enviromental surfaces, only 7.7% (1/13) of the isolates were non-biofilm formers, while the rest were biofilm formers [46.2% (6/13) strong, 23% (3 /13) moderate, and 23% (3/13) weak] (Fig. 2).
There was no statistically significant difference in biofilm-forming abilities between MRSA isolates recovered from personnel and those recovered from environmental surfaces (χ2 = 7.733, df = 3, P = 0.0519). Similarly, no significant differences were found in biofilm production between isolates recovered from clinical contact surfaces and those recovered from housekeeping surfaces (χ2 = 2.829, df = 3, P = 0.4188).
Discussion
A very limited number of studies, none of which was performed in Egypt, investigated carriage frequency, antibiotic resistance, virulence properties, and genetic diversity of MRSA strains isolated from personnel and environmental surfaces from dental health-care personnel (DHCPs), dental patients, and dental environment. In an effort to fill this knowledge gap, we phenotypically and genotypically characterized MRSA isolates from six different wards at a university outpatient dental clinic in Egypt. We elected to restrict screening of personnel samples to hand and nasal swab specimens, while the environmental surfaces chosen included those commonly encountered within and outside of the patient care area. The recovered isolates were initially identified based on resistance to cefoxitin and oxacillin, and were further confirmed by mecA gene detection. Cefoxitin demonstrated a 100% sensitivity for MRSA detection in our hands, as compared to 73.5% in the case of oxacillin, which missed the detection of 10 (26.47%) of the mecA-positive isolates (Table 2, IDs: 2, 11, 15, 20, 25, 35, 68, 70, 71, 93). The superiority of cefoxitin for MRSA identification has been reported by a number of authors35–37.
In the current study, MRSA represented 3.9% of all recovered bacterial isolates (34/863) and 30.4% (34/112) of all recovered S. aureus strains. A 6.6% (12/182) MRSA prevalence was observed among DHCPs, which is in line with the 6.1% prevalence reported among health care workers in the Middle East38. This rate is higher than the average global prevalence of MRSA carriage by DHCPs of 4.6%38. The MRSA nasal carriage rate among the outpatients screened in the current study (10.2%, 9/88) was lower than the 32% rate reported in a sample of Egyptian outpatients attending primary health care centers [48]. Similarly, the MRSA hand carriage rate among our outpatients (9.8%, 6/61) was much lower than the 47.4% rate obsereved among those attending a dermatology clinic39. We detected higher nasal carriage rates of MRSA in outpatients (11.1%, 3/27) than DHCPs (8.2%, 5/61). This is in agreement with the results of a previous study that showed a higher prevalence of nasal MRSA colonization in patients compared with health care workers (5.1% vs. 4.8%)40. Overall, our results related to the hand and nasal carriage of MRSA among medical and non-medical personnel, as well as MRSA colonization on surfaces in the clinic environment are consistent with results reported in a number of studies41–45. The differences between prevalence rates in our study and others could be attributed to differences in study design, sample size, patient characteristics, and specimen types tested.
Excluding that of prosthodontics, all the investigated wards had one or more of their surfaces positive for MRSA. The absence of MRSA in the tested surfaces of the prosthodontics ward may be attributed to the nature of dental procedures performed in this particular ward, being mostly non-invasive46. Supporting this suggestion is the finding that MRSA-positivity of patients’ sink faucets in our study occurred exclusively in the prosthetic dentistry and periodontics wards, where bleeding of patients is common, due to the invasive dental procedures performed in both wards46. As might be expected, surfaces with more patient contact (clinical contact surfaces) had higher rates of MRSA colonization than surfaces with less patient contact (housekeeping surfaces). Door knobs were the most contaminated among the investigated housekeeping surfaces. Given the absence of MRSA in housekeeping surfaces that are mainly touched by DHCPs (disinfectant containers, dentist/nurse hand washing sink, and nurses’ desks), it may be reasonable to assume that door knobs in the present study were mainly contaminated by patients’ hand contact. Looking at the possibility of MRSA transmission among the various dental wards and/or among the various specimen categories, only three pairs of isolates showed the same antibiogram (Table 2, IDs: 26 and 44; 63 and 65; and 30 and 95). However, differences were found in the SCCmec types and spa types carried by the two isolates within each of these pairs.
The majority (n = 24; 70.6%) of our MRSA isolates showed multidrug resistance pattern to most of the antimicrobials used. While this may be a reflection of the excessive, unjustified use of broad-spectrum antibiotics in Egypt47, this pattern is usually associated with HA-MRSA, because antibiotic resistance in CA-MRSA strains is often limited to β-lactams48, 49. Surprisingly, 14 (51.9%) of the multidrug-resistant isolates in our study carried SCCmec type IV, which is commonly found in CA-MRSA. The emergence and spread of these multidrug-resistant CA-MRSA isolates could also be the result of the selective pressure of excessive and inappropriate antibiotic usage in our community. In the case of antibiotics known for their potent anti-MRSA activities, the isolates showed a relatively high rate of resistance to vancomycin and linezolid (29.4% and 26.4%, respectively), while there was virtually no resistance to imipenem. Overall, nineteen and thirteen antibiotic resistance profiles were observed among MRSA isolates from personnel and environmental surfaces, respectively. This difference might reflect the presence of strong selective pressure from antibiotic usage in the personnel group.
The investigated isolates were characterized using SCCmec and spa molecular typing tools. Based on the former typing method, the classical nosocomial SCCmec types I and II represented 38.2% and 2.9%, respectively, whereas SCCmec type III was completely absent. On the other hand, SCCmec type IV (which is usually considered a CA-MRSA marker) was the most predominant type. However, the multidrug resistance profiles and the relatively low prevalence of the PVL gene (14.7%) seen in our isolates are also common in HA-MRSA strains. The predominance of SCCmec type IV in this study is in agreement with other studies conducted on community-derived and hospital-derived MRSA isolates in the neighbouring territories of Jordan and Gaza50–52. Taken collectively, these findings suggest that in Egypt, and probably other neighbouring regions, the population structure of MRSA in the community is starting to mirror that found in the hospital setting, making the boundaries between these two categories so blurred. A similar observation has been made in other countries, which may be attributed to increased MRSA colonization rates in the community, or increased prevalence of nosocomial MRSA53–59.
In the current study, spa-CC223 was the main spa clonal complex (58.8% of the total MRSA isolates). Interestingly, spa-CC223 has been reported as the second most predominant spa-CC in a study conducted in Kuwait60. Additionally, we identified 8 different spa types among the tested MRSA isolates, 6 of which (t223, t267, t084, t380, t8506, and t1339) have been previously reported in other Arab countries (Table 4). The limited diversity and the high frequency of the spa type t223 are in agreement with previous data from general population studies as well as from hospital-based studies in the Arab region50, 61, 62. This may suggest that certain MRSA clones are more successful than others at surviving, colonizing, and spreading in this geographical region, which is consistent with what has been reported in Europe63. The other two remaining spa types identified in the current study, namely t14339 and t3689, have been previously reported in isolates from Ireland and Denmark, respectively. Herein, the detection of these two types may be due to the accquision of their respective clones during international travel, or may be a function of the study location in a clinic within a private university, where Arab and non-Arab students from different nationalities are enrolled, some of which have received their secondary education in European countries. Interestingly, the spa types from environmental surface isolates were far more clonally conserved than the spa types from personnel (4 and 8 different spa types, respectively; Table 5). We have also observed differences in the antimicrobial resistance profiles of the strains recovered from the two specimen categories. Taken together, these observations may reflect the different reservoirs of strains to which the two categories are exposed.
Contrary to other studies with similar sample sizes, MRSA isolates in this study showed a limited genetic diversity, with CC80-MRSA-IV-t267 (17.7% of all tested MRSA isolates) and CC22-MRSA-IV-t223 (14.7% of all tested MRSA isolates) being the most predominant clones. While this limited genetic diversity may be attributed to the monocentric nature of the study, the predominance of CC80 and CC22 among the identified clonal complexes is consistent with what has been described previously in studies from Egypt and other neighbouring countries50, 52, 64, 65. Additionally, five (14.7%) of the current MRSA isolates (Table 2, IDs: 5, 11, 13, 111, and 112) were identified as tst-positive CC22-IV isolates, which are expected to belong to the ‘Middle Eastern variant’ of EMRSA-15. Isolates with similar characteristics have been also reported in studies from Egypt, Italy, the Gaza Strip, Saudi Arabia, Jordan, Kuwait, and the United Arab Emirates50, 61, 62, 66–69. Interestingly, the strains reported in Italy and Gaza appeared to be endemic in the tested health care units66, and both showed the spa type t223, as with the majority of our isolates, suggesting the dissemination of this clone into hospital settings in the Arab and Mediterranean regions. It is noteworthy that the Italian strain exhibited an antimicrobial profile that was different from most of our isolates, because the former was a non-multiresistant MRSA strain. A similar diversity has been shown in a recent study, in which six variants of CC22-MRSA-IV have been detected in the Gulf region70. Therefore, the two tst-negative CC22-MRSA-IV isolates recovered in the current study (Table 2, IDs: 65, and 103) might be either tst deletion mutants of the ‘Middle Eastern variant’ strain or derived from imported European UK-EMRSA-15/Barnim epidemic strain. The latter possibility is supported by the spa type (t3689) possessed by of one of the two isolates (Table 2, ID: 65), since it is a common type in Denmark, but not in the Arab region (Table 4). Further investigations are necessary to track the origin of the seven CC22-MRSA-IV isolates described herein, as well as to assess whether they represent a true HA-MRSA clone, such as the classic EMRSA-15, or, alternatively, a CA-MRSA clone that might have spread into the tested dental clinic via the DHCPs or the patients. Future studies should determine risk factors, geographical abundance, transmission patterns, population dynamics, and clinical implications for CC22 strains harbouring the tst gene, given the high rate of endemicity characterizing these strains50, 62, 66, 71.
An interesting aspect of the current study is the genotypic characterization of the MRSA isolates, which were examined for the presence or absence of five genes (mecA, mecC, vanA, tst, and PVL) with antimicrobial or virulence-related functions. Given the correlation between antimicrobial resistance and antibiotic consumption72, 73, and given the widespread use of non-prescription antimicrobial agents in Egypt74, it may not be surprising that mecA-positive MRSA strains were recovered from hand and nasal specimens of the outpatients examined in the current study. Additionally, the mecA-positive MRSA isolates included strains recovered from DHCPs and environmental surfaces. Both specimen categories are exposed to high antibiotic pressure, caused by being in daily contact with patients receiving antibiotics, with ample opportunity to acquire antibiotic-resistant bacteria, and thus antibiotic resistance genes43. Contrary to mecA, the mecC gene was absent from all isolates. This finding supports previous findings, in which mecC-positive MRSA strains have been reported almost exclusively in Europe75–77.
When the prevalence of toxin-encoding genes was investigated, five of the tested MRSA isolates were found positive for the PVL gene, four of which (80%) were recovered from the hand swabs of DHCPs (including two nurses and two dentists, Table 2). Similarly, seventeen of the tested MRSA isolates were positive for the tst gene, five of which (29.4%) were recovered from both nasal and hand swabs of DHCPs (including four nurses and one dentist, Table 2). The hand and nasal colonization of DHCPs with MRSA isolates expressing these toxins is of public health interest, since DHCPs can serve as sources of transmission of these isolates in the community, especially among patients. The clinical implication of this becomes partciulary apparent when considering the carriage of more than one toxin-encoding gene, since three of the investigated isolates carried the gene for PVL in combination with the gene for tst. One of these three also harboured the vanA gene (Table 2, IDs: 11, 111, and 112). While the carriage of both PVL and tst genes has been reported in a limited number of studies, the majority of these studies50, 51, 64, as well as the current one, have been conducted on isolates recovered from the Arab region. This is an alarming observation that needs to be prioritized in the formulation of national and regional health care policies, especially considering the large population exchange between these countries.
The prevalence rate of PVL-positive isolates in the present study (14.7%) is comparable to two other studies that have indicated prevalence rates of 15% and 19% among hospital-isolated MRSA strains from Egypt78, 79. Similar to previous studies that have shown an association between PVL-producing genes and specific MLST CCs80–82, the majority (80%, 4/5) of the PVL-positive isolates in this study were associated with CC22.
In the case of the tst gene, the MLST lineage that showed the strongest association with tst-positive strains was also CC22 (70.6%, 12/17) and, to a lesser extent, CC80 (23.5%, 4/17). This fits well with previous studies that have reported the occurrence of tst-positive CC22 MRSA strains in Jordan51, 62, Kuwait61, Saudi Arabia68, and the United Arab Emirates69, as well as tst-positive strains belonging to CC80 in Jordan51, 64.
Some MRSA strains are able to produce biofilm on both mucosal and inanimate surfaces, making them difficult to eradicate83. Therefore, one of the aims of the present study was to evaluate the biofilm-forming ability of the tested isolates. The results showed that among the eight MRSA isolates recovered from nasal swabs, one (12.5%) isolate was classified as strong biofilm producer, three (37.5%) were moderate, one (12.5%) was weak, and three (37.5%) were non-biofilm producers. This is consistent with the general idea that a dispersed mode of growth is favoured over a biofilm mode during S. aureus nasal colonization84. Interestingly, three of the five isolates that are likely to be related to the ‘Middle Eastern variant’ of EMRSA-15 were strong biofilm producers (Table 2, IDs: 5, 11, and 13). Collectively, the detection of multiple virulence and antimicrobial resistance genes suggests the pathogenic potential of the MRSA isolates in the current study, especially when combined with their ability to form biofilms, and thus their potential to resist disinfectants or sanitizers.
This is the first study to provide an overview of MRSA clones currently circulating among patients, DHCPs, and environmental surfaces in dental clinics in Egypt. The main findings of this study include: (i) the limited genetic diversity of MRSA isolates within dental clinics in Egypt (ii) the detection of five tst-positive and two tst-negative CC22-IV isolates that are likely to be linked to the epidemic EMRSA-15 clone; (iii) the combined occurrence of tst and PVL in three of the isolates; (iv) the high level of multidrug resistance in the isolates; (v) the predominance of SCCmec type IV-harbouring MRSA isolates in the population; (vi) the blurring of traditional distinctions between CA-MRSA and HA-MRSA based on SCCmec type and antibiotic resistance in the community, which may suggest the infiltration of CA-MRSA into the hospitals in the area; and (vii) the detection of isolates with spa types (t14339 and t3689) that have never been reported before in any Arab country. In conclusion, the results suggest that personnel and dental clinic surfaces may serve as sources for transmission of MRSA. They can also act as important reservoirs for antibiotic resistance genes. Results reinforce the need for continuous national and regional MRSA surveillance programmes in order to keep track of the emerging clones. Strict antibiotic policy and infection control measures should be implemented to reduce the incidence of MRSA infection in dental clinics and other health care settings.
Materials and Methods
Study design and sample collection
In the present cross-sectional, monocentric study, a total of 1300 samples were collected from six different dental wards of a university outpatient dental clinic in Egypt, between January and May 2013. Samples collected were obtained from: (i) the hands and anterior nares of patients and DHCPs; and (ii) environmental surfaces within the clinic. The six dental wards included the endodontics, operative dentistry, periodontics, prosthetic dentistry, prosthodontics, and dental surgery wards19.
All samples collected in this study (whether from personnel or environmental surfaces) were obtained during the working hours of the clinic. Participants were chosen randomly to differentiate CA-infections from HA-infections. A written informed consent was obtained from each subject. The study protocol was approved from the Ethics Committee of the Faculty of Pharmacy, October University for Modern Sciences and Arts (MSA). All methods were performed in accordance with the relevant guidelines and regulations. Hand swabs were collected during working days, immediately after removing the gloves (if applicable) and before washing. For sampling, the palms and periungual areas were vigorously rubbed with sterile saline-moistened cotton swabs. Paired nasal swabs were collected from each participant according to a previously described method85.
The environmental surfaces investigated in this study were categorized into two groups: (i) the clinical-contact surfaces (that is, surfaces that are touched frequently during dental procedures), which included: dental light arms, dentists’ chairs, dentists’ drills, dentists’ tool racks, patients’ sink faucets, and X-ray switches; and (ii) the housekeeping surfaces (surfaces outside of the patient care area), which included: dentist/nurse hand-washing sinks, disinfectant containers, door knobs, floors away from the dentists’ chairs, light switches, and nurses’ desks.
One set of environmental surface samples from each of the investigated wards included all the above-mentioned areas from both clinical contact and housekeeping surfaces. These samples were collected at the end of the clinic hours (before general cleaning for the next day) and following the CDC guidelines for environmental infection control86. Briefly, each sample was collected by applying a sterile water-moistened swab firmly over an approximate area of 5 × 20 cm of the specific object. In wards with two to twelve chairs, two chairs were randomly chosen, and samples were taken from both chairs, while in wards with two dental chairs, samples were collected from both chairs.
Identification of S. aureus and screening for methicillin resistance
Swabs (collected from hands, anterior nares, or environmental surfaces) were inoculated into 2 ml of double strength brain heart infusion broth (BHI; Difco, USA), and incubated at 37 °C for 18–24 h. A volume of 100 μl was withdrawn from cultures showing growth, plated onto mannitol salt agar (Difco, USA), and the plates were incubated aerobically at 37 °C for 24 h. The yellow-colored colonies on mannitol salt agar were collected for further identification using standard microbiological methods. These methods included colony morphology on blood agar, Gram stain, in addition to catalase and coagulase tests. Strains with ambiguous biochemical results were analyzed by 16 S rRNA gene sequencing as described elsewhere87.
All S. aureus isolates were screened for methicillin resistance using oxacillin (1 µg) and cefoxitin (30 µg) disc diffusion tests. Briefly, bacterial cultures were adjusted to the 0.5 McFarland turbidity standard, which is equivalent to 1.5 × 108 CFU/ml, and inoculated (using a sterile cotton swab) on the surface of a Mueller Hinton Agar (MHA) (Oxoid, UK) in the case of cefoxitin, or MHA supplemented with 2% sodium chloride in the case of oxacillin. Zone diameters were measured and interpreted according to the guidelines of the Clinical Laboratory Standard Institute (CLSI)88. All MRSA isolates were stored at −20 °C in BHI containing 15% glycerol for further characterization.
Molecular typing methods
SCCmec typing
A multiplex PCR with five primer-pairs (Table 8) was used as previously described89 to discriminate between SCCmec types I, II, III, IV, and V. The SCCmec type was determined on the basis of the band pattern obtained. Isolates with no visible bands, or with a band pattern that was not in agreement with one of the five predicted band patterns, were classified as non-typeable.
Table 8.
PCR primers used in this study.
| Target gene (Primer specificity) | Primer sequence (5’ → 3’ direction)* | Amplicon size (bp) | Annealing temperature (°C) | Reference |
|---|---|---|---|---|
| mecA | Forward: GAAGATGGCTATCGTGTCACA Reverse: GGAACTTGTTGAGCAGAGGTT | 307 | 52 | Current study |
| mecC | Forward: GGGTTCAGCCAGATTCATTTGT Reverse: GTACTGTTGCTTCGTTCAATGG | 138 | 52 | Current study |
| spa | 1113_ Forward: TAAAGACGATCCTTCGGTGAGC 1514_ Reverse: CAGCAGTAGTGCCGTTTGCTT | Variable (180–670) | 52 | 91 |
| PVL | Forward: GCTGGACAAAACTTCTTGGAATAT Reverse: GATAGGACACCAATAAATTCTGGATTG | 83 | 50 | 94 |
| VanA | Forward: GGGAAAACGACAATTGC Reverse: GTACAATGCGGCCGTTA | 732 | 50 | 95 |
| SCCmec I | CIF2 F2: TTCGAGTTGCTGATGAAGAAGG CIF2 R2: ATTTACCACAAGGACTACCAGC | 495 | 47 | 96 |
| SCCmec II | KDP F1: AATCATCTGCCATTGGTGATGC KDP R1: CGAATGAAGTGAAAGAAAGTGG | 284 | 47 | 96 |
| SCCmec III | RIF5 F10: TTCTTAAGTACACGCTGAATCG RIF5 R13: GTCACAGTAATTCCATCAATGC | 414 | 47 | 96 |
| SCCmec I, II, IV | DCS F2: CATCCTATGATAGCTTGGTC DCS R1: CTAAATCATAGCCATGACCG | 342 | 47 | 96 |
| SCCmec V | ccrC F2: GTACTCGTTACAATGTTTGG ccrC R2: ATAATGGCTTCATGCTTACC | 449 | 47 | 89 |
| tst | Forward: ACCCCTGTTCCCTTATCATC Reverse: TTTTCAGTATTTGTAACGCC | 326 | 47 | 97 |
*F: forward primer; R: reverse primer.
spa typing and Based Upon Repeat Patterns (BURP) analysis
Using the primers listed in Table 8, all the investigated MRSA isolates (n = 34) were subjected to a PCR assay for amplification of the polymorphic repeat region (X region) of the spa gene as described elsewhere90. The spa amplicons were then purified using a Qiagen DNA purification kit (Qiagen GmbH, Hilden, Germany) and sequenced by Macrogen® (Seoul, South Korea) using capillary electrophoresis. spa typing was conducted as described by Harmsen et al.91, and the resulting spa types were then clustered into related spa clonal complexes (spa-CCs) using the BURP algorithm implemented in the Ridom StaphType software version 2.2.1 (Ridom GmbH, Würzburg, Germany). The default parameters of the BURP algorithm (exclusion of spa types shorter than 5 repeats and clustering of spa types if cost is less or equal to 5) were applied in this analysis, in order to prevent the formation of spa clusters that are too large or non-specific92. The spa type that could not be assigned to a cluster was considered as a singleton. Due to the high concordance between spa typing and MLST24, the MLST clonal complexes (CC) corresponding to the respective spa-CCs were deduced from the data on the Ridom SpaServer (http://spaserver.ridom.de, last accessed on 15 November 2016) and by literature search.
Antimicrobial susceptibility testing
Excluding vancomycin and linezolid, the antimicrobial susceptibility of the isolates to a panel of commonly used antibiotics was determined using the Kirby Bauer disc diffusion method on MHA plates according to CLSI guidelines and breakpoints88. The antimicrobial discs used, which were all obtained from Oxoid (UK), included: cefaclor (CE; 30 µg), cefoxitin (FOX; 30 μg), ceftriaxone (CRO; 30 μg), chloramphenicol (C; 30 μg), ciprofloxacin (CIP; 5 μg), clindamycin (CD; 2 µg), doxycycline (DO; 30 µg), erythromycin (E; 15 μg), gentamicin (CN; 10 μg), imipenem (IMP; 10 µg), neomycin (NE; 30 µg), and oxacillin (OX; 1 μg). The susceptibilities of the isolates to vancomycin and linezolid (both from Sigma Aldrich, Germany) were determined using the agar dilution method following the CLSI guidelines and interpretative criteria88. Throughout the study, the antimicrobial susceptibility tests were quality controlled using S. aureus ATCC 43300 (methicillin-resistant strain) and S. aureus ATCC 29213 (methicillin-sensitive strain).
Detection of mecA, mecC, vanA, tst, and PVL-encoding genes
Presumptive MRSA isolates were confirmed by polymerase chain reaction (PCR) using primers targeting the mecA and the mecC genes. Additionally, all the MRSA isolates were subjected to a PCR assay for detecting the lukF/S-PV genes encoding the PVL toxin and the tst gene encoding the TSST-1 toxin, while only those found to be phenotypically resistant or intermediately resistant to vancomycin were tested for the presence of vanA gene. The primers used in these assays (Table 8) were synthesized by Eurofins MWG Operon (Ebersberg, Germany). Genomic DNA extraction and purification was done using the GeneJET® Genomic DNA purification kit (Thermo Scientific, USA) according to the instructions of the manufacturer. Each PCR amplification cycle consisted of an initial denaturation step at 95 °C for 10 minutes, followed by denaturation at 95 °C for 30 s, annealing at 47–52 °C (depending on primers used, Table 8) for 30 s, and extension at 72 °C for 1 min for each kb of DNA amplified. This cycle was repeated 35 times followed by a final extension step at 72 °C for 10 minutes. The final volume of the reaction mixture for each PCR assay was 25 µl, and amplifications were performed using the Biometra TAdvanced thermal cycler (Biometra, Göttingen, Germany). All PCR-based assays employed known positive and negative controls. After amplification, 10 µl of each PCR reaction was separated on a 2% (w/v) agarose gel, stained with ethidium bromide (0.5 mg/ml), and visualized under a Gel Doc EZ Imager (Bio-Rad Laboratories, USA).
Biofilm formation assay
The ability of the MRSA isolates to form biofilm onto polystyrene microtiter plates was evaluated as described previously with slight modifications93. Briefly, overnight bacterial cultures in trypticase soy broth (TSB, Difco, USA) were diluted in the same medium to match the 0.5 McFarland turbidity standard, followed by further dilution (1:100) in TSB supplemented with 2% (w/v) glucose and 2% (w/v) NaCl. A volume of 200 µl of this diluted bacterial suspension was cultured in triplicates in microtiter wells (96 wells; Nunc, Denmark), while negative control wells contained uninoculated medium. The plates were incubated at 37 °C for 24 h. Following incubation, the plates were washed carefully three times with 200 μl of tryptone water (Difco, USA) to remove nonadherent planktonic cells, and the plates were subsequently dried at room temperature. The established biofilm was stained with 100 μl/well of 0.1% membrane filtered crystal violet solution (Sigma Aldrich, Germany) at room temperature for 2 min. The wells of the microtiter plates were then washed twice with sterile pyrogen-free water, and finally a mixture of ethanol:acetone (4:1, v/v) was used to elute bound crystal violet. The eluted crystal violet was diluted 1:10 with the same mixture of solvents, and the optical density was determined spectrophotometrically at λ = 545 nm using microplate ELISA reader (Stat Fax®2100). The isolates were classified as biofilm non-producers, weak, moderate, and strong biofilm producers based on previously published criteria34.
Statistical analysis
Categorical variables were compared using the Chi-square test (χ2) or Fisher’s exact two-tailed test, as appropriate, with P values of < 0.05 as the level of significance. These statistical analyses were carried out using the GraphPad Prism (version 6; GraphPad Software Inc.; USA).
Acknowledgements
We would like thank Assistant lecturer Mai Abd El Wahed (Microbiology department-MSA University) for assisting in specimen collection. We gratefully acknowledge Dr. Edet E Udo, and Dr. Samar S. Boswihi, at the Department of Microbiology, Faculty of Medicine, Kuwait University, Kuwait, for their help in clustering the spa types by BURP analysis.
Author Contributions
Dr. Ahmed, S. Khairalla, Dr. Reham Wasfi, and Dr. Hossam, M. Ashour contributed to the design of the study, performance of experiments, analysis of the results, and writing of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stefani S, Goglio A. Methicillin-resistant Staphylococcus aureus: related infections and antibiotic resistance. Int J Infect Dis. 2010;14(Suppl 4):S19–22. doi: 10.1016/j.ijid.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. The Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 3.Liu GY. Molecular pathogenesis of Staphylococcus aureus infection. Pediatr Res. 2009;65:71R–77R. doi: 10.1203/PDR.0b013e31819dc44d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyllie D, Paul J, Crook D. Waves of trouble: MRSA strain dynamics and assessment of the impact of infection control. J Antimicrob Chemother. 2011;66:2685–2688. doi: 10.1093/jac/dkr392. [DOI] [PubMed] [Google Scholar]
- 5.Aucken HM, Ganner M, Murchan S, Cookson BD, Johnson AP. A new UK strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-17) resistant to multiple antibiotics. J Antimicrob Chemother. 2002;50:171–175. doi: 10.1093/jac/dkf117. [DOI] [PubMed] [Google Scholar]
- 6.Amorim ML, et al. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J Clin Microbiol. 2007;45:2881–2888. doi: 10.1128/JCM.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/AAC.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel, R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res, 41–47 (2005). [DOI] [PubMed]
- 9.Garcia-Alvarez L, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wielders C, Fluit A, Brisse S, Verhoef J, Schmitz F. mecA gene is widely disseminated in Staphylococcus aureus population. Journal of clinical microbiology. 2002;40:3970–3975. doi: 10.1128/JCM.40.11.3970-3975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Zorn B, Courvalin P. vanA-mediated high level glycopeptide resistance in MRSA. The Lancet infectious diseases. 2003;3:67–68. doi: 10.1016/S1473-3099(03)00510-3. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T, et al. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shore AC, Coleman DC. Staphylococcal cassette chromosome mec: recent advances and new insights. Int J Med Microbiol. 2013;303:350–359. doi: 10.1016/j.ijmm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Wu Z, Li F, Liu D, Xue H, Zhao X. Novel Type XII Staphylococcal Cassette Chromosome mec Harboring a New Cassette Chromosome Recombinase, CcrC2. Antimicrob Agents Chemother. 2015;59:7597–7601. doi: 10.1128/AAC.01692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalyani K, Jayakumar K, Sunilkumar J. Prevalence of methicillin-resistant Staphylococcus aureus among health care workers of Shri Satya Sai Medical College and Hospital-a tertiary care centre. J Dent Med Sci. 2012;3:23–27. [Google Scholar]
- 18.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts MC, Soge OO, Horst JA, Ly KA, Milgrom P. Methicillin-resistant Staphylococcus aureus from dental school clinic surfaces and students. Am J Infect Control. 2011;39:628–632. doi: 10.1016/j.ajic.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Dukic VM, Lauderdale DS, Wilder J, Daum RS, David MZ. Epidemics of community-associated methicillin-resistant Staphylococcus aureus in the United States: a meta-analysis. PLoS One. 2013;8:e52722. doi: 10.1371/journal.pone.0052722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wertheim HF, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto T, et al. Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance. J Infect Chemother. 2010;16:225–254. doi: 10.1007/s10156-010-0045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li V, et al. Cost-effectiveness and efficacy of spa, SCCmec, and PVL genotyping of methicillin-resistant Staphylococcus aureus as compared to pulsed-field gel Electrophoresis. PLoS One. 2013;8:e79149. doi: 10.1371/journal.pone.0079149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strommenger B, et al. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol. 2006;44:2533–2540. doi: 10.1128/JCM.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araujo MW, Andreana S. Risk and prevention of transmission of infectious diseases in dentistry. Quintessence Int. 2002;33:376–382. [PubMed] [Google Scholar]
- 26.Mehta, S., Mehta, A. & Lodha, S. Occupational hazards in dentistry. Guident7 (2014).
- 27.Miller RL, Micik RE, Abel C, Ryge G. Studies on dental aerobiology. II. Microbial splatter discharged from the oral cavity of dental patients. J Dent Res. 1971;50:621–625. doi: 10.1177/00220345710500031701. [DOI] [PubMed] [Google Scholar]
- 28.Kurita, H., Kurashina, K. & Honda, T. Nosocomial transmission of methicillin-resistant Staphylococcus aureus via the surfaces of the dental operatory. Br Dent J201, 297–300 discussion 291, doi:10.1038/sj.bdj.4813974 (2006). [DOI] [PubMed]
- 29.Martin MV, Hardy P. Two cases of oral infection by methicillin-resistant Staphylococcus aureus. Br Dent J. 1991;170:63–64. doi: 10.1038/sj.bdj.4807419. [DOI] [PubMed] [Google Scholar]
- 30.Dancer SJ. Importance of the environment in meticillin-resistant Staphylococcus aureus acquisition: the case for hospital cleaning. Lancet Infect Dis. 2008;8:101–113. doi: 10.1016/S1473-3099(07)70241-4. [DOI] [PubMed] [Google Scholar]
- 31.French GL, et al. Tackling contamination of the hospital environment by methicillin-resistant Staphylococcus aureus (MRSA): a comparison between conventional terminal cleaning and hydrogen peroxide vapour decontamination. J Hosp Infect. 2004;57:31–37. doi: 10.1016/j.jhin.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths R, Fernandez R, Halcomb E. Reservoirs of MRSA in the acute hospital setting: a systematic review. Contemp Nurse. 2002;13:38–49. doi: 10.5172/conu.13.1.38. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Alcalde, J. et al. Gloves, gowns and masks for reducing the transmission of meticillin-resistant Staphylococcus aureus (MRSA) in the hospital setting. Cochrane Database Syst Rev. CD007087, doi:10.1002/14651858.CD007087.pub2 (2015). [DOI] [PMC free article] [PubMed]
- 34.Stepanović S, Ćirković I, Ranin L. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Letters in Applied Microbiology. 2004;38:428–432. doi: 10.1111/j.1472-765X.2004.01513.x. [DOI] [PubMed] [Google Scholar]
- 35.Anand KB, Agrawal P, Kumar S, Kapila K. Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol. 2009;27:27–29. [PubMed] [Google Scholar]
- 36.Felten A, Grandry B, Lagrange PH, Casin I. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): a disk diffusion method with cefoxitin and moxalactam, the Vitek 2 system, and the MRSA-screen latex agglutination test. J Clin Microbiol. 2002;40:2766–2771. doi: 10.1128/JCM.40.8.2766-2771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skov R, et al. Evaluation of a cefoxitin 30 microg disc on Iso-Sensitest agar for detection of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2003;52:204–207. doi: 10.1093/jac/dkg325. [DOI] [PubMed] [Google Scholar]
- 38.Albrich WC, Harbarth S. Health-care workers: source, vector, or victim of MRSA? Lancet Infect Dis. 2008;8:289–301. doi: 10.1016/S1473-3099(08)70097-5. [DOI] [PubMed] [Google Scholar]
- 39.Sobhy N, Aly F, Abd El Kader O, Ghazal A, Elbaradei A. Community-acquired methicillin-resistant Staphylococcus aureus from skin and soft tissue infections (in a sample of Egyptian population): analysis of mec gene and staphylococcal cassette chromosome. Braz J Infect Dis. 2012;16:426–431. doi: 10.1016/j.bjid.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Amorim ML, et al. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization among patients and healthcare workers in a Portuguese hospital: a pre-intervention study toward the control of MRSA. Microbial Drug Resistance. 2009;15:19–26. doi: 10.1089/mdr.2009.0881. [DOI] [PubMed] [Google Scholar]
- 41.Bisaga A, Paquette K, Sabatini L, Lovell EO. A prevalence study of methicillin-resistant Staphylococcus aureus colonization in emergency department health care workers. Ann Emerg Med. 2008;52:525–528. doi: 10.1016/j.annemergmed.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 42.Davis KA, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clinical Infectious Diseases. 2004;39:776–782. doi: 10.1086/422997. [DOI] [PubMed] [Google Scholar]
- 43.Klingenberg C, Glad GT, Olsvik R, Flaegstad T. Rapid PCR detection of the methicillin resistance gene, mecA, on the hands of medical and non-medical personnel and healthy children and on surfaces in a neonatal intensive care unit. Scand J Infect Dis. 2001;33:494–497. doi: 10.1080/00365540110026485. [DOI] [PubMed] [Google Scholar]
- 44.Kurashige EJ, Oie S, Furukawa H. Contamination of environmental surfaces by methicillin-resistant Staphylococcus aureus (MRSA) in rooms of inpatients with MRSA-positive body sites. Braz J Microbiol. 2016;47:703–705. doi: 10.1016/j.bjm.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuen JW, Chung TW, Loke AY. Methicillin-resistant Staphylococcus aureus (MRSA) contamination in bedside surfaces of a hospital ward and the potential effectiveness of enhanced disinfection with an antimicrobial polymer surfactant. Int J Environ Res Public Health. 2015;12:3026–3041. doi: 10.3390/ijerph120303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campo J, et al. Oral complication risks after invasive and non-invasive dental procedures in HIV-positive patients. Oral Dis. 2007;13:110–116. doi: 10.1111/j.1601-0825.2006.01262.x. [DOI] [PubMed] [Google Scholar]
- 47.Ibrahim OM, Saber-Ayad M. Antibiotic misuse in different hospital wards (a pilot study in an Egyptian hospital) Asian J Pharm Clin Res. 2012;5:95–97. [Google Scholar]
- 48.Defres S, Marwick C, Nathwani D. MRSA as a cause of lung infection including airway infection, community-acquired pneumonia and hospital-acquired pneumonia. Eur Respir J. 2009;34:1470–1476. doi: 10.1183/09031936.00122309. [DOI] [PubMed] [Google Scholar]
- 49.Hunter PA, et al. Antimicrobial-resistant pathogens in animals and man: prescribing, practices and policies. J Antimicrob Chemother. 2010;65(Suppl 1):i3–17. doi: 10.1093/jac/dkp433. [DOI] [PubMed] [Google Scholar]
- 50.Al Laham N, et al. MRSA clonal complex 22 strains harboring toxic shock syndrome toxin (TSST-1) are endemic in the primary hospital in Gaza, Palestine. PLoS One. 2015;10:e0120008. doi: 10.1371/journal.pone.0120008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Bakri AG, et al. The epidemiology and molecular characterization of methicillin-resistant staphylococci sampled from a healthy Jordanian population. Epidemiol Infect. 2013;141:2384–2391. doi: 10.1017/S0950268813000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biber A, et al. A typical hospital-acquired methicillin-resistant Staphylococcus aureus clone is widespread in the community in the Gaza strip. PLoS One. 2012;7:e42864. doi: 10.1371/journal.pone.0042864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Djoudi F, et al. Panton-Valentine leukocidin positive sequence type 80 methicillin-resistant Staphylococcus aureus carrying a staphylococcal cassette chromosome mec type IVc is dominant in neonates and children in an Algiers hospital. New Microbiol. 2013;36:49–55. [PubMed] [Google Scholar]
- 54.Egea AL, et al. New patterns of methicillin-resistant Staphylococcus aureus (MRSA) clones, community-associated MRSA genotypes behave like healthcare-associated MRSA genotypes within hospitals, Argentina. Int J Med Microbiol. 2014;304:1086–1099. doi: 10.1016/j.ijmm.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Espadinha D, et al. Extensive dissemination of methicillin-resistant Staphylococcus aureus (MRSA) between the hospital and the community in a country with a high prevalence of nosocomial MRSA. PLoS One. 2013;8:e59960. doi: 10.1371/journal.pone.0059960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saiman L, et al. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin Infect Dis. 2003;37:1313–1319. doi: 10.1086/379022. [DOI] [PubMed] [Google Scholar]
- 57.Sax H, et al. Control of a cluster of community-associated, methicillin-resistant Staphylococcus aureus in neonatology. J Hosp Infect. 2006;63:93–100. doi: 10.1016/j.jhin.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 58.Skov R, Jensen K. Community-associated meticillin-resistant Staphylococcus aureus as a cause of hospital-acquired infections. Journal of Hospital Infection. 2009;73:364–370. doi: 10.1016/j.jhin.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Valsesia G, Rossi M, Bertschy S, Pfyffer GE. Emergence of SCCmec type IV and SCCmec type V methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leukocidin genes in a large academic teaching hospital in central Switzerland: external invaders or persisting circulators? J Clin Microbiol. 2010;48:720–727. doi: 10.1128/JCM.01890-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boswihi SS, Udo EE, Al-Sweih N. Shifts in the Clonal Distribution of Methicillin-Resistant Staphylococcus aureus in Kuwait Hospitals: 1992-2010. PLoS One. 2016;11:e0162744. doi: 10.1371/journal.pone.0162744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Udo EE, Boswihi SS, Al-Sweih N. High prevalence of toxic shock syndrome toxin-producing epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) strains in Kuwait hospitals. New Microbes New Infect. 2016;12:24–30. doi: 10.1016/j.nmni.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aqel AA, Alzoubi HM, Vickers A, Pichon B, Kearns AM. Molecular epidemiology of nasal isolates of methicillin-resistant Staphylococcus aureus from Jordan. J Infect Public Health. 2015;8:90–97. doi: 10.1016/j.jiph.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 63.Grundmann H, et al. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harastani HH, Tokajian ST. Community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV (CC80-MRSA-IV) isolated from the Middle East: a heterogeneous expanding clonal lineage. PLoS One. 2014;9:e103715. doi: 10.1371/journal.pone.0103715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hefzy, E. M. & Hassan, G. M. Rapid Molecular Identification of Hospital-acquired Methicillin Resistant Staphylococcus aureus (HA-MRSA) Lineages. The Egyptian Journal of Medical Microbiology (EJMM)25 (2016).
- 66.Geraci DM, et al. Methicillin-resistant Staphylococcus aureus colonization: a three-year prospective study in a neonatal intensive care unit in Italy. PLoS One. 2014;9:e87760. doi: 10.1371/journal.pone.0087760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monecke S, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monecke S, et al. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC microbiology. 2012;12:146. doi: 10.1186/1471-2180-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber, S. et al. Genetic fingerprinting of MRSA from Abu Dhabi. ECCMID: Vienna (2010).
- 70.Senok A, et al. Diversity of methicillin-resistant Staphylococcus aureus CC22-MRSA-IV from Saudi Arabia and the Gulf region. International Journal of Infectious Diseases. 2016;51:31–35. doi: 10.1016/j.ijid.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 71.Geraci DM, et al. tst1-positive ST22-MRSA-IVa in healthy Italian preschool children. Infection. 2014;42:535–538. doi: 10.1007/s15010-013-0583-z. [DOI] [PubMed] [Google Scholar]
- 72.Borg MA, et al. Antibiotic consumption as a driver for resistance in Staphylococcus aureus and Escherichia coli within a developing region. American journal of infection control. 2010;38:212–216. doi: 10.1016/j.ajic.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 73.Monsen T, Ronnmark M, Olofsson C, Wistrom J. Antibiotic susceptibility of staphylococci isolated in blood cultures in relation to antibiotic consumption in hospital wards. Scand J Infect Dis. 1999;31:399–404. doi: 10.1080/00365549950163860. [DOI] [PubMed] [Google Scholar]
- 74.Al-Tawfiq JA, Stephens G, Memish ZA. Inappropriate antimicrobial use and potential solutions: a Middle Eastern perspective. Expert review of anti-infective therapy. 2010;8:765–774. doi: 10.1586/eri.10.56. [DOI] [PubMed] [Google Scholar]
- 75.Deplano A, Vandendriessche S, Nonhoff C, Denis O. Genetic diversity among methicillin-resistant Staphylococcus aureus isolates carrying the mecC gene in Belgium. J Antimicrob Chemother. 2014;69:1457–1460. doi: 10.1093/jac/dku020. [DOI] [PubMed] [Google Scholar]
- 76.Paterson GK, Harrison EM, Holmes MA. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22:42–47. doi: 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paterson GK, et al. Prevalence and characterization of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J Antimicrob Chemother. 2014;69:907–910. doi: 10.1093/jac/dkt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Enany S, Yaoita E, Yoshida Y, Enany M, Yamamoto T. Molecular characterization of Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus isolates in Egypt. Microbiol Res. 2010;165:152–162. doi: 10.1016/j.micres.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 79.Shady HMA, Bakr AEA, Hashad ME, Alzohairy MA. Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Brazilian Journal of Infectious Diseases. 2015;19:68–76. doi: 10.1016/j.bjid.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aires-de-Sousa M, Conceicao T, de Lencastre H. Unusually high prevalence of nosocomial Panton-Valentine leukocidin-positive Staphylococcus aureus isolates in Cape Verde Islands. J Clin Microbiol. 2006;44:3790–3793. doi: 10.1128/JCM.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rijnders MI, et al. Population structure of Staphylococcus aureus strains isolated from intensive care unit patients in the netherlands over an 11-year period (1996 to 2006) J Clin Microbiol. 2009;47:4090–4095. doi: 10.1128/JCM.00820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raggi, C. et al. Methicillin resistance, biofilm formation and resistance to benzalkonium chloride in Staphylococcus aureus clinical isolates. Clinical Microbiology: Open Access (2013).
- 84.Krismer B, Peschel A. Does Staphylococcus aureus nasal colonization involve biofilm formation? Future microbiology. 2011;6:489–493. doi: 10.2217/fmb.11.37. [DOI] [PubMed] [Google Scholar]
- 85.Shibabaw A, Abebe T, Mihret A. Nasal carriage rate of methicillin resistant Staphylococcus aureus among Dessie Referral Hospital health care workers; Dessie, Northeast Ethiopia. Antimicrob Resist Infect Control. 2013;2:25. doi: 10.1186/2047-2994-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sehulster L, Chinn RY. Cdc & Hicpac. Guidelines for environmental infection control in health-care facilities. Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC) MMWR Recomm Rep. 2003;52:1–42. [PubMed] [Google Scholar]
- 87.Becker K, et al. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J Clin Microbiol. 2004;42:4988–4995. doi: 10.1128/JCM.42.11.4988-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.CLSI. (Clinical and Laboratory Standards Institute, PA, USA, 2014).
- 89.Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frenay HM, et al. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur J Clin Microbiol Infect Dis. 1996;15:60–64. doi: 10.1007/BF01586186. [DOI] [PubMed] [Google Scholar]
- 91.Harmsen D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mellmann A, et al. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC microbiology. 2007;7:98. doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elkhatib WF, Khairalla AS, Ashour HM. Evaluation of different microtiter plate-based methods for the quantitative assessment of Staphylococcus aureus biofilms. Future Microbiol. 2014;9:725–735. doi: 10.2217/fmb.14.33. [DOI] [PubMed] [Google Scholar]
- 94.Stegger M, et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251) Clin Microbiol Infect. 2012;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 95.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:1434. doi: 10.1128/jcm.33.5.1434-1434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38:1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cuny C, et al. State-wide surveillance of antibiotic resistance patterns and spa types of methicillin-resistant Staphylococcus aureus from blood cultures in North Rhine-Westphalia, 2011–2013. Clinical Microbiology and Infection. 2015;21:750–757. doi: 10.1016/j.cmi.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 99.Zarfel G, et al. Increase of genetic diversity and clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in South-East Austria. FEMS Microbiology Letters. 2016;363:fnw137. doi: 10.1093/femsle/fnw137. [DOI] [PMC free article] [PubMed] [Google Scholar]


