To effectively combat invading pathogens, immune cells must rapidly switch from roughly spherical resting cells to polarized migratory ones, which then move in a directed fashion to the site of infection. The dramatic metamorphosis of leukocytes into polarized cells and their subsequent migration are two of the most fascinating phenomena in cell biology. Polarization and migration require the spatial and temporal control of signal transduction molecules so that substrate attachment and membrane extension occur at the cell front, while detachment and membrane retraction happen at the rear. How do cells coordinate signaling molecules to perform contrasting functions at opposite poles? It has long been appreciated that there is polarization in the protein machinery involved in cell migration. However, it is becoming evident that lipids are also distributed nonuniformly, and the distribution of lipids is an important factor for directional migration. In a paper in this issue of PNAS, Gómez-Moutón et al. (1) provide further evidence for the importance of specialized lipid domains in establishing and maintaining the polarity of motile cells. In particular, they show that both the leading edge and the uropod of polarized T lymphocytes are enriched in lipid components that partition into raft-like lipid domains. An interesting finding is that the distribution of certain lipid raft components differs at the two poles. Thus, ganglioside GM3 is enriched at the leading edge, whereas GM1 is concentrated at the uropod. Treatments such as cholesterol depletion, which disrupts plasma membrane organization, prevent the development of a polarized morphology and cell migration.
Although it is now understood that lipids are distributed nonrandomly in the plasma membrane and that this has important consequences for cell signaling and other functions (2–4), the precise nature of these lipid inhomogeneities (microdomains) remains somewhat enigmatic—partly because the lipid microdomains are apparently in a size range (10–300 nm) that is below the resolution of optical microscopy. In the current view of the plasma membrane, certain lipids and proteins assemble into dynamic, sub-μm-sized lateral organizations that function to facilitate signal transduction events (2, 4, 5). Regions of the plasma membrane that are enriched in sphingolipids and cholesterol are thought to exist in a liquid-ordered phase (6, 7) that confers detergent resistance to these structures (8) and allows for their ready isolation by flotation on sucrose density gradients (9). One model (3) is that signaling molecules are recruited to these small “rafts” from a largely liquid-disordered membrane.
Gómez-Moutón et al. provide further evidence for the importance of specialized lipid domains in establishing and maintaining the polarity of motile cells.
Because the plasma membrane contains up to 50 mole % cholesterol and also a very high amount of sphingomyelin in the outer leaflet, it might be expected that a high fraction of the lipids in the plasma membrane are resistant to extraction by cold nonionic detergents (10, 11). In fact, when cells are treated with cold Triton X-100 and the residual membranes are imaged by fluorescence microscopy and electron microscopy (11), most of the area of the cell remains covered by detergent-resistant membrane. In addition to cholesterol and sphingomyelin, glycosphingolipids and glycosylphosphatidylinositol (GPI)-anchored proteins are in the detergent-resistant lipid pools. In contrast to the lipids, most transmembrane proteins that are not linked to the cytoskeleton are solubilized by cold nonionic detergent treatment, but a small subset of proteins are insoluble. The fact that only a small fraction of membrane proteins are detergent resistant may give rise to the mistaken impression that rafts are a small fraction of the plasma membrane. A second oversimplification is that there are just two types of membrane domains: rafts and non-rafts. The report by Gómez-Moutón et al. (1) on rafts in polarized T cells adds to mounting evidence dispelling both of these misconceptions.
It was shown recently that rafts could be subdivided based on their susceptibility to solubilization in nonionic detergents (12); a subset of raft proteins in an epithelial cell line was found to be resistant to solubilization by the detergent Lubrol, but susceptible to solubilization by TritonX-100. Additionally, in epithelial cells, approximately half of CD44-containing lipid rafts floated to the low-density fraction of sucrose gradients, whereas the other half of CD44-containing lipid rafts only floated after disruption of the actin cytoskeleton (13). These findings imply the existence of two kinds of CD44-containing lipid rafts, which are distinguished by their association to the actin cytoskeleton or not. Now, Gómez-Moutón et al. show that the leading edge and the uropod of T cells contain raft domains with different compositions. Images of polarized T cells, fluorescently labeled for markers of each type of raft domain, show that GM3-enriched rafts localize to the cell front (lamella), whereas GM1-enriched rafts localize to the cell rear (uropod; ref. 1). Together, these raft domains constitute a significant portion of the total cell surface, in line with previous reports demonstrating that a large fraction (≈40–70%) of the plasma membrane is in detergent-resistant (i.e., liquid-ordered) structures (10, 11). Clearly, a binary model of the plasma membrane, in which rafts comprise a very minor fraction of the total cell surface, cannot accommodate these findings. Rather, the plasma membrane may more closely resemble a dense assembly of rafts of different types. Various signals may recruit certain types of rafts into larger assemblies (flotillas; Fig. 1). These larger assemblies are easily seen by optical microscopy.
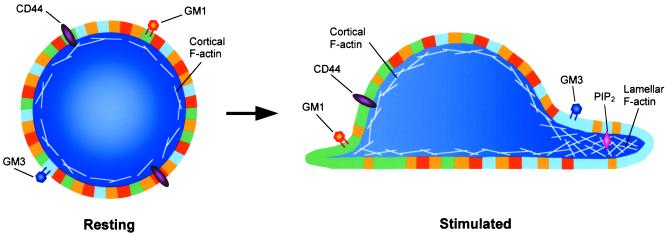
Figure 1.
The plasma membranes of cells are composed of many types of submicroscopic disordered (yellow regions) and more ordered (all other regions) membrane domains, which are depicted here at much larger scale relative to the cells than their putative size. Ordered domains are resistant to solubilization by nonionic detergents and comprise a large fraction of the cell surface. In resting leukocytes (Left), all types of membrane domains, which are below the resolution of light microscopy, are evenly distributed around the cell periphery. Following stimulation (Right), two types of ordered membrane domains (or rafts) segregate to either pole of the cell, forming large assemblies (or flotillas), which can be easily visualized by light microscopy. For ease of illustration, these flotillas are shown as uniform patches of membrane. However, in actuality they are more likely to be composed of ordered domains intercalated with disordered ones. In T cells, the flotilla at the front of the cell (blue region) is marked by the ganglioside GM3, whereas the flotilla at the rear (green region) contains GM1. Fore and aft flotillas may also have other compositional differences in transmembrane proteins (e.g., CD44) and/or lipids (e.g., PIP2), which impart unique functions to each end of the cell.
Gómez-Moutón et al. (1) demonstrate that the association of proteins with distinct membrane domains dictates their redistribution to the appropriate ends of polarizing T cells and that the segregation of the two types of domains to opposite cell poles requires an intact actin cytoskeleton. These data suggest a model in which actin-driven redistribution of ordered domains causes the redistribution of both transmembrane and lipid-linked ordered domain-associated proteins. Such a model is supported by several reports that describe an intimate connection between ordered domains and the underlying actin cytoskeleton (13–16). Although the exact molecular basis for an ordered domain/F-actin interaction has not yet been determined, several candidate molecules can be identified. Most relevant for the Gómez-Moutón et al. study is the transmembrane receptor CD44, which is found predominantly at the uropod of polarized leukocytes and is associated with ordered domains in several cell types, including T cells (1, 13, 14, 17). CD44 may provide a bridge between ordered domains and the actin cytoskeleton (13, 14) by acting through moesin, a member of the ezrin–radixin–moesin family of actin-binding proteins (18). CD44 that is concomitantly linked to the actin cytoskeleton and associated with ordered domains may be actively redistributed to the cell uropod, dragging some lipid-linked domain components (e.g., GPI-linked proteins) passively along while excluding other lipids and proteins. The feasibility of such a mechanism for organizing plasma membrane components has been verified by co-patching experiments in which aggregation of one ordered domain component caused the clustering and co-redistribution of other ordered domain components (19, 20). It is unclear how the two types of rafts would be directed to opposite poles of a T lymphocyte in such a model. Perhaps some proteins preferentially associate with rafts of a certain composition, and in this way movement of transmembrane proteins could move different raft components in opposite directions.
Although there is good evidence that the actin cytoskeleton can control domain organization (1, 14, 15), there is reason to believe that the reverse may also be true. Remodeling of the actin cytoskeleton is a fundamental step in cell polarization that must be tightly regulated in space and time. This is most clearly illustrated by the actin-driven extension of a single lamella exclusively at the cell front. This precise spatial regulation of actin dynamics, coupled with reports that membrane domains are distributed asymmetrically on the surface of polarized cells (1, 14, 21), leads to the possibility that membrane domains may control the organization of the underlying actin network. It is well established that Rho GTPases play a central role in actin remodeling (22). Upon activation, the Rho GTPases are recruited to the plasma membrane by means of C-terminal acylations, which may dictate localization to or exclusion from certain membrane domains. Interestingly, a regulator of the Rho GTPases, Rho GDP dissociation inhibitor (Rho GDI) binds directly to the ordered domain marker, CD44 (23), further implicating ordered domains in controlling actin cytoskeletal rearrangements. Another possibility is that ordered domain-mediated actin reorganization occurs via localized generation of polyphosphatidyl inositol (4, 5) bisphosphate (PIP2), which was found to be enriched in isolated ordered domains and dependent on membrane cholesterol (24). PIP2 can mediate actin polymerization by affecting the function of a variety of actin-associated proteins (reviewed in ref. 25). PIP2 was shown to act downstream of the Rho family member Rac in human neutrophils (26, 27), providing further support for the idea that Rho GTPase targeting is important for spatially regulating actin polymerization. Actin-driven segregation of specialized ordered domains may activate signal transduction events in one region of the cell, leading to spatially restricted actin rearrangement and membrane protrusion in that region, while inhibiting signaling in another region of the cell and preventing lamellar extension there.
The report by Gómez-Moutón et al. takes a step toward a more detailed view of plasma membrane organization that can help to explain complex biological processes. However, it is clear that our understanding of membrane organization is still rudimentary. How many distinct types of membrane domains coexist in a plasma membrane? What is the role of membrane proteins in determining the properties of their surrounding lipids (and vice versa)? What is the significance of the fact that most membrane proteins are detergent soluble, whereas the majority of plasma membrane lipids are detergent resistant? Do most proteins reside in a minor fraction of the plasma membrane? If the plasma membrane is densely populated by very small coexisting domains, the boundary regions must be a large fraction of the total area of the membrane. Do these boundary regions play a role in processes such as recruitment of signaling proteins to raft domains?
In summary, the view of membrane organization depicted by the raft hypothesis has provided a useful starting point for understanding how heterogeneities in the plasma membrane could partake in cell signaling. However, it is necessary to move toward a more detailed and realistic model of membrane organization to understand the mechanisms for regulating biological processes.
Acknowledgments
We thank Dr. Robert J. Eddy for helpful discussions and for his contribution of the artwork shown Fig. 1.
Footnotes
See companion article on page 9642.
References
- 1.Gómez-Moutón C, Abad J L, Mira E, Lacalle R A, Gallardo E, Jimenez-Baranda S, Illa I, Bernad A, Manes S, Martinez-A C. Proc Natl Acad Sci USA. 2001;98:9642–9647. doi: 10.1073/pnas.171160298. . (First Published August 7, 2001; 10.1073/pnas.171160298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown D A, London E. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 3.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 4.Sheets E D, Holowka D, Baird B. Curr Opin Chem Biol. 1999;3:95–99. doi: 10.1016/s1367-5931(99)80017-9. [DOI] [PubMed] [Google Scholar]
- 5.Langlet C, Bernard A-M, Drevot P, He H-T. Curr Opin Immunol. 2000;12:250–255. doi: 10.1016/s0952-7915(00)00084-4. [DOI] [PubMed] [Google Scholar]
- 6.Brown D A, London E. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 7.Ge M, Field K A, Aneja R, Holowka D, Baird B, Freed J H. Biophys J. 1999;77:925–933. doi: 10.1016/S0006-3495(99)76943-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed S N, Brown D A, London E. Biochemistry. 1997;36:10944–10953. doi: 10.1021/bi971167g. [DOI] [PubMed] [Google Scholar]
- 9.Brown D A, Rose J K. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 10.Fridriksson E K, Shipkova P A, Sheets E D, Holowka D, Baird B, McLafferty F W. Biochemistry. 1999;25:8056–8063. doi: 10.1021/bi9828324. [DOI] [PubMed] [Google Scholar]
- 11.Mayor S, Maxfield F R. Mol Biol Cell. 1995;6:929–944. doi: 10.1091/mbc.6.7.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roper K, Corbeil D, Huttner W B. Nat Cell Biol. 2000;2:582–592. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 13.Oliferenko S, Paiha K, Harder T, Gerke T, Schwarzler C, Schwarz H, Beug H, Gunthert U, Huber L A. J Cell Biol. 1999;146:843–854. doi: 10.1083/jcb.146.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foger N, Marhaba R, Zoller M. J Cell Sci. 2000;114:1169–1178. doi: 10.1242/jcs.114.6.1169. [DOI] [PubMed] [Google Scholar]
- 15.Holowka D, Sheets E D, Baird B. J Cell Sci. 2000;113:1009–1019. doi: 10.1242/jcs.113.6.1009. [DOI] [PubMed] [Google Scholar]
- 16.Harder T, Simons K. Eur J Immunol. 1999;29:556–562. doi: 10.1002/(SICI)1521-4141(199902)29:02<556::AID-IMMU556>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Neame S J, Uff C R, Sheikh H, Wheatley S C, Isacke C M. J Cell Sci. 1995;108:3127–3135. doi: 10.1242/jcs.108.9.3127. [DOI] [PubMed] [Google Scholar]
- 18.Bretscher A. Curr Opin Cell Biol. 1999;11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- 19.Harder T, Scheiffele P, Verkade P, Simons K. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas J L, Holowka D, Baird B, Webb W W. J Cell Biol. 1994;125:795–802. doi: 10.1083/jcb.125.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manes S, Mira E, Gomez-Mouton C, Lacalle R A, Keller P, Labrador J P, Martinez-A C. EMBO J. 1999;18:6211–6220. doi: 10.1093/emboj/18.22.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machesky L, Hall A. Trends Cell Biol. 1996;6:304–310. doi: 10.1016/0962-8924(96)10026-x. [DOI] [PubMed] [Google Scholar]
- 23.Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Hirao M, Sato N, et al. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pike L J, Miller J M. J Biol Chem. 1998;273:22298–22304. doi: 10.1074/jbc.273.35.22298. [DOI] [PubMed] [Google Scholar]
- 25.Sechi A, Wehland J. J Cell Sci. 2000;113:3685–3695. doi: 10.1242/jcs.113.21.3685. [DOI] [PubMed] [Google Scholar]
- 26.Zigmond S, Joyce M, Borleis J, Bokoch G, Devreotes P. J Cell Biol. 1997;138:363–374. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glogauer M, Hartwig J, Stossel T. J Cell Biol. 2000;150:785–796. doi: 10.1083/jcb.150.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]