Abstract
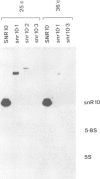
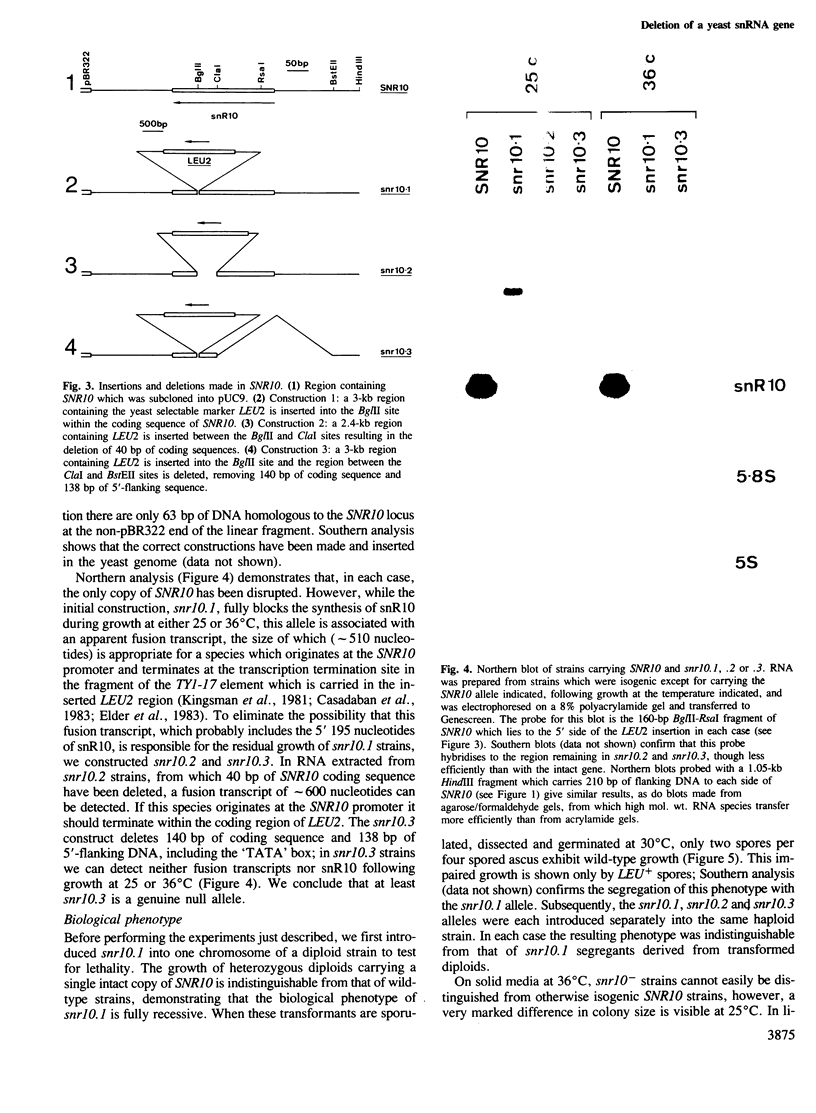
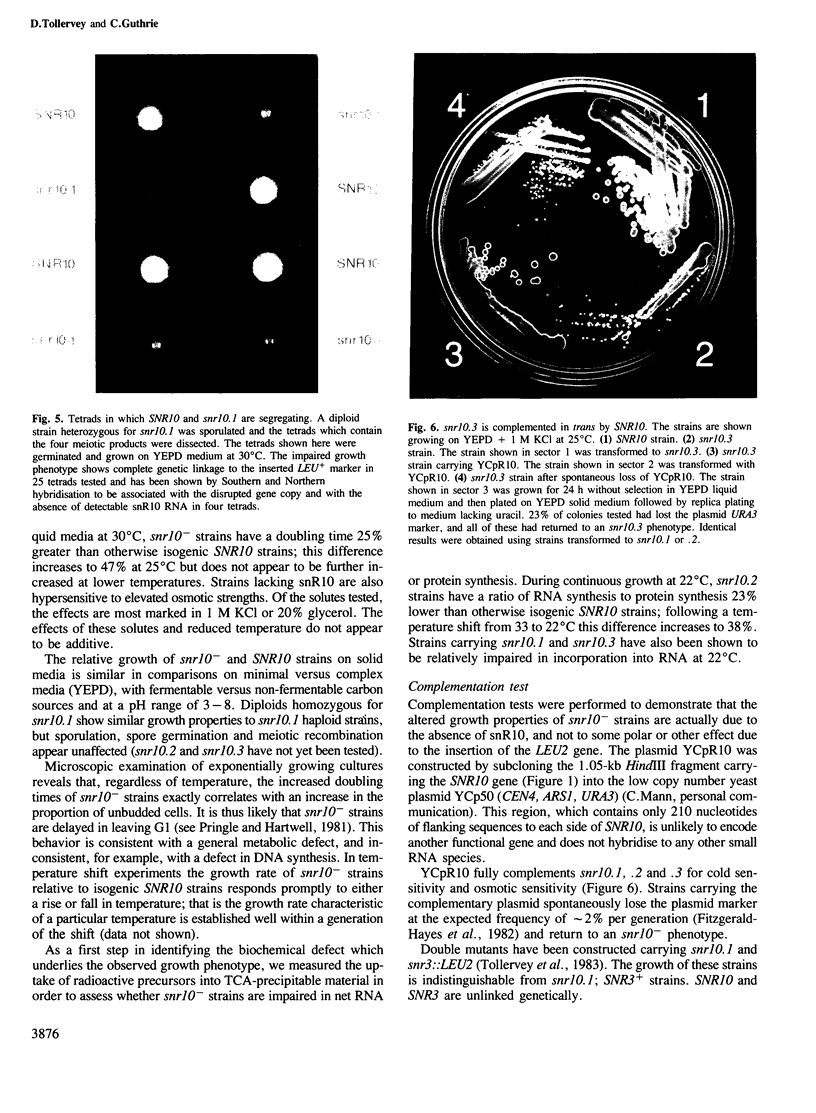
We have cloned and sequenced the single copy gene SNR10 which encodes the yeast small nuclear RNA, snR10. This species does not show obvious primary sequence homology to any previously identified small nuclear RNA. As an inital step towards determining the function of snR10, we have introduced insertions and deletions into the chromosomal copy of the gene. Strains lacking an intact copy of SNR10 are viable but considerably imparied in growth, particularly at elevated osmotic strengths or low temperatures; at 25 degrees C the doubling time of snr10- strains is 47% greater than that of otherwise isogenic SNR10 strains. As judged by the incorporation of radioactive precursors, snr10- strains are impaired in net RNA synthesis at low temperatures. The identification of a leaky, conditional phenotype associated with the deletion of this small nuclear RNA gene was entirely unexpected since the defect in snR10 synthesis is complete and non-conditional.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachellerie J. P., Michot B., Raynal F. Recognition signals for mouse pre-rRNA processing. A potential role for U3 nucleolar RNA. Mol Biol Rep. 1983 May;9(1-2):79–86. doi: 10.1007/BF00777477. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bringmann P., Appel B., Rinke J., Reuter R., Theissen H., Lührmann R. Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and for their association by intermolecular base pairing. EMBO J. 1984 Jun;3(6):1357–1363. doi: 10.1002/j.1460-2075.1984.tb01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Busch H., Reddy R., Rothblum L., Choi Y. C. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Chen M. W., Anné J., Volckaert G., Huysmans E., Vandenberghe A., De Wachter R. The nucleotide sequences of the 5 S rRNAs of seven molds and a yeast and their use in studying ascomycete phylogeny. Nucleic Acids Res. 1984 Jun 25;12(12):4881–4892. doi: 10.1093/nar/12.12.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch R. J., Kanaya S., Earl P. L. A model for the involvement of the small nucleolar RNA (U3) in processing eukaryotic ribosomal RNA. Mol Biol Rep. 1983 May;9(1-2):75–78. doi: 10.1007/BF00777476. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Fradin A., Jove R., Hemenway C., Keiser H. D., Manley J. L., Prives C. Splicing pathways of SV40 mRNAs in X. laevis oocytes differ in their requirements for snRNPs. Cell. 1984 Jul;37(3):927–936. doi: 10.1016/0092-8674(84)90427-6. [DOI] [PubMed] [Google Scholar]
- Futcher A. B., Cox B. S. Copy number and the stability of 2-micron circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol. 1984 Jan;157(1):283–290. doi: 10.1128/jb.157.1.283-290.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Stunnenberg H. G., Birnstiel M. L. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3' histone mRNA termini. Cell. 1983 Oct;34(3):823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- Georgiev O., Birnstiel M. L. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3' termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 1985 Feb;4(2):481–489. doi: 10.1002/j.1460-2075.1985.tb03654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W., Sankoff D., Cedergren R. J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984 Jul 25;12(14):5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Nashimoto H., Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci U S A. 1969 Jun;63(2):384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. U4 and U6 RNAs coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res. 1984 Apr 11;12(7):3283–3293. doi: 10.1093/nar/12.7.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hori H., Lim B. L., Osawa S. Evolution of green plants as deduced from 5S rRNA sequences. Proc Natl Acad Sci U S A. 1985 Feb;82(3):820–823. doi: 10.1073/pnas.82.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen E. G., Hellung-Larsen P., Frederiksen S. Synthesis of low molecular weight RNA components A, C and D by polymerase II in alpha-amanitin-resistant hamster cells. Nucleic Acids Res. 1979 Jan;6(1):321–330. doi: 10.1093/nar/6.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R., Sprague G. F., Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci U S A. 1983 May;80(10):3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda S., Gotoh O., Seno T., Takeishi K. Nucleotide sequence of Dictyostelium small nuclear RNA Dd8 not homologous to any other sequenced small nuclear RNA. J Biol Chem. 1983 Sep 10;258(17):10606–10613. [PubMed] [Google Scholar]
- Kato N., Harada F. Nucleotide sequence of nuclear 5.4 S RNA of mouse cells. Biochim Biophys Acta. 1984 Jun 16;782(2):127–131. doi: 10.1016/0167-4781(84)90015-0. [DOI] [PubMed] [Google Scholar]
- Kingsman A. J., Gimlich R. L., Clarke L., Chinault A. C., Carbon J. Sequence variation in dispersed repetitive sequences in Saccharomyces cerevisiae. J Mol Biol. 1981 Feb 5;145(4):619–632. doi: 10.1016/0022-2836(81)90306-5. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985 Oct;42(3):725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell. 1983 Jun;33(2):509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Murphy J. T., Burgess R. R., Dahlberg J. E., Lund E. Transcription of a gene for human U1 small nuclear RNA. Cell. 1982 May;29(1):265–274. doi: 10.1016/0092-8674(82)90111-8. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Mount S. M., Steitz J. A., Sharp P. A. Splicing of messenger RNA precursors is inhibited by antisera to small nuclear ribonucleoprotein. Cell. 1983 Nov;35(1):101–107. doi: 10.1016/0092-8674(83)90212-x. [DOI] [PubMed] [Google Scholar]
- Prestayko A. W., Tonato M., Busch H. Low molecular weight RNA associated with 28 s nucleolar RNA. J Mol Biol. 1970 Feb 14;47(3):505–515. doi: 10.1016/0022-2836(70)90318-9. [DOI] [PubMed] [Google Scholar]
- Rose J. K., Lodish H. F. Translation in vitro of vesicular stomatitis virus mRNA lacking 5'-terminal 7-methylguanosine. Nature. 1976 Jul 1;262(5563):32–37. doi: 10.1038/262032a0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Galli G., Busslinger M., Birnstiel M. L. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984 Dec 1;3(12):2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Wise J. A., Guthrie C. A U4-like small nuclear RNA is dispensable in yeast. Cell. 1983 Dec;35(3 Pt 2):753–762. doi: 10.1016/0092-8674(83)90108-3. [DOI] [PubMed] [Google Scholar]
- Wise J. A., Tollervey D., Maloney D., Swerdlow H., Dunn E. J., Guthrie C. Yeast contains small nuclear RNAs encoded by single copy genes. Cell. 1983 Dec;35(3 Pt 2):743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- Wooley J. C., Cone R. D., Tartof D., Chung S. Y. Small nuclear ribonucleoprotein complexes of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6762–6766. doi: 10.1073/pnas.79.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]