Abstract
A variety of genomic and proteomic tools have been used to study cancer metabolism and metabolomics in order to understand how cancer cells survive in their environment. Throughout the past decade, mass spectrometry has been routinely used for large-scale protein identification of complex biological mixtures. In this review, we discuss some recent developments in cancer metabolism by proteomic analysis using mass spectrometric techniques, focusing on pyruvate kinase, L-lactate dehydrogenase, Warburg effect, glutamine metabolism and oxidative stress.
Keywords: Cancer metabolism, Warburg effect, oxidative stress, mass spectrometry, review
Living organisms are able to carry out a highly integrated network of chemical reactions, known as metabolism, in order to extract energy and by reducing power from their environment, to synthesize the building blocks of their macromolecules. Living organisms also face a succession of environmental challenges as they grow and develop. In response to the imposed conditions, organisms are equipped with adaptive traits that are maintained and evolved by means of natural selection. By employing recent scientific tools in biochemistry, genomics, and cell physiology, a valid description of the mechanisms of adaptation has been achieved at the molecular and cellular level. For instance, receptor methylation is responsible for adaptation in bacterial chemotaxis, and β2-adrenergic receptor phosphorylation is involved in rapid adaption of cellular exposure to a high concentration of adrenaline (1). In addition to the post-translational modifications (PTMs), gene regulation is another essential mechanism for an organism to increase the versatility and adaptability, by allowing the cell to express proteins when needed, such as the lac operon regulation system in the genome of E. coli, discovered by Jacob and Monod in 1961, in which some enzymes involved in lactose metabolism are expressed in the presence of lactose and absence of glucose (2). Thus, a systematic study of cell metabolism will reveal how cells survive and reproduce in their habitats.
Cancer cells are cells that grow and divide at an unregulated, quickened pace, due to damaged or changed genetic material DNA, and it is well-known that cancer cells have a unique metabolism compared to normal cells. Using slices of living tissues, Otto Warburg studied the energy metabolism of a tumor and first reported that cancer cells produced large amounts of lactate even in aerobic conditions (3). Since then, the Warburg effect has been observed in several tumors in which the cells predominantly produced energy by a high rate of glycolysis followed by lactate fermentation in the cytosol, rather than by a comparatively low rate of glycolysis followed by oxidation of pyruvate in mitochondria like most normal cells, even in the presence of oxygen (4–6). Recently, cancer metabolism has been extensively explored to understand the processes that allow cancer cells to grow and reproduce, maintain cellular structures, and respond to environments (4–12). For this purpose, various methods, such as nuclear magnetic resonance (NMR) spectrometry and gas chromatography-mass spectrometry (GC-MS), have been developed to characterize the altered metabolites and gene regulation in metabolic pathways in cancer cells, facilitating the discovery and development of groundbreaking therapies (13–16). Notably, many drugs have been developed, targeting the specific needs of cancer cells (4, 17, 18). Today, liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) is routinely used for large-scale protein identifications and global profiling of PTMs in complex biological mixtures (19–22). Here, we attempt to discuss the cancer metabolism based on authors’ experience regarding proteomic analysis of pancreatic cancer cells by LC-MS/MS.
PKM2
Pyruvate kinase (PK) catalyzes the last step within the glycolytic sequence, the de-phosphorylation of phosphoenolpyruvate to pyruvate, and is responsible for net energy production within the glycolytic pathway. There are 4 isozymes of PK in mammals: L, R, M1 and M2. L type is major isozyme in liver and kidney; R is found in red cells; M1 is the main form in muscle and brain. The human PK isozyme M2 (PKM2), a splice variant of M1, is expressed in lung tissues as well as in all cells with high rates of nucleic acid synthesis, including all proliferating cells, such as embryonic cells, adult stem cells and especially tumor cells (23, 24). Notably, elevated levels of PKM2 have been observed in numerous cancerous cells, and PKM2 has been investigated as a potential tumor marker for diagnostic assays (25, 26). Recently, acknowledging exclusive expression of PKM2 in tumor tissues, Christofk reported that an exchange in the expression of PKM1 to PKM2 is causative for the aerobic glycolysis (Warburg effect) during tumorigenesis (27). Consequently, they suggested that selective targeting of PKM2 by small-molecule inhibitors is feasible for cancer therapy (28). Interestingly, several mechanisms have been proposed for the PKM2 activity regulation in tumor cells: (1) the ratio of inactive dimeric form PKM2 to the active tetrameric form regulates the proportions of glucose carbons that are channeled to synthetic processes or used for glycolytic energy production (23, 24); (2) PKM2 activity is regulated by phosphotyrosine signaling (29); (3) Anastasiou et al. demonstrated that PKM2 activity is also regulated by oxidative stress. PKM2 is specifically oxidized by hydrogen peroxide (H2O2) on cysteine 358 in cancer cells, which diminishes PKM2 activity, decreases pyruvate formation and increases flux of glycolytic metabolites into the pentose phosphate pathway (30); (4) Hitosugi showed that PKM2 activity in cancer can be tuned by phosphorylation (31).
In our proteomic analysis of pancreatic cancer cells by mass spectrometry, we found that PKM2 is indeed up-regulated in cancer cells (32, 33), consistent with previous report (23–25). The MS result also shows that PKM2 is actually an abundant protein in both normal pancreatic duct cells and cancer cells, which is discrepant from some researchers’ observation that PKM2 is replaced by tissue-specific isoforms during tissue differentiation in development and PKM1 is switched to PKM2 during tumorigenesis (23–25, 27). Recently, using a quantitative MS technique, Bluemlein demonstrated that PKM2 is the prominent isoform in several analyzed cancer samples and matched control tissues, which also challenges the conclusion that PKM2 is exclusively expressed in cancer cells and there is a switch of PKM1 to PKM2 during cancer development (34). Therefore, a great deal of caution should be exercised in developing the drug target to PKM2 since PKM2 is abundantly expressed in normal cells.
LDH-A
L-lactate dehydrogenase (LDH) converts pyruvate, the final product of glycolysis to lactate when oxygen is absent or in short supply, and it performs the reverse reaction during the Cori cycle in the liver (35). The enzymes are homo- or hetero-tetramers composed of M (LDH-A) and H (LDH-B) protein subunits encoded by the LDHA and LDHB genes respectively: LDH-1 (4H) in the heart and red blood cells, LDH-2 (3H1M) in the reticuloendothelial system, LDH-3 (2H2M) in the lungs, LDH-4 (1H3M) in the kidneys, placenta and pancreas, and LDH-5 (4M) in the liver and striated muscle (36). LDH plays a key role in cancer cells’ aerobic glycolysis since it is the alternative supplier of NAD+ in the absence of mitochondrial oxidation, and Fan et al. showed that tyrosine phosphorylation of LDH-A is important for NADH/NAD+ redox homeostasis in cancer cells (37). Separately, it has been reported that the lactate, produced by LDH, causes chronic acidification of the intratumoral microenvironment, which in turn helps to drive cancer cell metastasis (38–40). Moreover, overexpression of LDH-A has been implicated in the pathogenesis and progression of many cancer types (41–45). When LDH-A is inhibited by small-molecule inhibitor or is knocked-down using RNA interference, cancer cell proliferation is severely impaired (46, 47). Consequently, some potential therapeutic drugs have been developed to target LDH-A in order to interfere with tumor growth and invasiveness (17, 48–50).
Apparently, previous research on LDH is mainly focused on LDH-A. Surprisingly, our proteomic analysis of pancreatic cancer cells by MS reveals a different story of LDH. We demonstrated that the expression level of LDH-A in pancreatic cancer cells is not significantly changed compared to normal duct cells, but LDH-B is strikingly increased in the cancer cells (32, 33). Thus, the MS result offers proof-of-concept for targeting LDH-B as a therapeutic strategy in cancer, particularly pancreatic cancer. Notably, Hussien et al. recently confirmed that breast cancer cell line MCF-7 expresses mainly LDH-B (51), and Zha et al. reported that LDH-B is an important target of mTORC1 (52).
LDH is subjected to PTMs such as phosphorylation and acetylation (53, 54), and tyrosine phosphorylation of LDH-A is important for redox homeostasis in cancer cells (37). Recently, we characterized the PTMs of LDH from pancreatic cancer cells and identified multiple O-methylated residues from both LDH-A and LDH-B, providing important biochemical information toward further understanding of LDH modifications (56). Since PTMs of proteins via methylation plays an important role in a number of central processes in the cell (57), it will be interesting to further investigate how the methylation of LDH can affect its catalytic activity, localization in the cell, protein stability and the ability to form tetramers or complexes with other molecules.
Glutamine Metabolism
Glucose and glutamine are the two molecules catabolized in substantial quantities in both normal and cancer cells because they supply most of the carbon, nitrogen, free energy, and reduce equivalent necessary for cell growth and division (35). Notably, many cultured cancer cells exhibit increased glutamine consumption (58), and glioblastoma cells in culture convert as much as 90% of glucose and 60% of glutamine into lactate or alanine by 13C-NMR spectroscopy measurements. The glutamate derived from glutamine is converted to α-ketoglutarate via the activity of glutamate dehydrogenase, and is further catabolized into lactate with NADPH production via malic enzyme, or it is used to produce alanine via the activity of alanine aminotransferase (59). Separately, it has been reported that glutaminase, which converts glutamine to glutamate in the first step of glutaminolysis, is up-regulated in several cancer cells (60). Moreover, Metallo et al. demonstrated that cells grown under hypoxia rely almost exclusively on the reductive carboxylation of glutamine-derived α-ketoglutarate for de novo lipogenesis (61). Thus, glutamine’s relevance in tumor cell metabolism has drawn much attention in cancer research since it plays important roles in nucleotide biosynthesis, hexosamine biosynthesis and glycosylation reactions, amino acids synthesis, glutathione production, Krebs cycle, and generation of reducing equivalent such as NADPH (62, 63). It has been shown that glutamine metabolism is regulated by c-Myc, and some compounds have been designed to impair the glutamine addiction of cancer cells (58).
Interestingly, cytidine triphosphate (CTP) synthase is extremely over-expressed in pancreatic cancer cells, as has been shown by LC-MS/MS analysis (33). The enzyme interconverts uridine triphosphate (UTP) and CTP, and catalyzes the last committed step in pyrimidine nucleotide biosynthesis with the consumption of ATP (35). The generated ADP from this chemical reaction provides further substrate for aerobic glycolysis. Up-regulated CTP synthase activity has been widely seen in human and rodent tumors, and the glutamine analog DON has been used as an anticancer agent by acting as an irreversible inhibitor (64, 65). Separately, glutamyl-prolyl tRNA synthase (GPRS), a bi-functional aminoacyl-tRNA synthetase which catalyzes the aminoacylation of glutamate and proline tRNA species (66), is strikingly up-regulated in pancreatic cancer cells (33). In contrast, glutamate dehydrogenase 1 (GDH1) and isocitrate dehydrogenase-1 (IDH1), the two enzymes involved in reductive carboxylation of glutamine-derived α-ketoglutarate for de novo lipogenesis (61), are identified in pancreatic cancer cells without apparent up-regulation. Furthermore, the abundance of the enzyme alanine aminotransferase (or named glutamic-pyruvate transaminase), which catalyzes the production of alanine from glutamate, is relatively low in cells and no significant change can be detected by the MS analysis. Thus, the MS results demonstrate that the glutamine metabolism is indeed altered in pancreatic cancer cells, but the glutaminolysis may be different from previous reports (59, 61). It is likely that the high rate of glutamine uptake in pancreatic cancer cells results from its role as a nitrogen donor in nucleotide and amino acid biosynthesis to meet the need of fast growing cancer cells (33).
Warburg Effect
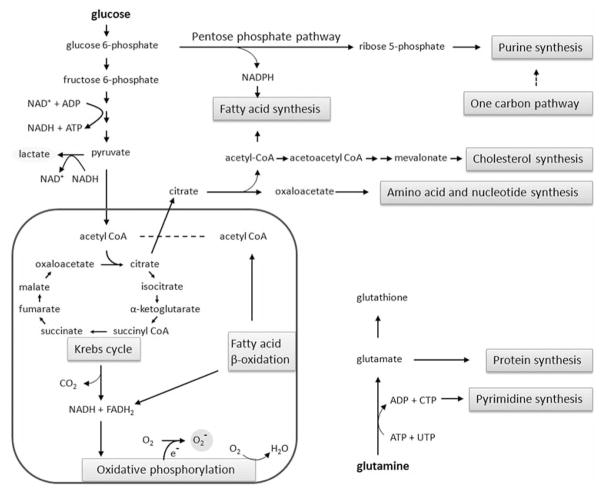
In mitochondrion, Krebs cycle (also known as the citric acid cycle, the tricarboxylic acid cycle) and oxidative phosphorylation are essential metabolic pathways that convert carbohydrates, fats, and proteins to carbon dioxide (CO2) and water (H2O), to generate adenosine triphosphate (ATP) for intracellular energy transfer with the consumption of oxygen (O2) (35). Our proteomic analysis showed that several enzymes in Krebs cycle and the components of oxidative phosphorylation are anomalous and down-regulated in pancreatic cancer cells (32, 33). Interestingly, carbonic anhydrase II, which catalyzes the rapid interconversion of CO2 and H2O to bicarbonate and proton, is diminished in pancreatic cancer cells, further indicating that Krebs cycle function is down-regulated and less oxygen is consumed in cancer cells. Notably, protein levels of several enzymes involved in fatty acid β-oxidation in mitochondrion are also reduced, whereas the ATP citrate lyase and the fatty acid synthase (FAS); the enzymes involved in fatty acid synthesis, are rigorously escalated in pancreatic cancer cells (33), consistent with a previous report (67). The MS analysis also identified that NADPH, which is needed for fatty acid synthesis, is likely acquired from the up-regulated pentose phosphate pathway in pancreatic cancer cells (33). Hence, these proteomic data demonstrated that the prominent function of mitochondrion is severely altered and there is a shift in energy production from aerobic respiration to anaerobic lactate fermentation in pancreatic cancer cells, consistent with the observed Warburg effect.
The Warburg effect has been observed in many cancer cells, particularly in solid tumors. It is known that mutations in oncogenes and tumor suppressor genes are responsible for malignant transformation (68, 69), and it is also documented that in the rapidly growing solid tumor, oxygen level is low due to poor vascularization (4–6). Proteomic data suggest that the Warburg effect is a consequence of an adaptation to low-oxygen environments within tumors, and the tumor-derived cell lines can maintain their metabolic phenotype in culture under regular condition with ample oxygen, due to irreversible gene mutations in the cells. Notably, K-Ras has been shown to play a critical role in pancreatic cancer initiation and maintenance (70, 71).
In addition to the observed Warburg effect, proteomic analysis by MS revealed many metabolic proteins that are up-regulated involved in cholesterol synthesis, amino acid synthesis, purine and pyrimidine synthesis, nucleotide-sugar synthesis, mevalonate pathway and one carbon metabolism, and down-regulated involving in glycogenesis, gluconeo-genesis, creatine biosynthesis, heme biosynthesis and energy homeostasis. Particularly, adenylate kinase 1 and 3, which are important for ATP/ADP ratio regulation, and nicotinamide phosphoribosyltransferase, which is involved in NAD+ biosynthesis, are down-regulated in pancreatic cancer cells, indicating that the energy homeostasis is altered in cancer cells (32, 33). Notably, serine and glycine biosynthetic pathways recently have been reported to be up-regulated in several cancers (72, 73).
Oxidative Stress
Reactive oxygen species (ROS) are broadly defined as oxygen-containing, reactive chemical species (74–76). An increase of ROS levels in cells is due either to an elevation of ROS production or to a decline of ROS-scavenging capacity (76). It has been reported that ROS generation is increased in cancer cells compared to normal cells (76–79). An elevation of ROS production in cancer cells is associated with metastasis (80), promoting the metabolic changes required for proliferation by PKM2 modification (30), and serving as signaling molecules to activate the transcription of genes involved in cellular hypoxic adaptation (81). Consequently, the concept that cancer cells have increased levels of ROS, even under hypoxia condition, has been accepted by many researchers, and it is believed that a moderate increase in ROS can promote cancer cell growth (63, 76, 82, 83). Notably, it is also recognized that excessive amounts of ROS can cause oxidative damage to lipids, proteins and DNA (63, 76, 84). If the increase of ROS reaches a certain threshold level that is incompatible with cellular survival, ROS may exert a cytotoxic effect, leading to the death of malignant cells (76, 85). Therefore, it has been proposed that cancer cells have developed several mechanisms to adapt to ROS stress, involving multiple pathways to activate redox-sensitive transcription factors, such as NF-κB, Nrf2, c-Jun and HIF-1, which lead to the increased expression of antioxidant molecules such as SOD, catalase, thioredoxin and the GSH antioxidant system (76). There is also a report suggesting that survival of detached mammary epithelial cells depends on NADPH and the neutralization of cellular ROS (86). Taken together, the current prevailing theory about ROS in cancer highlights that cancer cells are able to delicately maintain a higher level of ROS; however, this theory is apparently paradoxical: on the one hand, it recognizes that elevation of ROS is a result of decline of ROS-scavenging capacity; on the other hand, it supports that elevation of ROS triggers increased expression of antioxidant molecules through adaptation mechanism.
A major source of ROS is the mitochondria (74–76). The NADH and FADH2 formed in glycolysis, fatty acid oxidation and the Krebs cycle are energy-rich molecules because each contains a pair of electrons having a high transfer potential (NADH → NAD+ + H+ + 2 e−, and FADH2 → FAD + 2 H+ + 2 e−). When these electrons are donated to molecular oxygen by a series of electron carriers (½ O2 + 2 H+ + 2 e− → H2O), through a process known as oxidative phosphorylation, a large amount of free energy is liberated, which can be used to generate ATP. This is the major source of ATP in aerobic organisms. Electron leakage from the mitochondrial respiratory chain may react with molecular oxygen, resulting in the formation of superoxide anion (O2 + e− → O2−), which can subsequently be converted to other ROS. Basically, the formation of superoxide anion needs both electron and oxygen. In anaerobic lactate fermentation, the NADH generated from glycolysis will be largely used for reduction of pyruvate (pyruvate + NADH + H+ → lactate + NAD+). Since enzymes in Krebs cycle and β-oxidation are down-regulated in pancreatic cancer cells (32, 33), likely due to cell’s adaptation to hypoxia, it is expected that less NADH and FADH2 will be generated in mitochondria, and less superoxide anion can be produced due to the availability of electron and oxygen in hypoxic environment. On the other hand, the levels of ROS-scavenging enzymes such as superoxide dismutase (SOD), glutathione peroxidase and peroxiredoxin have been shown to be significantly altered in malignant cells and in primary cancer tissues (87–92), and we also confirmed that several antioxidant proteins are coincidentally down-regulated in pancreatic cancer cells (32, 33). Some researchers argue that ROS levels in cancer cells are therefore elevated due to reduced ROS-scavenging capacity; however, we think that the down-regulated levels of antioxidant proteins is a reflection of cancer cell’s adaptation to the environment, since living organism can use gene regulation mechanisms to increase the versatility and adaptability, by allowing cells to express proteins when needed and allowing cells to lessen proteins when not needed. Our proteomic analysis data fit nicely with the hypothesis that, in rapidly growing solid tumors, the oxygen level is low due to poor vascularization, and hypoxia leads pancreatic cancer cells in reprogramming of metabolic pathways through the pyruvate fermentation, the Krebs cycle, fatty acid β-oxidation and oxidative phosphorylation deviation in order to minimize the oxygen consumption. As a result, the oxidative stress in pancreatic cancer cells is lower than that of normal duct cells, and the tumor cells express less antioxidant proteins, due to cells adaptation to low-oxygen environment. Cancer cells in hypoxia will be more resistant to radiation therapy, since oxygen is needed for radiation therapy to generate free radicals that are able to damage the DNA of cancerous cells. The MS results also suggest that the tumor-derived cell lines are able to maintain their metabolic phenotype in culture under normoxic condition, little superoxide anion is produced in the mitochondria, and consequently, less SOD2 and other antioxidant proteins are expressed (Figure 1).
Figure 1.
The hypothesis that oxidative stress in pancreatic cancer cells is lower than that of normal duct cells. The formation of superoxide needs both oxygen and electron which is leaked from the oxidation of NADH and FADH2 by the respiratory chain in mitochondria. Enzymes in Krebs cycle and β-oxidation are down-regulated in pancreatic cancer cells in order to minimize the oxygen consumption, due to cells’ adaptation to hypoxia. As a result, less pyruvate will flux into mitochondria, and cancer cells adopt anaerobic lactate fermentation and elevated pentose phosphate pathway for biosynthesis of building blocks. Since the NADH generated from glycolysis will be largely consumed for the reduction of pyruvate and since less NADH and FADH2 will be generated by Krebs cycle and β-oxidation, less superoxide anion can be produced due to the availability of electron and oxygen in a hypoxic environment. Consequently, cancer cells express little manganese superoxide dismutase (SOD2) in mitochondria. Due to irreversible gene mutations in the cancer cells, the tumor-derived cell lines can maintain their metabolic phenotype in culture under regular conditions with ample oxygen, exhibiting aerobic glycolysis (Warburg effect).
It has been proposed that targeting cancer cells by ROS-mediated mechanisms can be a therapeutic approach (76). One approach is to increase ROS-scavenging capacity using antioxidants, thereby abrogating ROS signaling and suppressing tumor growth; however, the effectiveness is questionable since the oxidative stress in cancer cells is low and cancer cells have already down-regulated antioxidant proteins due to adaptation. Another approach is to treat cancer cells with pharmacological agents that have pro-oxidant properties, raising oxidative stress over the threshold of toxicity as antioxidant systems become overwhelmed; however, the lack of oxygen may limit the endogenous ROS production in hypoxic conditions.
Lysosomal Enzymes
Lysosomes are cellular organelles that contain acid hydrolase enzymes to break-down waste materials and cellular debris. Lysosomal dysfunction is linked with several diseases, including cancer and neurodegenerative disorders, and lysosomes are involved in autophagy which is essential to support cancer cell growth and metabolism (93, 94). It has been proposed that the increased expression and altered trafficking of lysosomal enzymes in cancer cells participate in tissue invasion (95–97). For instance, the aspartic protease cathepsin D has been found to be over-expressed and secreted at high levels by human epithelial breast cancer cells, and this protein has been used as a marker of poor prognosis in breast cancer (98). Intriguingly, proteomics analysis of lysosomal enzymes in pancreatic cancer cells is discrepant from the observations in several other cancers, since several lysosomal proteases are actually decreased in pancreatic cancer cells, implicating that lysosomes are impaired (32, 33). Certainly, the down-regulation of lysosomal enzymes in cancer cells is a reflection of cancer cells’ adaption to their environment; however, the exact mechanisms still need to be investigated.
Conclusion
All assays have limits in regard to analytical sensitivity, precision, and the dynamic range for quantification. In the past decade, mass spectrometry has been used to investigate tumor-specific changes in the proteomes of human cancers and normal cells, including metabolic alterations (99–105). Instead of focusing on one or several proteins, which is not uncommon in traditional cancer biology research, proteomic analysis by MS provides a comprehensive picture of changes of protein expression and changes of PTMs in the cells (32, 33, 55, 56, 106–108). The vast amount of information obtained from MS and other techniques will certainly facilitate our understanding over cancer cells’ adaptation and survival in local environment, providing a biochemical foundation of therapeutic applications. It is expected that more and more MS experiments will be carried out to understand the metabolism in various cancers.
Acknowledgments
The Authors wish to thank the support from the College of Science in George Mason University.
References
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Garland Science. Taylor & Francis Group; New York: 2007. Molecular Biology of the Cell. [Google Scholar]
- 2.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 3.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 4.Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira LM. Cancer metabolism: The Warburg effect today. Exp Mol Pathol. 2010;89:372–380. doi: 10.1016/j.yexmp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: A tumor’s dilemma? Biochim Biophys Acta. 2010;1807:552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Keijer J, Bekkenkamp-Grovenstein M, Venema D, Dommels YE. Bioactive food components, cancer cell growth limitation and reversal of glycolytic metabolism. Biochim Biophys Acta. 2010;1807:697–706. doi: 10.1016/j.bbabio.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Solaini G, Sgarbi G, Baracca A. Oxidative phosphorylation in cancer cells. Biochim Biophys Acta. 2010;1807:534–542. doi: 10.1016/j.bbabio.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–44. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 14.Griffin JL, Kauppinen RA. Tumour metabolomics in animal models of human cancer. J Proteome Res. 2007;6:498–505. doi: 10.1021/pr060464h. [DOI] [PubMed] [Google Scholar]
- 15.Muñoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weljie AM, Jirik FR. Hypoxia-induced metabolic shifts in cancer cells: moving beyond the Warburg effect. Int J Biochem Cell Biol. 2011;43:981–989. doi: 10.1016/j.biocel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Dang CV, Hamaker M, Sun P, Le A, Gao P. Therapeutic targeting of cancer cell metabolism. J Mol Med (Berl) 2011;89:205–212. doi: 10.1007/s00109-011-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 19.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 20.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 21.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 23.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Seminars in Cancer Biology. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 26.Hardt PD, Mazurek S, Toepler M, Schlierbach P, Bretzel RG, Eigenbrodt E, Kloer HU. Faecal tumour M2 pyruvate kinase: a new, sensitive screening tool for colorectal cancer. Brit J Cancer. 2004;91:980–984. doi: 10.1038/sj.bjc.6602033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 28.Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, Xian J, Cantley LC. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol. 2010;79:1118–1124. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosinebinding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 30.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, Xie J, Gu TL, Polakiewicz RD, Roesel JL, Boggon TJ, Khuri FR, Gilliland DG, Cantley LC, Kaufman J, Chen J. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou W, Capello M, Fredolini C, Piemonti L, Liotta LA, Novelli F, Petricoin EF. Proteomic analysis of pancreatic ductal adenocarcinoma cells reveals metabolic alterations. J Proteome Res. 2011;10:1944–1152. doi: 10.1021/pr101179t. [DOI] [PubMed] [Google Scholar]
- 33.Zhou W, Capello M, Fredolini C, Racanicchi L, Piemonti L, Liotta LA, Novelli F, Petricoin EF. Proteomic analysis reveals Warburg effect and anomalous metabolism of glutamine in pancreatic cancer cells. J Proteome Res. 2012;11:554–563. doi: 10.1021/pr2009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bluemlein K, Grüning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stryer L. Biochemistry. W. H. Freeman and Company; New York: 1995. [Google Scholar]
- 36.Miura S. Lactic dehydrogenase isozymes of the brain. I. Electrophoretic studies on regional distribution and ontogenesis. Folia Psychiatr Neurol Jpn. 1966;20:337–347. doi: 10.1111/j.1440-1819.1966.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 37.Fan J, Hitosugi T, Chung TW, Xie J, Ge Q, Gu TL, Polakiewicz RD, Chen GZ, Boggon TJ, Lonial S, Khuri FR, Kang S, Chen J. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Mol Cell Biol. 2011;31:4938–4950. doi: 10.1128/MCB.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez-Zaguilán R, Seftor EA, Seftor RE, Chu YW, Gillies RJ, Hendrix MJ. Acidic pH enhances the invasive behavior of human melanoma cells. Clin Exp Metastasis. 1996;14:176–186. doi: 10.1007/BF00121214. [DOI] [PubMed] [Google Scholar]
- 39.Brizel DM, Schroeder T, Scher RL, Walenta S, Clough RW, Dewhirst MW, Mueller-Klieser W. Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2001;51:349–353. doi: 10.1016/s0360-3016(01)01630-3. [DOI] [PubMed] [Google Scholar]
- 40.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 41.Goldman RD, Kaplan NO, Hall TC. Lactic dehydrogenase in human neoplastic tissues. Cancer Res. 1964;24:389–99. [PubMed] [Google Scholar]
- 42.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koukourakis MI, Giatromanolaki A, Sivridis E, Bougioukas G, Didilis V, Gatter KC, Harris AL Tumour and Angiogenesis Research Group. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br J Cancer. 2003;89:877–885. doi: 10.1038/sj.bjc.6601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang L, Scolyer RA, Murali R, McCarthy SW, Zhang XD, Thompson JF, Hersey P. Lactate dehydrogenase 5 expression in melanoma increases with disease progression and is associated with expression of Bcl-XL and Mcl-1, but not Bcl-2 proteins. Mod Pathol. 2010;23:45–53. doi: 10.1038/modpathol.2009.129. [DOI] [PubMed] [Google Scholar]
- 45.Kayser G, Kassem A, Sienel W, Schulte-Uentrop L, Mattern D, Aumann K, Stickeler E, Werner M, Passlick B, zur Hausen A. Lactate-dehydrogenase 5 is overexpressed in non-small cell lung cancer and correlates with the expression of the transketolase-like protein 1. Diagn Pathol. 2010;5:22. doi: 10.1186/1746-1596-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 48.Xie H, Valera VA, Merino MJ, Amato AM, Signoretti S, Linehan WM, Sukhatme VP, Seth P. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol Cancer Ther. 2009;8:626–635. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y, Deck JA, Hunsaker LA, Deck LM, Royer RE, Goldberg E, Vander Jagt DL. Selective active site inhibitors of human lactate dehydrogenases A4, B4, and C4. Biochem Pharmacol. 2001;62:81–89. doi: 10.1016/s0006-2952(01)00636-0. [DOI] [PubMed] [Google Scholar]
- 50.Granchi C, Bertini S, Macchia M, Minutolo F. Inhibitors of lactate dehydrogenase isoforms and their therapeutic potentials. Curr Med Chem. 2010;17:672–697. doi: 10.2174/092986710790416263. [DOI] [PubMed] [Google Scholar]
- 51.Hussien R, Brooks GA. Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines. Physiol Genomics. 2011;43:255–264. doi: 10.1152/physiolgenomics.00177.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zha X, Wang F, Wang Y, He S, Jing Y, Wu X, Zhang H. Lactate dehydrogenase B is critical for hyperactive mTOR-mediated tumorigenesis. Cancer Res. 2011;71:13–18. doi: 10.1158/0008-5472.CAN-10-1668. [DOI] [PubMed] [Google Scholar]
- 53.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 54.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 55.Zhou W, Capello M, Fredolini C, Piemonti L, Liotta LA, Novelli F, Petricoin EF. Mass spectrometry analysis of the post-translational modifications of α-enolase from pancreatic ductal adenocarcinoma cells. J Proteome Res. 2010;9:2929–2936. doi: 10.1021/pr901109w. [DOI] [PubMed] [Google Scholar]
- 56.Zhou W, Capello M, Fredolini C, Racanicchi L, Piemonti L, Liotta LA, Novelli F, Petricoin EF. MS analysis reveals O-methylation of L-lactate dehydrogenase from pancreatic ductal adenocarcinoma cells. Electrophoresis. 2012;33:1850–1854. doi: 10.1002/elps.201200017. [DOI] [PubMed] [Google Scholar]
- 57.Erce MA, Pang CN, Hart-Smith G, Wilkins MR. The methylproteome and the intracellular methylation network. Proteomics. 2012;12:564–586. doi: 10.1002/pmic.201100397. [DOI] [PubMed] [Google Scholar]
- 58.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erickson JW, Cerione RA. Glutaminase: a hot spot for regulation of cancer cell metabolism? Oncotarget. 2010;1:734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeBerardinis RJ, Cheng T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 64.Kizaki H, Williams JC, Morris HP, Weber G. Increased cytidine 5’-triphosphate synthetase activity in rat and human tumors. Cancer Res. 1980;40:3921–3927. [PubMed] [Google Scholar]
- 65.Ahluwalia GS, Grem JL, Hao Z, Cooney DA. Metabolism and action of amino acid analog anti-cancer agents. Pharmacol Ther. 1990;46:243–271. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- 66.Cerini C, Kerjan P, Astier M, Gratecos D, Mirande M, Semeriva M. A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 1991;10:4267–4277. doi: 10.1002/j.1460-2075.1991.tb05005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 68.Bertram JS. The molecular biology of cancer. Mol Aspects Med. 2000;21:167–223. doi: 10.1016/s0098-2997(00)00007-8. [DOI] [PubMed] [Google Scholar]
- 69.Grandér D. How do mutated oncogenes and tumor suppressor genes cause cancer? Med Oncol. 1998;15:20–26. doi: 10.1007/BF02787340. [DOI] [PubMed] [Google Scholar]
- 70.Motojima K, Urano T, Nagata Y, Shiku H, Tsurifune T, Kanematsu T. Detection of point mutations in the Kirsten-ras oncogene provides evidence for the multicentricity of pancreatic carcinoma. Ann Surg. 1993;217:138–143. doi: 10.1097/00000658-199302000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 75.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 76.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature Rev Drug Discovery. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 77.Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 78.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–798. [PubMed] [Google Scholar]
- 79.Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–1785. doi: 10.1158/0008-5472.CAN-07-5259. [DOI] [PubMed] [Google Scholar]
- 80.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 81.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr Opin Cell Biol. 2009;21:894–899. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamanaka RB, Chandel NS. Warburg effect and redox balance. Science. 2011;334:1219–1220. doi: 10.1126/science.1215637. [DOI] [PubMed] [Google Scholar]
- 83.Afanas’ev I. Reactive oxygen species signaling in cancer: comparison with aging. Aging Dis. 2011;2:219–230. [PMC free article] [PubMed] [Google Scholar]
- 84.Perry G, Raina AK, Nunomura A, Wataya T, Sayre LM, Smith MA. How important is oxidative damage? Lessons from Alzheimer’s disease. Free Radic Biol Med. 2000;28:831–834. doi: 10.1016/s0891-5849(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 85.Fruehauf JP, Meyskens FL., Jr Reactive oxygen species: a breath of life or death? Clin Cancer Res. 2007;13:789–794. doi: 10.1158/1078-0432.CCR-06-2082. [DOI] [PubMed] [Google Scholar]
- 86.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979;39:1141–1149. [PubMed] [Google Scholar]
- 88.Cullen JJ, Weydert C, Hinkhouse MM, Ritchie J, Domann FE, Spitz D, Oberley LW. The role of manganese superoxide dismutase in the growth of pancreatic adenocarcinoma. Cancer Res. 2003;63:1297–1303. [PubMed] [Google Scholar]
- 89.Oberley TD, Oberley LW. Antioxidant enzyme levels in cancer. Histol Histopathol. 1997;12:525–535. [PubMed] [Google Scholar]
- 90.Hu Y, Rosen DG, Zhou Y, Feng L, Yang G, Liu J, Huang P. Mitochondrial manganese-superoxide dismutase expression in ovarian cancer: role in cell proliferation and response to oxidative stress. J Biol Chem. 2005;280:39485–39492. doi: 10.1074/jbc.M503296200. [DOI] [PubMed] [Google Scholar]
- 91.Saydam N, Kirb A, Demir O, Hazan E, Oto O, Saydam O, Güner G. Determination of glutathione, glutathione reductase, glutathione peroxidase and glutathione S-transferase levels in human lung cancer tissues. Cancer Lett. 1997;119:13–19. doi: 10.1016/s0304-3835(97)00245-0. [DOI] [PubMed] [Google Scholar]
- 92.Murawaki Y, Tsuchiya H, Kanbe T, Harada K, Yashima K, Nozaka K, Tanida O, Kohno M, Mukoyama T, Nishimuki E, Kojo H, Matsura T, Takahashi K, Osaki M, Ito H, Yodoi J, Murawaki Y, Shiota G. Aberrant expression of selenoproteins in the progression of colorectal cancer. Cancer Lett. 2008;259:218–230. doi: 10.1016/j.canlet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 93.Hafner Česen M, Pegan K, Spes A, Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp Cell Res. 2012;318:1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 94.Lozy F, Karantza V. Autophagy and cancer cell metabolism. Semin Cell Dev Biol. 2012;23:395–401. doi: 10.1016/j.semcdb.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allison AC. Lysosomes in cancer cells. J Clin Pathol Suppl (R Coll Pathol) 1974;7:43–50. [PMC free article] [PubMed] [Google Scholar]
- 96.Fehrenbacher N, Jäättelä M. Lysosomes as targets for cancer therapy. Cancer Res. 2005;65:2993–2995. doi: 10.1158/0008-5472.CAN-05-0476. [DOI] [PubMed] [Google Scholar]
- 97.Kroemer G, Jäättelä M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 98.Rochefort H. Cathepsin D in breast cancer: a tissue marker associated with metastasis. Eur J Cancer. 1992;28A:1780–1783. doi: 10.1016/0959-8049(92)90003-k. [DOI] [PubMed] [Google Scholar]
- 99.Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby PJ, Banks RE. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620–1632. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- 100.Friedman DB, Hill S, Keller JW, Merchant NB, Levy SE, Coffey RJ, Caprioli RM. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2004;4:793–811. doi: 10.1002/pmic.200300635. [DOI] [PubMed] [Google Scholar]
- 101.Bi X, Lin Q, Foo TW, Joshi S, You T, Shen HM, Ong CN, Cheah PY, Eu KW, Hew CL. Proteomic analysis of colorectal cancer reveals alterations in metabolic pathways: mechanism of tumorigenesis. Mol Cell Proteomics. 2006;5:1119–1130. doi: 10.1074/mcp.M500432-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 103.Choong LY, Lim S, Chong PK, Wong CY, Shah N, Lim YP. Proteome-wide profiling of the MCF10AT breast cancer progression model. PLoS ONE. 2010;5:e11030. doi: 10.1371/journal.pone.0011030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Skvortsov S, Schäfer G, Stasyk T, Fuchsberger C, Bonn GK, Bartsch G, Klocker H, Huber LA. Proteomics profiling of microdissected low- and high-grade prostate tumors identifies Lamin A as a discriminatory biomarker. J Proteome Res. 2011;10:259–268. doi: 10.1021/pr100921j. [DOI] [PubMed] [Google Scholar]
- 105.Drabovich AP, Pavlou MP, Dimitromanolakis A, Diamandis EP. Quantitative analysis of energy metabolic pathways in MCF-7 breast cancer cells by selected reaction monitoring assay. Mol Cell Proteomics. doi: 10.1074/mcp.M111.015214. Published on April 25, 2012. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 107.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nordone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzek R, Macneill J, Ren JM. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 108.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]