Abstract
Uridine diphosphate-glucuronosyltransferase (UGT) 2B7 is expressed mostly in the human liver, lung and kidney and can transfer endogenous glucuronide group into its substrate and impact the pharmacological effects of several drugs such as estriol, AZT and morphine. UGT2B7 and its allelic variants can dimerize with the homologous enzymes UGT1A1 and UGT1A9, as well as their allelic variants, and then change their enzymatic activities in the process of substrate catalysis. The current study was designed to identify this mechanism using morphine as the substrate of UGT2B7. Single-recombinant allozymes, including UGT2B7*1 (wild type), UGT2B7*71S (A71S, 211G>T), UGT2B7*2 (H268Y, 802C>T), UGT2B7*5 (D398N, 1192G>A), and double-recombinant allozymes formed by the dimerization of UGT1A9*1 (wild type), UGT1A9*2 (C3Y, 8G>A), UGT1A9*3 (M33T, 98T>C), UGT1A9*5 (D256N, 766G>A), UGT1A1 (wild type) with its splice variant UGT1A1b were established and incubated with morphine in vitro. Each sample was analyzed with HPLC-MS/MS. All enzyme kinetic parameters were then measured and analyzed. From the results, the production ratio of its aberrant metabolism and subsequent metabolites, morphine-3-glucuronide (M3G) and morphine-6-glucuronide (M6G), changes regioselectively. Double-recombinant allozymes exhibit stronger enzymatic activity catalyzing morphine than the single-recombinant alloyzymes. Compared to UGT2B7*1, UGT2B7*2 singles or doubles have lower Km values for M3G and M6G, whereas UGT2B7*5 allozymes perform opposite effects. The double allozymes of UGT1A9*2 or UGT1A9*5 with UGT2B7 tend to produce M6G. Interestingly, the majority of single or double allozymes significantly reduce the ratio of M3G to M6G. The UGT1A9*2-UGT2B7*1 double enzyme has the lowest M3G:M6G ratio, reflecting that more M6G would form in morphine glucuronide metabolism. This study demonstrates that UGT2B7 common SNPs and their dimers with UGT1A1 and UGT1A9 and their allelic variants can regioselectively affect the generation of two metabolites of morphine via altering the CLint ratios of M3G to M6G. These results may predict the effectiveness of morphine antinociception in individualized opioid treatment.
Keywords: UGT2B7, single nucleotide polymorphisms, dimerization, enzyme kinetics, morphine metabolism
Introduction
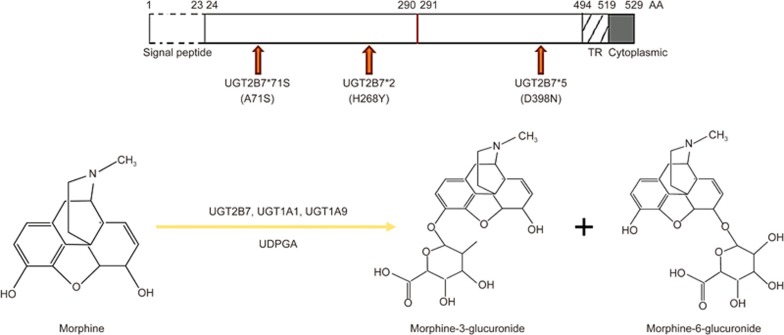
As a member of the uridine diphosphate-glucuronosyltransferse (UGT) family, UGT2B7 is expressed mostly in the human liver, lung and kidney and can transfer endogenic glucuronide group into its substrate1,2. This glucuronidation can impact the pharmacological effects of several drugs in vivo such as estriol3, 3'-azido-deoxythymidine (AZT) and morphine4. At the same time, its enzymatic activity can be inhibited by certain chemical compounds, such as ketoconazole5,6. UGT2B7 has many functional variants, some of which have been found among East Asians. It was reported that the UGT2B7*71S (211G>T, A71S) variant was found in 12% of 50 normal genome DNA samples extracted from the blood of healthy South Korean volunteers7. In addition, UGT2B7*2 (802C>T, H268Y) showed a frequency of 9.2% in a Chinese population8, and UGT2B7*5 (1192G>T, D398N) was found widely in a Japanese population9. However, some variants have the potential to induce serious maladies. For example, individuals with the 161 C>T variant in UGT2B7 may be susceptible to breast cancer10, and the UGT2B7*2 variant was found to induce tumor of the bladder11. UGT1A1*28 (TA 7/7) and *6 (A/A) lead to a severe irinotecan-induced toxicity in colorectal carcinoma patients, contributing to an obvious enhancement of the accumulation of its toxic metabolite, SN-38, in patient colon tissues 12,13. Therefore, variant UGTs may convert non-toxic drugs or endogenous compounds to toxic metabolites and then give rise to severe diseases.
Recent research has demonstrated that different UGTs and their allelic variants can be co-expressed in same genome of the same person in clinical practice. Mehlotra et al found that Hispanic-Americans can harbor both UGT2B7*71S and UGT1A9*314. Deng et al analyzed the DNA of 200 Chinese renal transplant recipients and observed that some of them harbor both UGT1A9*3 and UGT2B7*2 15. Accordingly, Yuan et al discovered that UGT2B7 can dimerize with its variants and affect enzymatic affinity to its substrates16. Liu et al recognized that UGT1A1, UGT1A9 and their variants can dimerize and form oligomers, then change each of their enzymatic activities and regioselectivities involved in quercetin glucuronidation17. One hypothesis is that some co-expression of UGT allelic variants in one patient can result in protein–protein interactions (PPI). If this hypothesis is correct, it would alter enzyme-substrate binding mechanisms for each UGT enzyme.
Morphine, a vital analgesic, can be converted to two main glucuronide metabolites, morphine-6-glucuronide (M6G) and morphine-3-glucuronide (M3G) by UGT2B7 (Figure 1). M6G has stronger antinociception and agonist activity on opioid receptors than morphine18,19. In addition, after morphine treatment, the antinociceptive effect of M6G may be antagonized by M3G20. Interestingly, the latter one does not contribute to the performance of morphine antinociception21. Therefore, to reduce pain, a decreased dose of M3G or an increased dose of M6G would be beneficial. It has been reported that patients with diverse UGT2B7 variants have a different analgesic response to the same dose of morphine22. In addition, Sawyer et al investigated that patients suffering pain who carried the UGT2B7 homozygous variant genotype 161C>T displayed lower enzymatic activity in morphine catalysis23. All these findings indicated that single-nucleotide polymorphisms (SNPs) of UGT2B7 would be a major factor in morphine glucuronidation.
Figure 1.
The allelic variant gene structures of UGT2B7 and a schematic diagram of morphine metabolism.
UGT2B7, UGT1A1 and UGT1A9 are all involved in the process of morphine metabolism24. UGT1A1 contributes to M6G generation25. As described above, UGT2B7, UGT1A1 and UGT1A9, as well as their allelic variants, can dimerize with each other. These double-recombinant allozymes, which are constructed via a Bac-to-Bac system and acquired from sf9 cells, can change their affinities between UGT2B7 and zidovudine16,17. This means the dimerization produced in vivo between each UGT enzyme would be another vital element intervening in the process of morphine metabolism.
To verify these hypotheses, in the present study, we analyzed the regioselectivity for morphine glucuronidation under the influence of common UGT2B7 SNP variants in a population and their dimers with UGT1A1 and UGT1A9 as well as their allelic variants. We then determined all the enzyme kinetic parameters of M3G and M6G production catalyzed by each single or double recombinant allozyme. The complete results were reviewed and used for specific analyses.
Materials and methods
Materials and reagents
Plasmids with UGT2B7 *1 (wild type), *71S (A71S, 211G>T), *2 (H268Y, 802C>T), *5 (D398N, 1192G>A); UGT1A9*1 (wild type), UGT1A9*2 (C3Y, 8G>A), UGT1A9*3 (M33T, 98T>C), UGT1A9*5 (D256N, 766G>A), and UGT1A1 (wild type) with its splicing variant UGT1A1b were constructed and ligated to the pFastBac vector before all genes were connected with HA or CFP tags, which are convenient for the detection of their expression. Then, all the plasmids were verified by double digestion and sequenced previously16,17. In brief, all vectors were transferred to E coli DH10α-competent cells for the acquisition of recombinant bacmid plasmids. UGT2B7, UGT1A1 and UGT1A9 plasmids were double-transfected into sf9 cells. Baculovirus was then collected on the third passage (all titers were more than 108 pfu/mL). After fluorescence resonance energy transfer (FRET) assays and co-immunoprecipitation (Co-IP) analyses, all allozymes were used as functional proteins16,17.
Morphine, M3G and M6G were obtained from Cerilliant Corporation (Round Rock, TX, USA). M6G-d3 was labeled by Biomag System Company (Changshu, China). UDPGA were purchased from Sigma Chemical Co (St Louis, MO, USA). The BCA assay kit was purchased from Sangon Biotech Company (Shanghai, China). Tris-HCl, NaCl, EDTA, MgCl2, perchloric acid and KH2PO4 were purchased from Sinopharm Chemical Regent Co (Beijing, China). Alamethicin was obtained from Sigma-Aldrich Company (Shanghai, China). HPLC-grade methanol and formic acid were purchased from Tedia Company (Fairfield, OH, USA). Ultrapure water (18.2 MΩ) was prepared using an ELGA PureLab Ultra system (High Wycombe, UK).
Incubation assay
The incubation mixture (100 μL total volume) consisted of 50 mmol/L potassium phosphate buffer (pH 7.4), 5 mmol/L MgCl2, 10mmol/L UDPGA, 0.1 mg/mL total recombinant proteins, alamethicin (50 μg/mg of protein based on the concentration of each allozyme) and 1–200 μmol/L morphine. The optimum conditions (protein concentration and incubation time) were ascertained by preliminary experiments. Then, the reaction was initiated by adding 5 μL of 10 mmol/L UDPGA to concentrations of 500 μmol/L in final 100 μL volume solutions following a 5 min preincubation at 37 °C. The incubation was performed at 37 °C for 50 min. The reaction was terminated by adding 200 μL of acetonitrile and spiked M6G-d3 as the internal standard at a final concentration of 10 ng/mL. After vortex and vibration, the mixture was centrifuged at 12 000×g for 15min, and the supernatants were then evaporated and dried in a centrifugal thickener (Labconco, USA). Residues were dissolved in a 100μL mobile phase (A) methanol: (B) 0.5‰ formic acid and purified water solution (2:98, v/v) by vortex. A 10 μL sample was injected into HPLC-MS/MS for further analysis.
Determination of morphine glucuronides
Agilent 1290 infinity LC system equipped with a G4220A quaternary pump, G4226A auto sampler, and G1330B 1290 thermostat; AB SCIEX 4000 plus triple quadrupole mass spectrometer (AB SCIEX Technologies) included an electrospray ionization source were performed for the HPLC-MS/MS analysis. The auto sampler was maintained at 4 °C, and the temperature for column compartment was set at 30 °C. Chro-matographic separations were achieved on an Agilent HILIC PLUS SB-C18 column (2.1 mm×50 mm, 3.5 μm). The mobile phase for simultaneously analyzing M3G and M6G with deuterated internal standard M6G-d3 (ISTD) consisted of the solutions we referred previously. The separation method was applied via a gradient elution mode of 95% A and 5% B at 0–1 min, 2% A and 98% B at 1–6 min, 95% A and 5% B at 6–7 min with a flow rate of 0.25 mL/min.
The mass spectrometer with an ESI source was operated in positive ionization mode. The mass spectrometer parameters were set as following: collision energy, 43 eV for M3G, M6G and M6G-d3 (ISTD); declustering potential, 85 V for all the three compounds; integral temperature of drying gas, 350 °C; drying gas flow, 8 L/min; temperature for ionization 550 °C. Data were acquired using Analyst 1.5.2 (AB SCIEX) in multiple reac-tion monitoring (MRM) mode by recording ion currents for the following transitions: m/z 462.1–286.1 for M3G and M6G, m/z 465.1–289.1 for M6G-d3 (ISTD). The method was fully validated according to the US FDA guidelines (http://www.fda.gov/downloads/Drugs/GuidanceCompliance RegulatoryInformation/Guidances/UCM368107.pdf).
Kinetic data analyses
The kinetic parameters were analyzed using GraphPad Prism, version 6.0 (GraphPad Software Inc, San Diego, CA, USA). The enzymatic kinetic parameters such as Km and Vmax for morphine glucuronidation were estimated by analyzing the Michaelis-Menten equation:
where V is the velocity of the reaction, [S] is the substrate concentration, Km is the Michaelis–Menten constant, and Vmax is the maximum velocity. CLint is the intrinsic clearance. Each value was determined as the ratio of Vmax/Km.
Statistical analysis
The data were expressed as the mean±standard deviation (SD) of at least three independent experiments in triplicate. The significant differences between the experimental groups were determined using the one-way analysis of variance (ANOVA) followed by a Dunnett's post hoc test (SPSS 13.0 software, SPSS Inc, Chicago, IL, USA). A value of P<0.05 was considered statistically significant.
Results
Kinetics analysis of morphine glucuronides catalyzed by four UGT2B7 SNPs
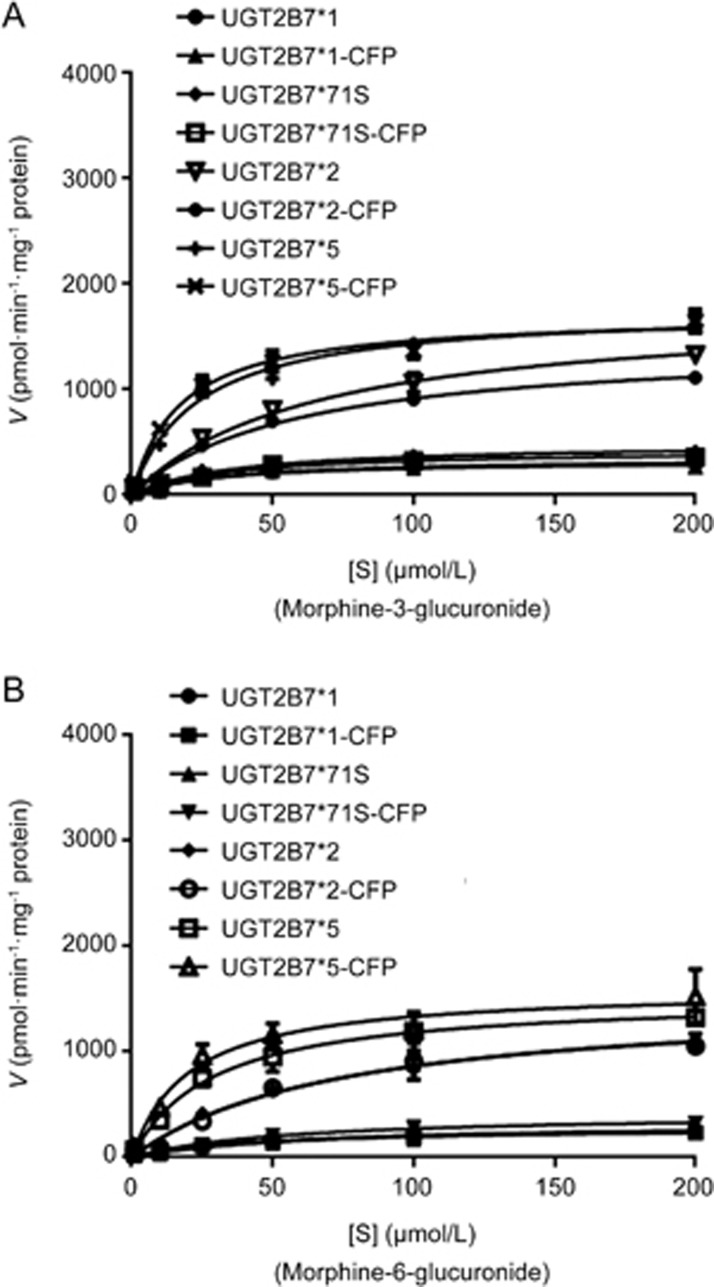
Human of UGT2B7*1 (wildtype) enzymes and three sense variants, *71S (A71S, 211G>T), *2 (H268Y, 802C>T), and *5 (D398N, 1192G>A) (Figure 1) were established and characterized and then used in vitro assay of morphine incubation 16. The results showed decreasing trends in the Km values of UGT2B7*5-HA to M3G and M6G by 31.39% and 15.41%, respectively, compared to HA-tagged UGT2B7*1. However, UGT2B7*2-HA had an opposite effect, in which the Km value to M3G increased by 89.83%, and that to M6G increased by 123.4% compared to HA-tagged UGT2B7*1 (Figure 2, Table 1). In addition, the CLint ratios of M3G to M6G mediated by UGT2B7*2 and UGT2B7*5 were all lower than those of other single allozymes.
Figure 2.
Common SNPs of UGT2B7 single allzoymes mediate the glucuronide metabolism of morphine. The Michaelis–Menten kinetic equation shows the enzymatic reaction curves for (A) M3G; (B) M6G.
Table 1. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT2B7*N single recombinant allozymes.
| UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT2B7*1-HA | 33.93±9.004 | 55.10±12.40 | 358.7±31.12 | 324.4±28.26 | 10.57 | 5.887 | 1.795 | 100.0 | 100.0 |
| UGT2B7*1-CFP | 29.78±5.759 | 42.49±6.399* | 328.7±19.85 | 282.0±15.02 | 11.04 | 6.637 | 1.663 | 104.4 | 112.7 |
| UGT2B7*71S-HA | 36.06±8.092 | 66.07±16.66* | 494.5±37.06 | 337.6±35.12 | 13.71 | 5.110 | 2.683 | 129.7 | 86.80 |
| UGT2B7*71S-CFP | 32.29±6.169 | 54.43±9.753 | 429.3±26.35 | 421.8±29.15 | 13.30 | 7.749 | 1.716 | 125.8 | 131.6 |
| UGT2B7*2-HA | 64.41±11.14** | 75.81±12.30** | 1772±125.3** | 1522±106.9** | 27.51** | 20.08* | 1.370 | 260.3 | 341.1 |
| UGT2B7*2-CFP | 57.55±9.407** | 71.75±11.81** | 1444±92.77** | 1486±103.8** | 25.09** | 20.71* | 1.211 | 237.4 | 351.8 |
| UGT2B7*5-HA | 23.28±3.728** | 28.70±4.436** | 1769±81.07** | 1525±72.68** | 75.99** | 53.14** | 1.430 | 718.9 | 902.7 |
| UGT2B7*5-CFP | 17.29±2.855** | 20.10±4.250** | 1718±72.90** | 1604±92.04** | 99.36** | 79.80** | 1.245 | 940.0 | 1356 |
Kinetics analysis of common SNPs of UGT2B7 mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (*P<0.05; **P<0.01).
Kinetics analysis of morphine glucuronides catalyzed by recombinant UGT2B7 allozymes with its allelic variants
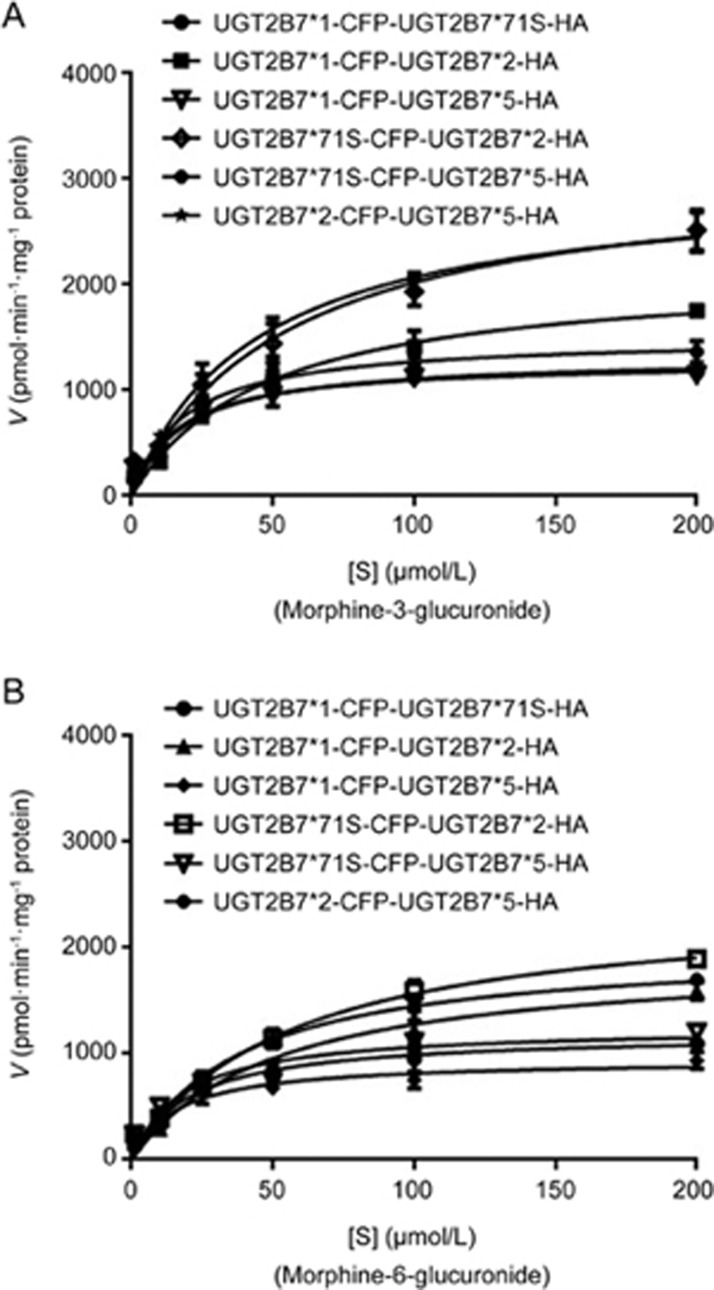
To further determine whether the dimerization of UGT2B7 and each of its allelic variants could impact the metabolism of morphine, all double allozymes were prepared and tested. The results indicated Km values of UGT2B7*5-HA mediated double allozymes to M3G or M6G declined substantially compared to HA-tagged UGT2B7*1 single allozyme (Figure 3, Table 2). In contrast, that of UGT2B7*2-HA double allozymes increased clearly to M3G, but not M6G. Notably, UGT2B7*2-UGT2B7*5 double integrally showed a moderate Km value to M3G and M6G compared to those of other UGT2B7-dependent singles or doubles. The Km and CLint of UGT2B7*71S doubles were slightly upregulated to both M3G and M6G compared to UGT2B7*1. However, CLint ratios of M3G to M6G mediated by UGT2B7 with its variants in double allozymes were all lower than those of their single variants.
Figure 3.
UGT2B7*N-UGT2B7*N double allozymes mediate the glucuronide metabolism of morphine. The Michaelis–Menten kinetic equation shows the enzymatic reaction curves for (A) M3G; (B) M6G.
Table 2. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT2B7*N-UGT2B7*N double recombinant allozymes.
| UGT2B7*N -UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT2B7*1-CFP -UGT2B7*71S-HA | 18.80±2.304** | 20.59±3.687** | 1319±42.82** | 1195±58.59** | 70.16** | 58.04** | 1.209 | 663.8 | 985.9 |
| UGT2B7*1-CFP -UGT2B7*2-HA | 47.25±6.493* | 50.59±9.884 | 2139±107.9** | 1926±141.4** | 45.27** | 38.07** | 1.189 | 428.3 | 646.7 |
| UGT2B7*1-CFP -UGT2B7*5-HA | 15.93±3.132** | 16.03±3.634** | 1269±62.21** | 946.4±53.63** | 79.66** | 59.04** | 1.349 | 753.6 | 1003 |
| UGT2B7*71S-CFP -UGT2B7*2-HA | 54.93±9.426** | 55.41±7.584 | 3131±207.8** | 2435±129.3** | 57.00** | 43.95** | 1.297 | 539.3 | 746.6 |
| UGT2B7*71S-CFP -UGT2B7*5-HA | 18.55±4.323** | 18.31±4.345** | 1506±92.56** | 1258±78.34** | 81.19** | 68.71** | 1.182 | 768.1 | 1167 |
| UGT2B7*2-CFP -UGT2B7*5-HA | 44.29±6.318* | 38.49±5.139** | 2986±148.5** | 2008±91.61** | 67.42** | 52.17** | 1.292 | 637.8 | 886.2 |
Kinetics analysis of UGT2B7*N-UGT2B7*N double recombinant allozymes mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (*P<0.05; **P<0.01).
Kinetics analysis of morphine glucuronides catalyzed by recombinant allozymes of UGT2B7 and UGT1A1
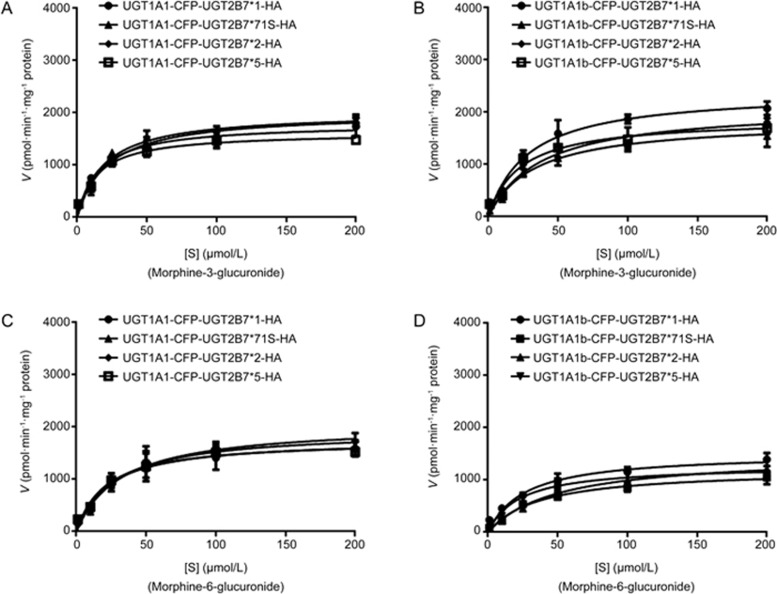
UGT1A1 and its alternatively spliced variant, UGT1A1b, dimerized with UGT2B7 and its allelic variants. In our test, we found that double allozymes formed between UGT1A1 or UGT1A1b and UGT2B7*N (including wildtype UGT2B7*1 and three variants, UGT2B7*71S, UGT2B7*2 and UGT2B7*5) showed relatively stronger enzymatic activities to catalyze morphine compared to UGT2B7*N singles (Figure 4, Table 3 and 4). The Km values of UGT1A1b double allozymes were all lower than those of UGT1A1 mediated ones. Although the Km values of M3G and M6G obtained from the double allozymes of UGT2B7*5 and UGT1A1 or UGT1A1b were slightly lower than the others, the CLint for all double allozymes consisting of UGT2B7*N and UGT1A1 or UGT1A1b were almost the same. In addition, the CLint ratio of M3G to M6G for the double of UGT1A1b-UGT2B7*2 was higher than that of the UGT2B7*2 single allozyme, which indicated that UGT1A1b played a minor role in forming M6G than UGT2B7*2. Compared to UGT2B7 double allozymes, UGT1A1 and UGT1A1b double allozymes exhibited relatively lower ratios of CLint for the formations of M6G.
Figure 4.
UGT1A1-UGT2B7*N and UGT1A1b-UGT2B7*N double allozymes mediate the glucuronide metabolism of morphine. The Michaelis–Menten kinetic equation shows the enzymatic reaction curves for (A, B) M3G; (C, D) M6G.
Table 3. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT1A1-UGT2B7*N double recombinant allozymes.
| UGT1A1 -UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT1A1-CFP -UGT2B7*1-HA | 15.55±2.364** | 22.35±3.616** | 1795±67.43** | 1768±80.68** | 115.4** | 79.11** | 1.459 | 1092 | 1344 |
| UGT1A1-CFP -UGT2B7*71S-HA | 18.97±3.297** | 27.23±5.693** | 2010±92.88** | 1939±122.7** | 106.0** | 71.21** | 1.489 | 1003 | 1210 |
| UGT1A1-CFP -UGT2B7*2-HA | 21.45±3.040* | 31.97±5.806** | 2004±78.93** | 2059±119.7** | 93.43** | 64.40** | 1.451 | 883.9 | 1094 |
| UGT1A1-CFP -UGT2B7*5-HA | 14.06±2.333** | 20.97±4.477** | 1625±64.17** | 1753±103.2** | 115.6** | 83.60** | 1.383 | 1094 | 1420 |
Kinetics analysis of UGT1A1-UGT2B7*N double recombinant allozymes mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (*P<0.05; **P<0.01).
Table 4. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT1A1b-UGT2B7*N double recombinant allozymes.
| UGT1A1b -UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT1A1b-CFP -UGT2B7*1-HA | 28.57±4.744 | 26.39±4.910** | 2403±122.8** | 1518±84.52** | 84.11** | 57.52** | 1.462 | 795.7 | 977.1 |
| UGT1A1b-CFP -UGT2B7*71S-HA | 32.71±5.229 | 36.74±5.956** | 1837±94.77** | 1207±65.78** | 56.16** | 32.85** | 1.710 | 531.3 | 558.0 |
| UGT1A1b-CFP -UGT2B7*2-HA | 37.61±7.334 | 49.76±9.057 | 2108±139.3** | 1485±101.0** | 56.05** | 29.84** | 1.878 | 530.3 | 506.9 |
| UGT1A1b-CFP -UGT2B7*5-HA | 22.66±4.575* | 22.41±4.898** | 1881±107.7** | 1283±79.15** | 83.01** | 57.25** | 1.450 | 366.3 | 972.5 |
Kinetics analysis of UGT1A1b-UGT2B7*N double recombinant allozymes mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (*P<0.05; **P<0.01).
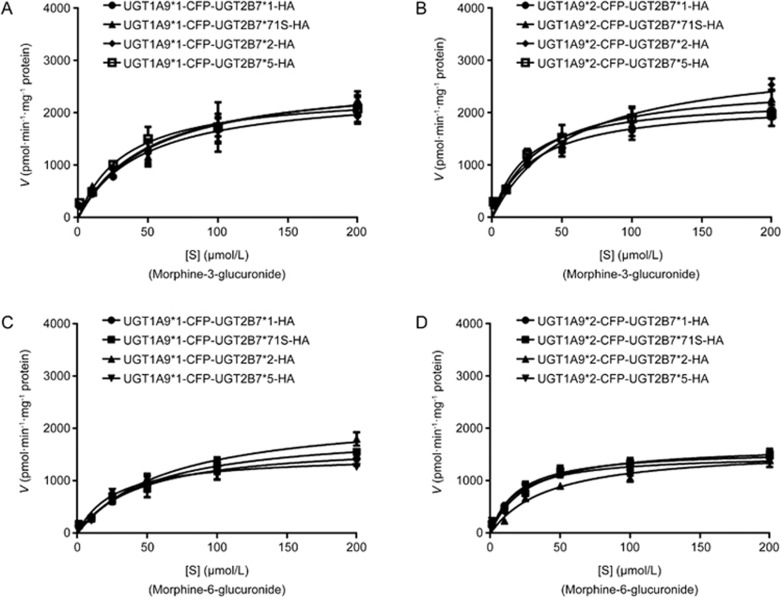
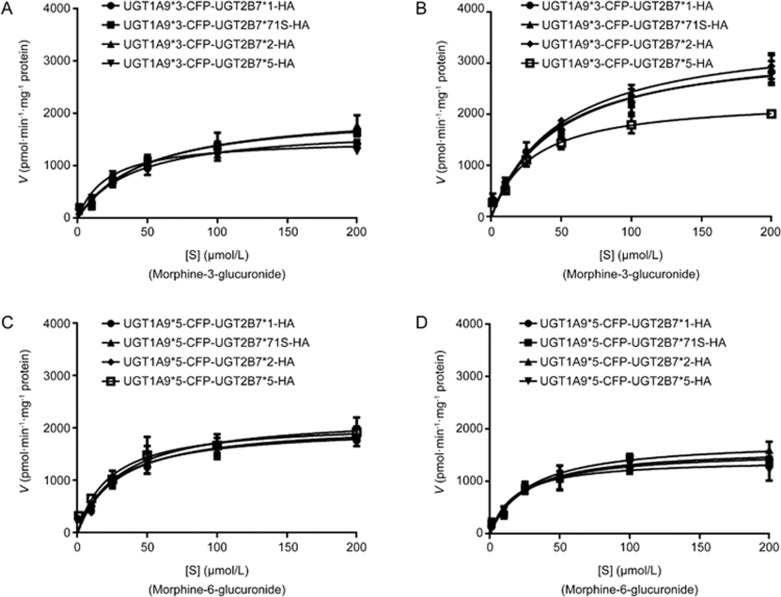
Kinetics analysis of morphine glucuronides catalyzed by recombinant allozymes of UGT2B7 and UGT1A9
Double allozymes of UGT1A9*1, *2, *3, *5 and UGT2B7 with their allelic variants were used to perform the test. From the results, the affinities of UGT1A9*2 and UGT1A9*5 double allozymes to morphine were found to be much higher than those of UGT1A9*1 and UGT1A9*3 (Figure 5 and 6; Table 5, 6, 7, 8). In addition, UGT1A9*2 and UGT1A9*5 mediated double allozymes tended to generate M6G based on the CLint ratios of the two metabolites. UGT1A9*3-UGT2B7*N double allozymes had much higher CLint ratios of M3G to M6G which illustrated that they owned higher enzymatic activities to catalyze morphine and form M6G than UGT1A9*3 single. Notably, the CLint ratio of UGT1A9*2-UGT2B7*1 double allozymes had the lowest value (only 0.9014) among all the singles or doubles, which could be attributed to an increase in the formation of M6G.
Figure 5.
UGT1A9*1-UGT2B7*N and UGT1A9*2-UGT2B7*N double allozymes mediate the glucuronide metabolism of morphine. The Michaelis–Menten kinetic equation shows the enzymatic reaction curves for (A, B) M3G; (C, D) M6G.
Figure 6.
UGT1A9*3-UGT2B7*N and UGT1A9*5-UGT2B7*N double allozymes mediate the glucuronide metabolism of morphine. The Michaelis–Menten kinetic equation shows the enzymatic reaction curves for (A, B) M3G; (C, D) M6G.
Table 5. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT1A9*1-UGT2B7*N double recombinant allozymes.
| UGT1A9*1 -UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT1A9*1-CFP -UGT2B7*1-HA | 28.57±4.744 | 43.96±6.479* | 2425±159.0** | 1731±91.54** | 52.50** | 39.38** | 1.333 | 496.7 | 668.9 |
| UGT1A9*1-CFP -UGT2B7*71S-HA | 49.58±11.01** | 53.32±9.603 | 2682±222.3** | 1972±136.0** | 54.09** | 36.98** | 1.463 | 511.7 | 628.2 |
| UGT1A9*1-CFP -UGT2B7*2-HA | 52.73±8.907** | 64.91±11.01* | 2717±175.0** | 2315±161.0** | 51.53** | 35.66** | 1.445 | 487.5 | 605.7 |
| UGT1A9*1-CFP -UGT2B7*5-HA | 33.67±8.646 | 26.56±5.348** | 2410±201.8** | 1495±90.29** | 71.58** | 56.29** | 1.272 | 677.2 | 956.2 |
Kinetics analysis of UGT1A9*1-UGT2B7*N double recombinant allozymes mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (*P<0.05; **P<0.01).
Table 6. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT1A9*2-UGT2B7*N double recombinant allozymes.
| UGT1A9*2 -UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT1A9*2-CFP -UGT2B7*1-HA | 29.63±3.937 | 19.34±3.161** | 2200±91.15** | 1593±69.72** | 74.25** | 82.37** | 0.9014 | 702.5 | 1399 |
| UGT1A9*2-CFP -UGT2B7*71S-HA | 34.48±5.827 | 25.89±3.624** | 2586±143.7** | 1693±70.44** | 75.00** | 65.39** | 1.147 | 709.6 | 1111 |
| UGT1A9*2-CFP -UGT2B7*2-HA | 56.03±12.57** | 41.48±7.671* | 3079±269.2** | 1624±105.3** | 54.95** | 39.15** | 1.404 | 519.9 | 665.0 |
| UGT1A9*2-CFP -UGT2B7*5-HA | 24.47±4.658* | 18.56±3.669** | 2284±126.7** | 1508±78.62** | 101.6** | 81.25** | 1.250 | 961.2 | 1380 |
Kinetics analysis of UGT1A9*2-UGT2B7*N double recombinant allozymes mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (*P<0.05; **P<0.01).
Table 7. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT1A9*3-UGT2B7*N double recombinant allozymes.
| UGT1A9*3 -UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT1A9*3-CFP -UGT2B7*1-HA | 43.96±6.959* | 39.60±6.245** | 3360±190.4** | 1751±95.29** | 76.43** | 44.22** | 1.728 | 723.1 | 751.1 |
| UGT1A9*3-CFP -UGT2B7*71S-HA | 47.19±8.718* | 46.73±9.841* | 3422±231.9** | 2036±156.8** | 72.52** | 43.57** | 1.664 | 686.1 | 740.1 |
| UGT1A9*3-CFP -UGT2B7*2-HA | 49.06±6.343* | 49.01±9.100 | 3642±175.1** | 2079±143.5** | 74.24** | 42.42** | 1.750 | 702.4 | 720.6 |
| UGT1A9*3-CFP -UGT2B7*5-HA | 29.01±4.053 | 21.09±3.808** | 2313±99.98** | 1519±75.79** | 79.73** | 72.02** | 1.107 | 754.3 | 1223 |
Kinetics analysis of UGT1A9*3-UGT2B7*N double recombinant allozymes mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (*P<0.05; **P<0.01).
Table 8. Kinetic parameters for morphine-3-glucuronide and morphine-6-glucuronide catalyzed by UGT1A9*5-UGT2B7*N double recombinant allozymes.
| UGT1A9*5 -UGT2B7*N |
Km (μmol/L) |
Vmax (pmol·min−1·mg−1 protein) |
CLint (μL·min−1·mg−1 protein) |
CLint (M3G)/ CLint (M6G) | % of 2B7*1HA |
||||
|---|---|---|---|---|---|---|---|---|---|
| M3G | M6G | M3G | M6G | M3G | M6G | M3G | M6G | ||
| UGT1A9*5-CFP -UGT2B7*1-HA | 26.03±4.453 | 23.55±4.242** | 2020±102.9** | 1590±82.29** | 77.60** | 67.52** | 1.149 | 734.2 | 1147 |
| UGT1A9*5-CFP -UGT2B7*71S-HA | 29.36±3.938 | 24.47±4.555** | 2095±87.31** | 1645±89.21** | 71.36** | 67.23** | 1.061 | 675.1 | 1142 |
| UGT1A9*5-CFP -UGT2B7*2-HA | 30.77±6.448 | 27.41±4.998** | 2245±148.6** | 1796±99.32** | 72.96** | 65.52** | 1.114 | 690.3 | 1113 |
| UGT1A9*5-CFP -UGT2B7*5-HA | 22.97±5.130* | 17.27±4.430** | 2108±134.1** | 1426±93.92** | 91.77** | 82.57** | 1.111 | 868.2 | 1403 |
Kinetics analysis of UGT1A9*5-UGT2B7*N double recombinant allozymes mediate the glucuronide metabolism of morphine. Data are the mean±SD of three independent determinations, the asterisks indicate differences that are statistically significant compared to 2B7*1HA (**P<0.01; *P<0.05).
Discussion
In this study, we used single or double UGT2B7, UGT1A1, UGT1A9 recombinant allozymes to incubate morphine in vitro and then quantified M3G and M6G to investigate the connections between SNPs or dimerization-induced UGT2B7 enzymatic activity changes and the regioselectivity for morphine glucuronidation. In addition, morphine antinociception and the CLint of M6G formed by single or double allozymes was also explored.
For single allozymes of UGT2B7 including its three allelic variants *2, *71S and *5, they had higher enzymatic activities to catalyze morphine and form M3G and M6G than UGT2B7*1 from the values of CLint. However, the Km values to each metabolite were different from that. There was a predominant increase trend from Km values of UGT2B7*2 to each M3G or M6G compared to UGT2B7*1. In contrast, UGT2B7*5 exhibited a reverse character. Since we found codons 71 and 268 of UGT2B7 are both located in the substrate binding domain, whereas the highly conserved codon 398 was located in the latter half of the UDPGA-binding domain16. These variant sites may lead to differentials of glucuronidation generated in each substrate. Wang et al have used UGT2B7*1, *71S and *2 to metabolize flurbiprofen (FPF) racemate and generate S- and R-FPF glucuronide metabolites. They observed UGT2B7*2 predominated to generate R-FPF glucuronide26. From this example, we found the UGT2B7*2 (H268Y, 802C>T) variant may result in regioselectively changing of its substrates' metabolites. Moreover, we noticed that UGT2B7*5 (D398N, 1192G>A) showed lower Km values when it catalyzed morphine and generated M3G and M6G compared to the other singles, which suggests that the codon 398 variants may mainly increase the binding of enzyme substrates. Additionally, the affinities of CFP-labeled enzymes have not been verified to be significantly different from HA-labeled enzymes based on these results. In other words, there were no effects of the fusion proteins on the change of each allozyme`s function.
UGT2B7 dimerized with its allelic variants can increase the CLint values for both M3G and M6G compared to UGT2B7 wild type single allozyme. After oligomerizing with UGT2B*5, the enzymatic activities of UGT2B7*1 or UGT2B7*71S were activated, but their activities took an opposite reaction after interacting with UGT2B7*2. The integral alteration tends for Km values of UGT2B7*2 or UGT2B7*5 to morphine were almost the same as zidovudine16. In addition, the Km to M3G and M6G was diminished during the catalysis by the UGT2B7*2-UGT2B7*5 double allozyme compared to the UGT2B7*2-HA single. As a result, UGT2B7*5 was an active allelic variant during morphine glucuronidation, but UGT2B7*2 was not.
The alternatively spliced variant of UGT1A1b has a frame shift mutation in intron 4 (from I1089 to I1122) of UGT1A1 between 1302bp and 1303bp which alters the C-terminal structure compared to the UGT1A1 wild type. We observed that the enzymatic activity of UGT1A1b declined remarkably compared to that of UGT1A1 due to a mutation that disrupted the hydrophobic packing of the protein27. Similar results were obtained for the double allozymes formed by UGT1A1 or UGT1A1b with UGT2B7*N. It is notable that compared to UGT2B7*N-UGT2B7*N double allozymes, UGT1A1 or UGT1A1b and UGT2B7*N double allozymes had noticeably lower Km values for M3G or M6G.
All single-expressed UGT2B7 and its variant allozymes showed higher CLint values for M3G and M6G than UGT1A9 wild type-related double allozymes in morphine glucuronidation did. For example, the UGT1A9*2-UGT2B7*1 double allozyme exhibited 2.09- and 1.41-fold higher clearance of M6G and M3G, respectively, compared to the UGT1A9*1-UGT2B7*1 double allozyme. Concurrently, the CLint values of M3G and M6G from UGT2B7*5-UGT1A9*N-formed double allozymes were much higher than those of the other UGT2B7*N-UGT1A9*N doubles. In contrast, UGT2B7*2 and UGT1A9*N double allozymes had few changes in CLint values, excepting for a sharp decrease in the CLint of the UGT1A9*2-UGT2B7*2 double allozyme. Since UGT1A9*2 exhibited a 1.66-fold higher enzymatic activity to catalyze mycophenolic acid glucuronidation than UGT1A9*1, so the mutant of UGT1A9*2(C3Y, 8G>A) would be an active variant in UGT1A9 28.
UGT subtypes including UGT1A9, UGT1A10, UGT2B7, UGT2B15, and UGT2B17 can stereo-selectively catalyze the glucuronidation of substrates and affect their metabolites` pharmacological effects26,29,30,31,32,33. However, there was no report on whether morphine antinociception can be enhanced via the selective alteration of its metabolite ratios by UGT2B7 allelic variants or dimerized with other UGTs. In our study, the CLint ratios of M3G to M6G for single or double allozymes were measured and analyzed. Since M6G has stronger antinociception effects than M3G, as we described previously18,19, a single or double allozyme with a low CLint ratio of M3G to M6G would improve morphine during analgesic treatment. Compared to the CLint ratio of M3G to M6G in the UGT2B7*1 single allozyme (1.795 and 1.663 for HA- and CFP-tagged UGT2B7*1, respectively), UGT1A9*2-UGT2B7*1 double allozyme exhibited the lowest ratio (0.9014), which indicates that morphine can be converted more M6G under such conditions.
In conclusion, UGT2B7 common SNPs and their dimers with UGT1A1 and UGT1A9 and their allelic variants can regioselectively affect the generation of two metabolites of morphine via altering the CLint ratios of M3G to M6G. These results may predict the effectiveness of morphine antinociception in individualized opioid treatment.
Author contribution
Zi-zhao YANG, Li LI, Lu WANG, Ling-min YUAN, Ming-cheng XU, Jing-kai GU, Hui-di JIANG, Lu-shan YU, and Su ZENG participated in research design; Zi-zhao YANG, Li LI, and Ling-min YUAN conducted experiments; Zi-zhao YANG, Li LI, and Lu WANG performed data analysis; Zi-zhao YANG and Su ZENG wrote or contributed to the writing of the manuscript.
Abbreviation
UGT, uridine diphosphate-glucuronosyltransferase; M3G, morphine-3-glucuronide; M6G, morphine-6-glucuronide; SNPs, single nucleotide polymorphisms; UDPGA, uridine diphosphate glucuronic acid; HA tag, human influenza hemagglutinin tag; CFP tag, enhanced green fluorescent protein C; FRET, fluorescence resonance energy transfer; Co-IP, co-immunoprecipitation.
Acknowledgments
This study was supported by the International Science & Technology Cooperation Program of China (No 2014DFE30050); Program for Zhejiang Leading Team of S&T Innovation Team (No 2011R50014); Natural Science Foundation of Zhejiang Province (No LQ15H310003); and Fundamental Research Funds for the Central Universities of China Ministry of Education (No 2016XZZX001-08).
References
- Ekström L, Johansson M, Rane A. Tissue distribution and relative gene expression of UDP-glucuronosyltransferases (2B7, 2B15, 2B17) in the human fetus. Drug Metab Dispos 2013; 41: 291–5. [DOI] [PubMed] [Google Scholar]
- Sadeque AJ, Usmani KA, Palamar S, Cerny MA, Chen WG. Identification of human UDP-glucuronosyltransferases involved in N-carbamoyl glucuronidation of lorcaserin. Drug Metab Dispos 2012; 40: 772–8. [DOI] [PubMed] [Google Scholar]
- Gall WE, Zawada G, Mojarrabi B, Tephly TR, Green MD, Coffman BL, et al. Differential glucuronidation of bile acids, androgens and estrogens by human UGT1A3 and 2B7. J Steroid Biochem Mol Biol 1999; 70: 101–8. [DOI] [PubMed] [Google Scholar]
- Bélanger AS, Caron P, Harvey M, Zimmerman PA, Mehlotra RK, Guillemette C. Glucuronidation of the antiretroviral drug efavirenz by UGT2B7 and an in vitro investigation of drug-drug interaction with zidovudine. Drug Metab Dispos 2009; 37: 1793–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Kitajima Y, Ishii Y, Nishimura Y, Mackenzie PI, Oguri K, et al. Inhibition of UDP-glucuronosyltransferase 2b7-catalyzed morphine glucuronidation by ketoconazole: dual mechanisms involving a novel noncompetitive mode. Drug Metab Dispos 2006; 34: 1277–82. [DOI] [PubMed] [Google Scholar]
- Nagaoka K, Hanioka N, Ikushiro S, Yamano S, Narimatsu S. The effects of N-glycosylation on the glucuronidation of zidovudine and morphine by UGT2B7 expressed in HEK293 cells. Drug Metab Pharmacokinet 2012; 27: 388–97. [DOI] [PubMed] [Google Scholar]
- Hwang MS, Lee SJ, Jeong HE, Lee S, Yoo MA, Shin JG. Genetic variations in UDP-glucuronosyltransferase 2B7 gene (UGT2B7) in a Korean population. Drug Metab Pharmacokinet 2010; 25: 398–402. [DOI] [PubMed] [Google Scholar]
- Lin GF, Guo WC, Chen JG, Qin YQ, Golka K, Xiang CQ, et al. An association of UDP-glucuronosyltransferase 2B7 C802T (His268Tyr) polymorphism with bladder cancer in benzidine-exposed workers in China. Toxicol Sci 2005; 85: 502–6. [DOI] [PubMed] [Google Scholar]
- Saeki M, Saito Y, Jinno H, Tanaka-Kagawa T, Ohno A, Ozawa S, et al. Single nucleotide polymorphisms and haplotype frequencies of UGT2B4 and UGT2B7 in a Japanese population. Drug Metab Dispos 2004; 32: 1048–54. [PubMed] [Google Scholar]
- Sawyer MB, Pituskin E, Damaraju S, Bies RR, Vos LJ, Prado CM, et al. A uridine glucuronosyltransferase 2B7 polymorphism predicts epirubicin clearance and outcomes in early-stage breast cancer. Clin Breast Cancer 2016; 16: 139–44. [DOI] [PubMed] [Google Scholar]
- Zimmermann A, Blaszkewicz M, Roth G, Seidel T, Dietrich H, Schutschkow O, et al. UDP-glucuronosyltransferase 2B7 C802T (His268Tyr) polymorphism in bladder cancer cases. J Toxicol Environ Health A 2008; 71: 911–4. [DOI] [PubMed] [Google Scholar]
- Xu JM, Wang Y, Ge FJ, Lin L, Liu ZY, Sharma MR, et al. Severe irinotecan-induced toxicity in a patient with UGT1A1 28 and UGT1A1 6 polymorphisms. World J Gastroenterol 2013; 19: 3899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JE, Saksena R, Jabbour SK, Nosher JL, Hermes-DeSantis E, Moss RA. The power of genes: a case of unusually severe systemic toxicity after localized hepatic chemoembolization with irinotecan-eluted microspheres for metastatic colon cancer. Ann Pharmacother 2014; 48: 1646–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Bockarie MJ, Zimmerman PA. Prevalence of UGT1A9 and UGT2B7 nonsynonymous single nucleotide polymorphisms in West African, Papua New Guinean, and North American populations. Eur J Clin Pharmacol 2007; 63: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XY, Wang CX, Wang XD, Bi HC, Chen X, Li JL, et al. Genetic polymorphisms of UGT1A8, UGT1A9, UGT2B7 and ABCC2 in Chinese renal transplant recipients and a comparison with other ethnic populations. Pharmazie 2013; 68: 240–4. [PubMed] [Google Scholar]
- Yuan L, Qian S, Xiao Y, Sun H, Zeng S. Homo- and hetero-dimerization of human UDP-glucuronosyltransferase 2B7 (UGT2B7) wild type and its allelic variants affect zidovudine glucuronidation activity. Biochem Pharmacol 2015; 95: 58–70. [DOI] [PubMed] [Google Scholar]
- Liu YQ, Yuan LM, Gao ZZ, Xiao YS, Sun HY, Yu LS, et al. Dimerization of human uridine diphosphate glucuronosyltransferase allozymes 1A1 and 1A9 alters their quercetin glucuronidation activities. Sci Rep 2016; 6: 23763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregori S, De Gregori M, Ranzani GN, Allegri M, Minella C, Regazzi M. Morphine metabolism, transport and brain disposition. Metab Brain Dis 2012; 27: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frances B, Gout R, Monsarrat B, Cros J, Zajac JM. Further evidence that morphine-6 beta-glucuronide is a more potent opioid agonist than morphine. J Pharmacol Exp Ther 1992; 262: 25–31. [PubMed] [Google Scholar]
- Faura CC, Olaso MJ, Garcia Cabanes C, Horga JF. Lack of morphine-6-glucuronide antinociception after morphine treatment. Is morphine-3-glucuronide involved? Pain 1998; 65: 25–30. [DOI] [PubMed] [Google Scholar]
- Ouellet DM, Pollack GM. Effect of prior morphine-3-glucuronide exposure on morphine disposition and antinociception. Biochem Pharmacol 1997; 53: 1451–7. [DOI] [PubMed] [Google Scholar]
- Bastami S, Gupta A, Zackrisson AL, Ahlner J, Osman A, Uppugunduri S. Influence of UGT2B7, OPRM1 and ABCB1 gene polymorphisms on postoperative morphine consumption. Basic Clin Pharmacol Toxicol 2014; 115: 423–31. [DOI] [PubMed] [Google Scholar]
- Sawyer MB, Innocenti F, Das S, Cheng C, Ramírez J, Pantle-Fisher FH, et al. A pharmacogenetic study of uridine diphosphate-glucuronosyltransferase 2B7 in patients receiving morphine. Clin Pharmacol Ther 2003; 73: 566–74. [DOI] [PubMed] [Google Scholar]
- Al Saabi A, Allorge D, Sauvage FL, Tournel G, Gaulier JM, Marquet P, et al. Involvement of UDP-glucuronosyltransferases UGT1A9 and UGT2B7 in ethanol glucuronidation, and interactions with common drugs of abuse. Drug Metab Dispos 2013; 41: 568–74. [DOI] [PubMed] [Google Scholar]
- Ohno S, Kawana K, Nakajin S. Contribution of UDP-glucuronosyltransferase 1A1 and 1A8 to morphine-6-glucuronidation and its kinetic properties. Drug Metab Dispos 2008; 36: 688–94. [DOI] [PubMed] [Google Scholar]
- Wang H, Yuan L, Zeng S. Characterizing the effect of UDP-glucuronosyltransferase (UGT) 2B7 and UGT1A9 genetic polymorphisms on enantioselective glucuronidation of flurbiprofen. Biochem Pharmacol 2011; 82: 1757–63. [DOI] [PubMed] [Google Scholar]
- Laakkonen L, Finel M. A molecular model of the human UDP-glucuronosyltransferase 1A1, its membrane orientation, and the interactions between different parts of the enzyme. Mol Pharmacol 2010; 77: 931–9. [DOI] [PubMed] [Google Scholar]
- Lévesque E, Delage R, Benoit-Biancamano MO, Caron P, Bernard O, Couture F, et al. The impact of UGT1A8, UGT1A9, and UGT2B7 genetic polymorphisms on the pharmacokinetic profile of mycophenolic acid after a single oral dose in healthy volunteers. Clin Pharmacol Ther 2007; 81: 392–400. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Yamanaka H, Fujiwara R, Katoh M, Yokoi T. Stereoselective glucuronidation of 5-(4′-hydroxyphenyl)-5-phenylhydantoin by human UDP-glucuronosyltransferase (UGT) 1A1, UGT1A9, and UGT2B15: effects of UGT-UGT interactions. Drug Metab Dispos 2007; 35: 1679–86. [DOI] [PubMed] [Google Scholar]
- Bichlmaier I, Siiskonen A, Finel M, Yli-Kauhaluoma J. Stereochemical sensitivity of the human UDP-glucuronosyltransferases 2B7 and 2B17. J Med Chem 2006; 45: 1818–27. [DOI] [PubMed] [Google Scholar]
- Sten T, Qvisen S, Uutela P, Luukkanen L, Kostiainen R, Finel M. Prominent but reverse stereoselectivity in propranolol glucuronidation by human UDP-glucuronosyltransferases 1A9 and 1A10. Drug Metab Dispos 2006; 34: 1488–94. [DOI] [PubMed] [Google Scholar]
- Tougou K, Gotou H, Ohno Y, Nakamura A. Stereoselective glucuronidation and hydroxylation of etodolac by UGT1A9 and CYP2C9 in man. Xenobiotica 2004; 34: 449–61. [DOI] [PubMed] [Google Scholar]
- Court MH, Duan SX, Guillemette C, Journault K, Krishnaswamy S, Von Moltke LL, et al. Stereoselective conjugation of oxazepam by human UDP-glucuronosyltransferases (UGTs): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9. Drug Metab Dispos 2002; 30: 1257–65. [DOI] [PubMed] [Google Scholar]