Abstract
Aberrant expression of miR-511 is involved in the development of cancer, but the role of miR-511 in hepatocellular carcinoma (HCC) is not well documented. In this study, we explored the molecular mechanisms of miR-511 in hepatocarcinogenesis. Our results of bioinformatics analysis suggested that B cell translocation gene 1 (BTG1), a member of anti-proliferative gene family, was one of the putative targets of miR-511. The expression levels of miR-511 were significantly higher in 30 clinical HCC tissues than in corresponding peritumor tissues, and were negatively correlated with those of BTG1 in the HCC tissues (r=-0.6105, P<0.01). In human hepatoma cell lines HepG2 and H7402, overexpression of miR-511 dose-dependently inhibited the expression of BTG1, whereas knockdown of miR-511 dose-dependently increased the expression of BTG1. Luciferase reporter gene assays verified that miR-511 targeted the 3′UTR of BTG1 mRNA. In the hepatoma cells, overexpression of miR-511 significantly decreased BTG1-induced G1 phase arrest, which was rescued by overexpression of BTG1. Furthermore, overexpression of miR-511 promoted the proliferation of the hepatoma cells, which was rescued by overexpression of BTG1. Conversely, knockdown of miR-511 inhibited cell proliferation, which was reversed by knockdown of BTG1. In conclusion, miR-511 promotes the proliferation of human hepatoma cells in vitro by targeting the 3′UTR of BTG1 mRNA.
Keywords: hepatocellular carcinoma, HepG2 cells, H7402 cells, miR-511, B cell translocation gene 1 (BTG1), cell proliferation, cell cycle
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer in the world1, and the second leading cause of cancer death, with 700 000 deaths annually2,3,4. Moreover, the dysregulation of various genes contributes to the development and progression of HCC, making it a sophisticated and intractable disease5,6,7. Despite extensive research efforts, the prognosis of HCC remains poor, and the overall 5-year survival rate worldwide is only approximately 3%8. Hence, the precise mechanisms of hepatocarcinogenesis urgently need to be defined at different molecular levels.
MicroRNAs (miRNAs), a class of endogenous non-coding small RNAs containing approximately 22 nucleotides, can post-transcriptionally regulate gene expression9,10. By directly binding to the 3′-untranslated region (3′UTR) of their target mRNAs, miRNAs can mediate the degradation and/or repress the translation of these mRNAs11,12. miRNAs are involved in a variety of biological processes, including cell growth, differentiation, apoptosis, cell cycle regulation, embryonic development and disease progression13,14,15, and accumulating evidence indicates that miRNA expression is significantly dysregulated in different human cancers and that miRNAs can act as oncogenes or tumor suppressor genes to regulate tumorigenesis, progression and metastasis12,16. Specifically, previous reports revealed that miR-511 participated in the propagation of several cancers, such as lymphoblastic leukemia, lung adenocarcinoma and ovarian epithelial tumors17,18,19. However, the role of miR-511 in HCC is not well documented.
B cell translocation gene 1 (BTG1), which was originally identified as a translocation partner of the c-Myc gene in a case of B-cell chronic lymphocytic leukemia, belongs to the BTG anti-proliferative protein family (also known as transducer of ErbB2)20,21. Human BTG1, which is localized to chromosome 12q22, is ubiquitously distributed in different tissues20,22,23. Its expression peaks in the G0/G1 phases of the cell cycle and decreases when the cells progress through G1 phase24,25. Additionally, it is an important cofactor affecting cell proliferation, differentiation, apoptosis, angiogenesis and survival. As a tumor suppressor, BTG1 is involved in the pathogenesis of several diseases, including breast cancer, multiple sclerosis, ovarian cancer and prostate cancer26,27,28,29,30,31,32,33. In hepatocellular carcinoma, BTG1 expression is decreased, and it has recently been reported that BTG1 ameliorates liver steatosis by decreasing stearoyl-CoA desaturase 1 (SCD1) levels and altering hepatic lipid metabolism34. However, the underlying regulatory mechanism of BTG1 in HCC remains poorly understood.
In the present study, we investigated the significance of miR-511 in liver cancer. Interestingly, our data show that miR-511 promotes the proliferation of hepatoma HepG2 and H7402 cells by directly targeting the 3′UTR of BTG1 mRNA. Our findings provide new insights into the mechanisms by which miR-511 modulates the development of HCC.
Materials and methods
Patient samples
The thirty HCC tissues and their corresponding nearby peritumorous liver tissues utilized in this study were obtained from the Tianjin First Central Hospital (Tianjin, China) after surgical resection. Written consent was obtained from each patient approving the use of their tissue for research purposes after surgery. All study procedures were in compliance with the regulations of the Institute of Research Ethics Committee at Nankai University (Tianjin, China). The medical records of the patients are listed in Supplementary Table S1.
Cell lines and cell culture
The human hepatoma cell lines HepG2 and H740216 were maintained in Dulbecco's modified Eagle's medium and RPMI medium 1640, respectively (Gibco, CA, USA). The media were supplemented with heat-inactivated 10% fetal bovine serum (FBS, Gibco, CA, USA), 100 U/mL penicillin and 100 mg/mL streptomycin in 5% CO2 at 37 °C.
Plasmids and construction of the 3′UTR of BTG1
The fragment containing the coding sequence (CDS) of BTG1 was cloned into the pcDNA3.1 vector, and a 326 bp fragment containing the target site of miR-511 in the 3′UTR region of BTG1 mRNA was cloned into the pGL3-control vector (Promega, Madison, WI, USA) immediately downstream of the stop codon of the luciferase gene to generate pGL3-BTG1-wt. A mutant construct of the BTG1 3′UTR (named as pGL3-BTG1-mut) in which 7 nucleotides in the core seed sequence of miR-511 were substituted was generated using overlapping extension PCR. The following primers were used to construct these vectors: pcDNA3.1-BTG1 forward, 5′-CCGGAATTCATGCATCCCTTCTACACC-3′, reverse, 5′-GCTCTAGAACCTGATACAGTCATCATAT-3′ pGL3-BTG1-wt forward, 5′-GCTCTAGATTTCAGTTTCTCCCAGACATA-3′, reverse, 5′-GGGGGCCGGCCAGATTCTGGTCACTTGCTACT-3′ pGL3-BTG1-mut forward, 5′-TGTATAAATGTACATTTTCTGTAACTAGTAAGCATGA-3′, reverse, 5′-TCATGCTTACTAGTTACAGAAAATGTACATTTATACA-3′.
RNA extraction, reverse-transcription and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells (or tissues) using TRIzol reagent (Invitrogen, UK), and first-strand cDNA was synthesized as reported previously35. To detect mature miR-511, total RNA was polyadenylated using poly (A) polymerase (Ambion, Austin, TX, USA) according to the manufacturer's protocol. Reverse transcription was performed using poly (A)-tailed total RNA and a reverse transcription primer with ImPro-II Reverse Transcriptase (Promega, Madison, WI, USA) as reported previously36. QRT-PCR was performed on a Bio-Rad sequence detection system according to the manufacturer's instructions using double-stranded DNA-specific Fast Start Universal SYBR Green Master Mix (Roche, Indianapolis, IN, USA). All experiments were conducted in duplicate in three independent assays. Relative fold-changes in transcription were calculated using the 2-ΔΔCt method37. β-Actin was used as an internal control for normalization, and U6 was used as an internal control to normalize miR-511 levels. The following primers were used: BTG1 forward, 5′-CATCTCCAAGTTTCTCCGCACC-3′, reverse, 5′-GCGAATACAACGGTAACCCGATC-3′ β-actin forward, 5′-CTTAGTTGCGTTACACCCTTTC-3′, reverse, 5′-CACCTTCACCGTTCCAGTTT-3′ miR-511 forward, 5′-ACUGACGUCUCGUUUUCUGUG-3′, reverse, 5′-GCGAGCACAGAATTAATACGAC-3′ U6 forward, 5′-AGAGCCTGTGGTGTCCG-3′, reverse, 5′-CATCTTCAAAGCACTTCCCT-3′.
Cell transfection
The cells were cultured in a 6-well or a 24-well plate for 24 h and then transfected with plasmids, miRNAs or small-interfering RNA (siRNAs). All transfections were performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. BTG1 siRNA and non-specific scrambled control oligonucleotides, miR-511 (or anti-miR-511), miRNA control and anti-miRNA control were synthesized by RiboBio (Guangzhou, China). The siRNA duplex sequences used were as follows: si-BTG1-1, 5′-AGCTGTAAGGAGGAACTTCdTdT-3′ si-BTG1-2, 5′-CAACCCAGAGTGTGAGTTCdTdT-3′.
Protein extraction and Western blot analysis
Total protein was extracted from the cells using radio-immunoprecipitation assay (RIPA, Solarbio, China) buffer according to the manufacturer's protocol. Western blotting was conducted as described previously1 using rabbit anti-BTG1 polyclonal antibody (Abgent, USA) and mouse anti-β-actin monoclonal antibody (Abcam, UK). All experiments were repeated three times.
Flow cytometry analysis
For cell cycle analysis, HepG2 and H7402 cells were harvested 48 h after transfection with the indicated reagents and washed twice with cold PBS. The washed cells were fixed in 75% ethanol at 4 °C overnight and then rinsed twice with PBS before being treated with 50 μg/mL propidium iodide solution (Sigma, St Louis, MO, USA) containing 50 μg/mL Rnase A (Sigma) at 37 °C for 60 min. Stained cells were analyzed on a FACScan flow cytometer (Becton Dickinson, Bedford, MA, USA).
MTT assays
HepG2 and H7402 cells were seeded onto 96-well plates (1000 cells/well) for 24 h before transfection, and MTT assays were used to assess cell proliferation every day from the first day until the third day after transfection according to a previously described protocol11.
Colony formation assays
Forty-eight hours after transfection with the indicated reagents, approximately 1000 viable treated cells were plated in 6-well plates and maintained in complete medium for approximately 2 weeks. Resultant colonies were fixed with methanol and stained with crystal violet. All assays were repeated at least three times. The colonies were counted using a dissecting microscope, and the colony formation efficiency was also calculated.
Luciferase reporter gene assays
HepG2 and H7402 cells were transferred into 24-well plates at a density of 3×104 cells per well. After 24 h, the cells were transiently co-transfected with the pRL-TK plasmid (Promega, Madison, WI, USA) containing the Renilla luciferase gene, which is used for internal normalization, and various constructs containing the seed sequence or mutant seed sequence of the BTG1 3′UTR. At 48 h post-transfection, a standard dual luciferase reporter gene assay was performed, and the results were normalized using pRL-TK. All experiments were performed at least three times.
Statistical analysis
Each experiment was repeated at least three times. Statistical significance was assessed by comparing mean value±standard deviation (SD) using Student's t test for independent groups. *P<0.05, **P<0.01, and ***P<0.001 indicated significant differences, and non-significant differences were denoted by NS. Pearson's correlation coefficient was used to determine the correlation between the levels of miR-511 and BTG1 mRNA in HCC tissues.
Results
miR-511 is up-regulated and its expression is inversely correlated with BTG1 expression in clinical HCC tissues
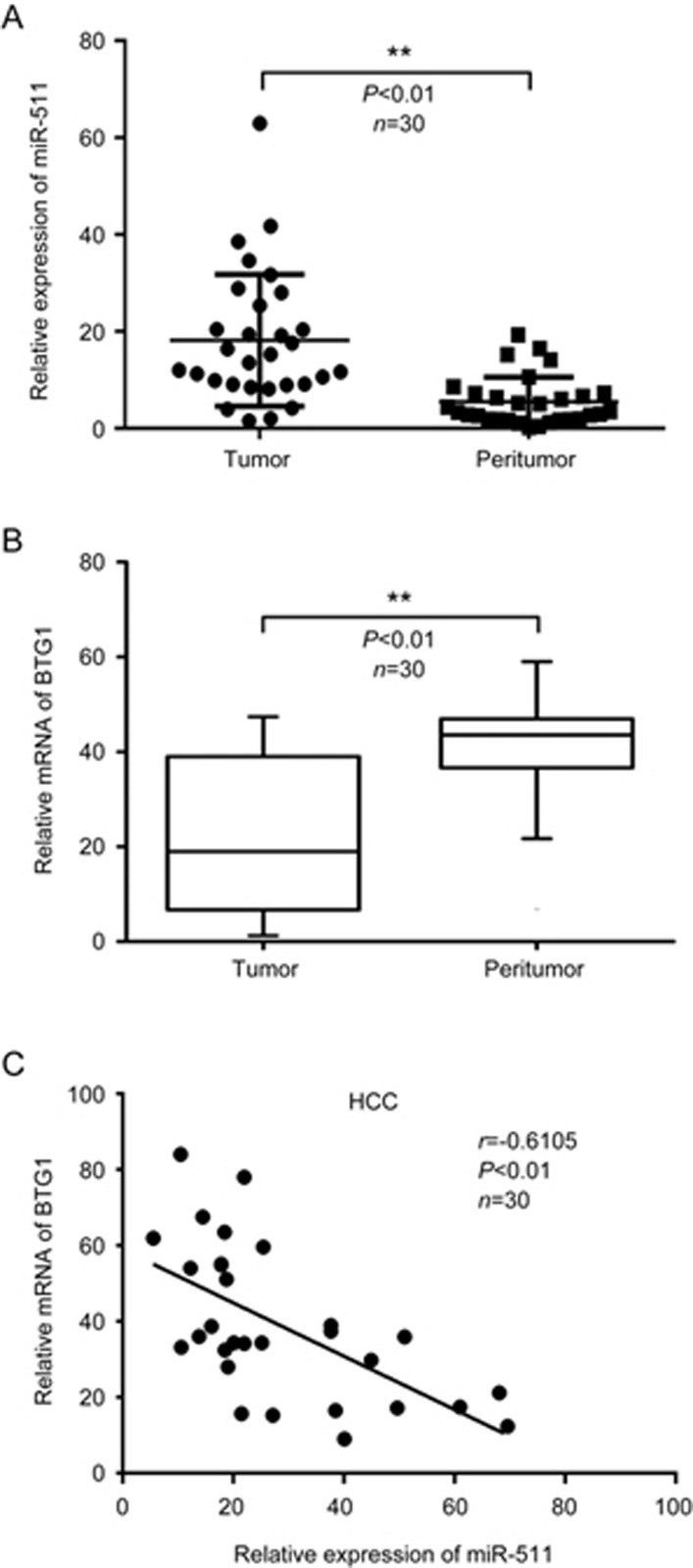
miR-511 reportedly participates in the propagation of several cancers17,18,19, indicating that miR-511 is crucial for tumorigenesis. However, the role of miR-511 in HCC is not well documented. In this study, a qRT-PCR analysis revealed that the expression levels of miR-511 were much higher in HCC tissues than in adjacent peritumor tissues (Figure 1A), suggesting that miR-511 plays important roles in hepatocarcinogenesis. To identify the underlying mechanism of miR-511, we screened its target genes using the miRNA prediction software programs TargetScan (http://www.targetscan.org/), miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/genepub.html) and Oncomine Main (https://www.oncomine.org/resource/login.html)16,38,39,40. Interestingly, we observed that BTG1, a member of an anti-proliferative gene family, was one of the putative targets with high predicted scores. Because BTG1 contributes to cell growth, differentiation, apoptosis, angiogenesis and survival in various cancers27,32,34,41, we focused on the investigation of BTG1. Interestingly, the expression levels of BTG1 were much lower in HCC tissues than in adjacent peritumor tissues (Figure 1B). Next, we evaluated the relationship between the expression levels of miR-511 and BTG1 in 30 clinical HCC tissues. As expected, we demonstrated that the expression levels of miR-511 had a significant negative correlation with those of BTG1 mRNA in the above HCC tissues (Pearson's correlation coefficient r=-0.6105, P<0.01) (Figure 1C), suggesting that miR-511 might target BTG1 mRNA in HCC tissues. Thus, we conclude that miR-511 is up-regulated and that its expression inversely correlates with that of BTG1 in clinical HCC tissues.
Figure 1.
miR-511 is up-regulated and its expression inversely correlates with BTG1 expression in clinical HCC tissues. (A and B) The expression levels of miR-511 and BTG1 mRNA were examined by qRT-PCR in 30 clinical HCC tissue and paired peritumor tissue samples. (C) Correlation of miR-511 levels with BTG1 mRNA levels was examined by qRT-PCR analysis in 30 clinical HCC tissue samples (Pearson's correlation coefficient, r=−0.6105). Data are presented as the mean±SD of three independent experiments. **P<0.01; Student's t test.
miR-511 suppresses the expression of BTG1 in hepatoma cells
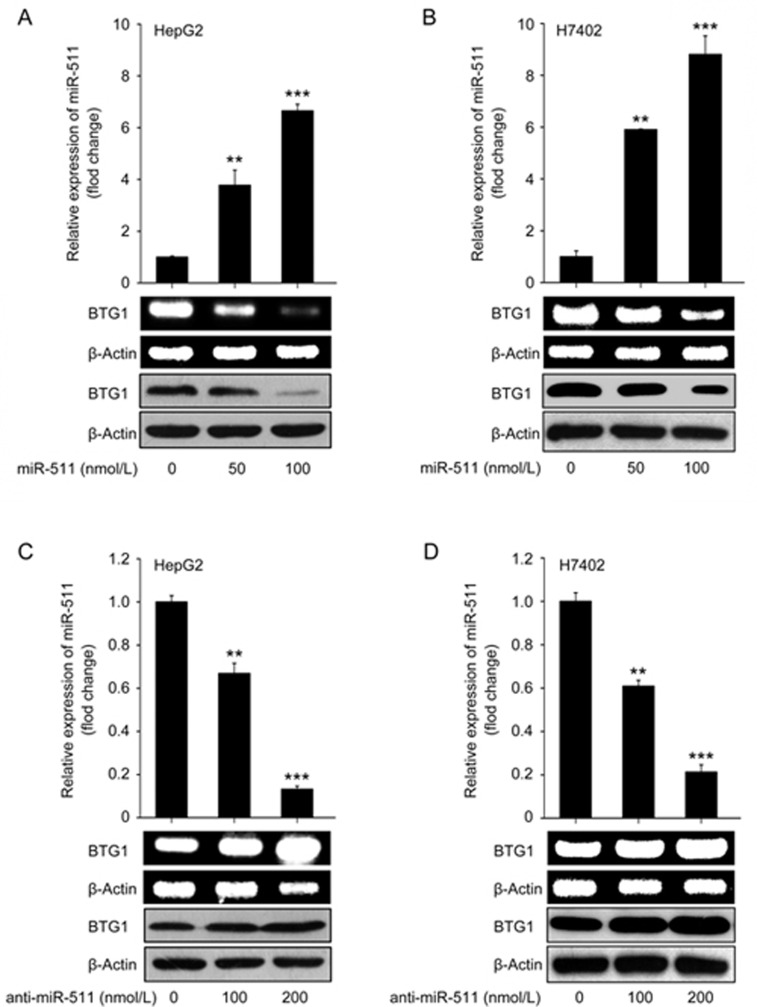
Next, we explored the effect of miRNA-511 on BTG1 expression in hepatoma cells by transiently transfecting HepG2 and H7402 cells with miR-511 (or anti-miR-511). Our data showed that the overexpression of miR-511 was able to dose-dependently down-regulate BTG1 at the mRNA and protein levels (Figure 2A and 2B). On the contrary, the expression levels of BTG1 were increased in HepG2 and H7402 cells when the endogenous miR-511 was impaired by anti-miR-511 (Figure 2C and 2D), indicating that miR-511 inhibits BTG1 expression. Transfection efficiency was validated by qRT-PCR (Figure 2A-2D). Taken together, we conclude that miR-511 is able to suppress the expression of BTG1 in hepatoma cell lines.
Figure 2.
miR-511 suppresses the expression of BTG1 in hepatoma cells. (A and B) The mRNA and protein levels of BTG1 were detected in HepG2 and H7402 cells transfected with miR-511 using RT-PCR and Western blotting, respectively. The transfection efficiency of miR-511 was determined by qRT-PCR. (C and D) The mRNA and protein levels of BTG1 were measured in HepG2 and H7402 cells transfected with anti-miR-511 by RT-PCR and Western blotting, respectively. The transfection efficiency of anti-miR-511 was assessed by qRT-PCR. Data are presented as the mean±SD of three independent experiments. *P<0.05, **P<0.01, ***P<0.001; Student's t test.
miR-511 inhibits the expression of BTG1 by directly targeting its mRNA 3′UTR
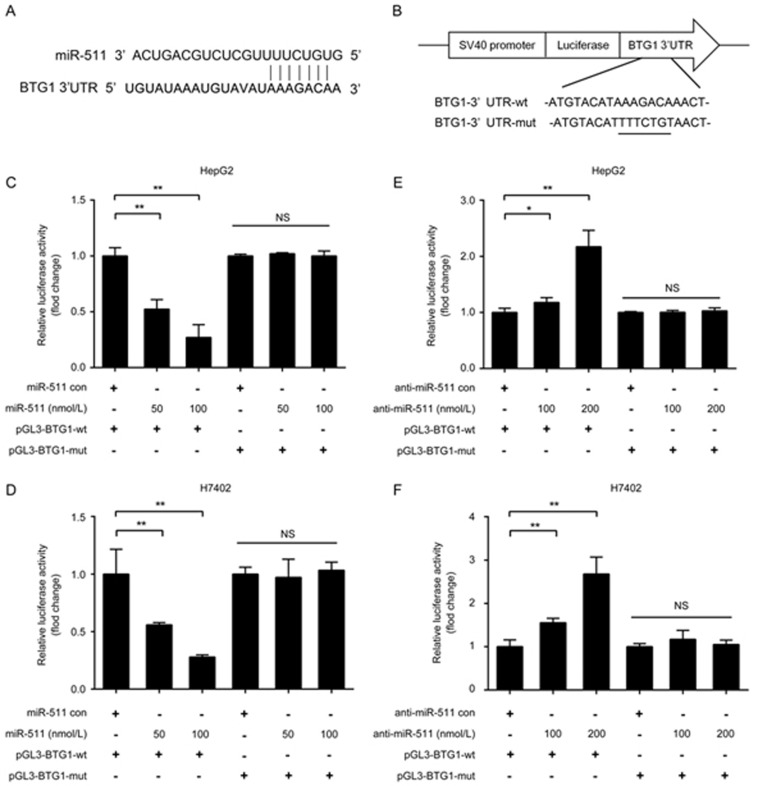
To gain insight into the mechanism by which miR-511 inhibits BTG1, we identified the miR-511 binding site in the 3′UTR of BTG1 mRNA using a bioinformatics analysis (Figure 3A). Accordingly, we cloned the 3′UTR of BTG1 mRNA (or its mutant) into the downstream region of the pGL3-control luciferase reporter gene vector (named pGL3-BTG1-wt or pGL3-BTG1-mut) (Figure 3B). Luciferase reporter gene assays indicated that miR-511 remarkably suppressed the luciferase activities of pGL3-BTG1-wt in HepG2 and H7402 cells in a dose-dependent manner, but luciferase activities were not suppressed when its seed sequence was mutated (Figure 3C and 3D). Moreover, abolishing endogenous miR-511 expression with anti-miR-511 resulted in the increase of luciferase activities of the pGL3-BTG1-wt but not of the pGL3-BTG1-mut (Figure 3E and 3F). Therefore, we conclude that miR-511 can inhibit the expression of BTG1 by directly targeting the 3′UTR of BTG1 mRNA in hepatoma cells.
Figure 3.
miR-511 inhibits the expression of BTG1 by directly targeting the 3′UTR of its mRNA. (A) A schematic representation of the predicted binding site of miR-511 in the 3′UTR of BTG1 mRNA. (B) The mutant seed region was generated at the 3′UTR of BTG1 mRNA as indicated. A BTG1 3′UTR fragment containing the wild-type or mutant miR-511-binding sequence was cloned into the downstream region of the pGL3-control luciferase reporter gene vector. (C and D) The effect of miR-511 on the pGL3-BTG1-wt and pGL3-BTG1-mut reporters in HepG2 and H7402 cells was measured using luciferase reporter gene assays. (E and F) The effect of anti-miR-511 on the pGL3-BTG1-wt and pGL3-BTG1-mut reporters in HepG2 and H7402 cells was examined using luciferase reporter gene assays. Data are presented as the mean±SD of three independent experiments. *P<0.05, **P<0.01, NS, no significance; Student's t test.
miR-511 promotes the proliferation of HCC cells by reducing BTG1-induced G1 phase arrest
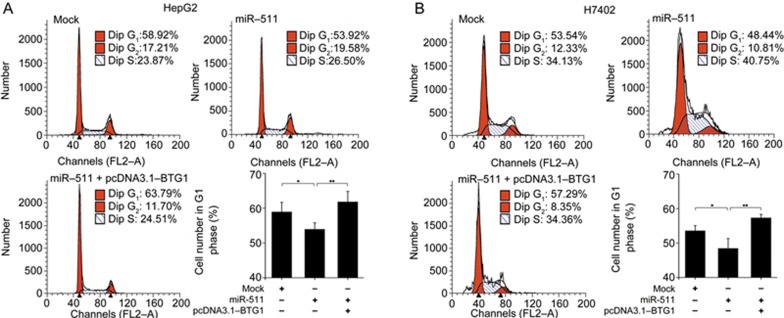
Given that the expression of BTG1 is highest in the G0/G1 phases of the cell cycle and is down-regulated when cells progress through the G1 phase20,24, we examined the impact of miR-511 on the cell cycle using flow cytometry. Interestingly, our results showed that the overexpression of miR-511 reduced the percentage of HepG2 and H7402 cells in G1 phase, but that the overexpression of BTG1 rescued the miR-511-induced decrease in cells in G1 phase (Figure 4A and 4B). Thus, we conclude that miR-511 promotes the proliferation of HCC cells by reducing BTG1-induced G1 phase arrest.
Figure 4.
miR-511 promotes the proliferation of HCC cells by inhibiting BTG1-induced G1 phase arrest. (A and B) Flow cytometry analysis was conducted to examine the effect of miR-511 (or miR-511/BTG1) on the cell cycle in HepG2 cells and H7402 cells. Data are presented as the mean±SD of three independent experiments. *P<0.05, **P<0.01; Student's t test.
miR-511 promotes the proliferation of HCC cells by inhibiting BTG1
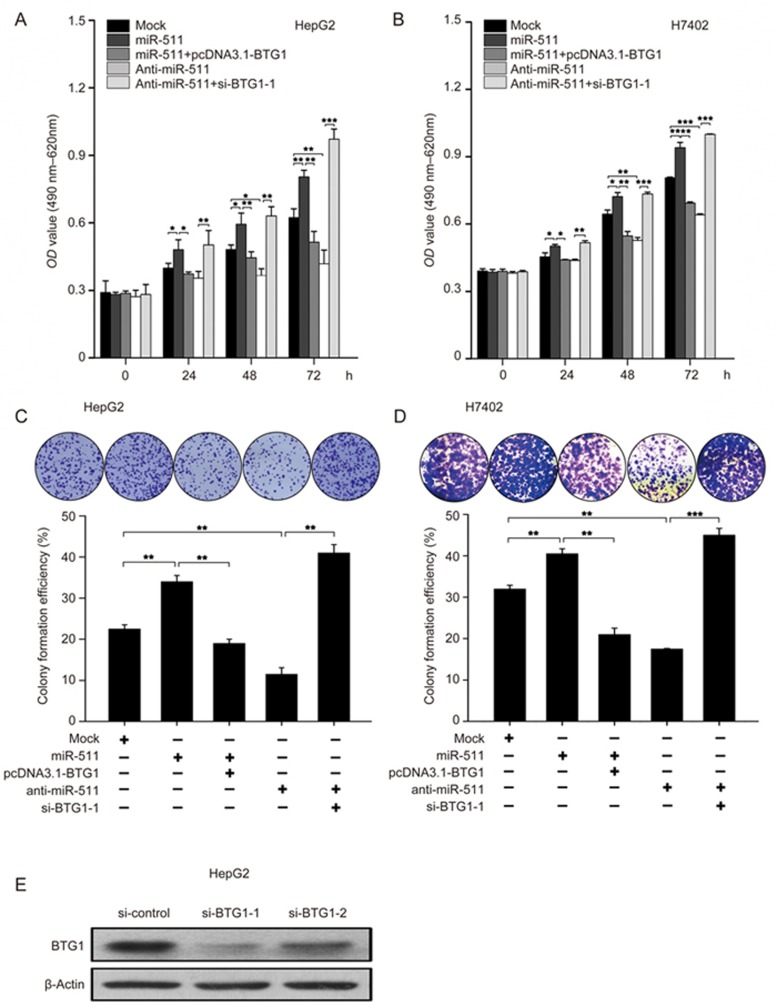
Next, we investigated the effect of miR-511 on the proliferation of hepatoma cells. MTT assays demonstrated that cell proliferation capacity was increased when HepG2 and H7402 cells were transfected with miR-511, but that overexpression of BTG1 could efficiently block the event mediated by miR-511 in the cells. Conversely, anti-miR-511 could decrease cell proliferation, and the co-transfection of anti-miR-511 and BTG1 siRNA abolished the anti-miR-511-induced attenuation of cell proliferation (Figure 5A and 5B). Moreover, these results were validated in colony formation assays (Figure 5C and 5D), which suggested that miR-511 promotes the proliferation of hepatoma cells by inhibiting BTG1. Furthermore, the efficiency of BTG1 siRNA transfection in HepG2 cells was validated by Western blotting (Figure 5E). Thus, we conclude that miR-511 promotes the proliferation of hepatoma cells by inhibiting BTG1.
Figure 5.
miR-511 promotes the proliferation of HCC cells by inhibiting BTG1. (A and B) The effect of miR-511 (or miR-511/BTG1, anti-miR-511, anti-miR-511/si-BTG1-1) on proliferation of HepG2 and H7402 cells was assessed using MTT assays. (C and D) The effect of miR-511 (or miR-511/BTG1, anti-miR-511, anti-miR-511/si-BTG1-1) on the proliferation of HepG2 and H7402 cells was assessed using colony-formation assays. The colony formation efficiency was also calculated. (E) The efficiency of BTG1 siRNA (si-BTG1-1 or si-BTG1-2) transfection in HepG2 cells was validated by Western blotting. Data are presented as the mean±SD of three independent experiments. *P<0.05, **P<0.01, ***P<0.001; Student's t test.
Discussion
HCC is one of the most malignant cancers worldwide and involves multiple alterations of signaling pathways and the dysregulation of oncogene or anti-oncogenes3,42. Moreover, miRNA networks have been reported to be associated with hepatocarcinogenesis via distinct mechanisms, such as the suppression of gene expression by a RNA-induced silencing complex, which mediates mRNA cleavage or translational repression by binding to the 3′UTR of target mRNAs43. The aberrant expression of miR-511 is crucial for various cancers, such as ovarian epithelial tumors, lymphoblastic leukemia and lung cancer17,18,19, but the role of miR-511 in HCC is poorly understood. Thus, we investigated the significance of miR-511 in hepatocarcinogenesis.
To better understand the significance of miR-511 in liver cancer, we evaluated the expression levels of miR-511 in 30 pairs of clinical HCC tissues and their adjacent peritumor tissues. Interestingly, we found that the expression levels of miR-511 were much higher in HCC tissues than in their adjacent peritumor tissues. To identify the mechanism of action of miR-511, we predicted the target genes of miR-511 using bioinformatics tools, which identified BTG1, a member of an anti-proliferative gene family, as a putative target gene. BTG1 reportedly plays important roles in cell growth, differentiation, apoptosis, angiogenesis and survival in various cancers5,6, and it negatively regulates cell proliferation and serves as a novel prognostic indicator of hepatocellular carcinoma20. Therefore, we focused on investigating BTG1. To this end, we examined the association between miR-511 and BTG1 expression in clinical HCC samples. As expected, the expression levels of miR-511 were negatively correlated with those of BTG1 mRNA in the above samples, which suggests that miR-511 may target BTG1. Moreover, we found that miR-511 inhibited the expression of BTG1 at the mRNA and protein levels in hepatoma cells by targeting the 3′UTR of BTG1 mRNA. It has been reported that BTG1 also plays crucial roles in cell cycle regulation. Specifically, its expression peaks in the G0/G1 phases of the cell cycle and decreases when the cells progress through G144,45,46. Furthermore, BTG1 negatively regulates the cell cycle in hepatocellular carcinoma47. Therefore, we examined the impact of miR-511 on the cell cycle in hepatoma cells. As expected, our results showed that miR-511 promotes the proliferation of HCC cells by reducing BTG1-induced G1 phase arrest, which supports the idea that miR-511 promotes the proliferation of HCC cells by modulating the cell cycle. Functionally, we demonstrated that miR-511 promoted the proliferation of hepatoma cells by inhibiting BTG1 in vitro.
Previous studies showed that miR-511 is overexpressed in B-cell acute lymphoblastic leukemia18, and miR-511 expression levels were negatively associated with the overall survival of patients with HCC48. Our data are consistent with these reports. However, some other studies reported that miR-511 is down-regulated in HCC tissues39,49,50. This discrepancy may be related to the heterogeneity of tumors. In this study, our data support the idea that miR-511 promotes the proliferation of hepatoma cells because its target gene, BTG1, is an important inhibitor of proliferation. Additionally, we partially elucidated the role of miR-511 in HCC in this study; miR-511 may also promote cell proliferation via other target genes, and other functions of miR-511 need to be further investigated. Therapeutically, miR-511 may serve as a potential target for the treatment of HCC.
In summary, in this study we report that miR-511 is able to promote the proliferation of hepatoma cells by targeting the 3′UTR of BTG1 mRNA. This finding provides new insights into the roles of miR-511 in hepatocarcinogenesis.
Author contribution
Xiao-dong ZHANG conceived the projects; Xiao-dong ZHANG, Jun-qi NIU and Li-hong YE designed the experiments and drafted the manuscript; Shu-qin ZHANG designed the experiments, performed the experiments and drafted the manuscript; Zhe YANG and Xiao-li CAI performed the experiments and revised the manuscript; Man ZHAO, Ming-ming SUN, and Jiong LI performed the experiments; Guo-xing FENG and Jin-yan FENG revised the manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No 81272218, 81372186 and 31670769), the National Basic Research Program of China (973 Program, No 2015CB553703 and 2015CB553905) and the Project of Prevention and Treatment of Key Infectious Diseases (No 2014ZX0002002-005).
Footnotes
Supplementary information is available on the Acta Pharmacologica Sinica's website.
Supplementary Information
Clinical characteristics of HCC samples.
References
- Zhang T, Zhang J, You X, Liu Q, Du Y, Gao Y, et al. Hepatitis B virus X protein modulates oncogene Yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology 2012; 56: 2051–9. [DOI] [PubMed] [Google Scholar]
- Sze KM, Chu GK, Lee JM, Ng IO. C-terminal truncated hepatitis B virus X protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology 2013; 57: 131–9. [DOI] [PubMed] [Google Scholar]
- Sun X, Niu X, Chen R, He W. Chen Kang R, et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology 2016; 64: 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZP, Xiao ZL, Yang Z, Li J, Feng GX, Chen FQ, et al. Hepatitis B virus X protein promotes human hepatoma cell growth via upregulation of transcription factor AP2alpha and sphingosine kinase 1. Acta Pharmacol Sin 2015; 36: 1228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene 2007; 26: 7499–507. [DOI] [PubMed] [Google Scholar]
- Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, et al. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet 1997; 16: 260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan FS, Ali I, Afridi UK, Ishtiaq M, Mehmood R. Epigenetic mechanisms regulating the development of hepatocellular carcinoma and their promise for therapeutics. Hepatol Int 2017; 11: 45–53. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang K, Yan Z, Xia Y, Li J, Shi L, et al. Axl expression stratifies patients with poor prognosis after hepatectomy for hepatocellular carcinoma. PLoS One 2016; 11: e0154767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004; 432: 226–30. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu N, Yang X, Tu L, Zhang X. Small RNA-mediated responses to low- and high-temperature stresses in cotton. Sci Rep 2016; 6: 35558–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen F, Zhao M, Yang Z, Zhang S, Ye L, et al. MiR-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mRNA 3′UTR. Biochem Biophys Res Commun 2016; 480: 455–60. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Gong L, Wang J, Tu Q, Yao L, Zhang JR, et al. miR-10b exerts oncogenic activity in human hepatocellular carcinoma cells by targeting expression of CUB and sushi multiple domains 1 (CSMD1). BMC Cancer 2016; 16: 806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersani F, Lingua MF, Morena D, Foglizzo V, Miretti S, Lanzetti L, et al. Deep sequencing reveals a novel miR-22 regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Res 2016; 76: 6095–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh SL, Das S. Tumour biology of obesity-related cancers: understanding the molecular concept for better diagnosis and treatment. Tumour Biol 2016; 37: 14363–80. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zheng XY. MicroRNA-490 inhibits tumorigenesis and progression in breast cancer. Onco Targets Ther 2016; 9: 4505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cui M, Sun BD, Liu FB, Zhang XD, Ye LH. MiR-506 suppresses proliferation of hepatoma cells through targeting YAP mRNA 3′UTR. Acta Pharmacol Sin 2014; 35: 1207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, et al. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology 2010; 57: 734–43. [DOI] [PubMed] [Google Scholar]
- Luna-Aguirre CM, de la Luz Martinez-Fierro M, Mar-Aguilar F, Garza-Veloz I, Trevino-Alvarado V, Rojas-Martinez A, et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark 2015; 15: 299–310. [DOI] [PubMed] [Google Scholar]
- Zhang HH, Pang M, Dong W, Xin JX, Li YJ, Zhang ZC, et al. miR-511 induces the apoptosis of radioresistant lung adenocarcinoma cells by triggering BAX. Oncol Rep 2014; 31: 1473–9. [DOI] [PubMed] [Google Scholar]
- Kanda M, Sugimoto H, Nomoto S, Oya H, Hibino S, Shimizu D, et al. B-cell translocation gene 1 serves as a novel prognostic indicator of hepatocellular carcinoma. Int J Oncol 2015; 46: 641–8. [DOI] [PubMed] [Google Scholar]
- Xiao F, Deng J, Yu J, Guo Y, Chen S, Guo F. A novel function of B-cell translocation gene 1 (BTG1) in the regulation of hepatic insulin sensitivity in mice via c-Jun. FASEB J 2016; 30: 348–59. [DOI] [PubMed] [Google Scholar]
- Cho JW, Kim JJ, Park SG, Lee DH, Lee SC, Kim HJ, et al. Identification of B-cell translocation gene 1 as a biomarker for monitoring the remission of acute myeloid leukemia. Proteomics 2004; 4: 3456–63. [DOI] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem 1996; 271: 15034–44. [DOI] [PubMed] [Google Scholar]
- Cortes U, Moyret-Lalle C, Falette N, Duriez C, Ghissassi FE, Barnas C, et al. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol Carcinog 2000; 27: 57–64. [PubMed] [Google Scholar]
- Rouault JP, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet 1996; 14: 482–6. [DOI] [PubMed] [Google Scholar]
- Camina-Tato M, Morcillo-Suarez C, Navarro A, Fernandez M, Horga A, Montalban X, et al. Genetic association between polymorphisms in the BTG1 gene and multiple sclerosis. J Neuroimmunol 2009; 213: 142–7. [DOI] [PubMed] [Google Scholar]
- Li W, Zou ST, Zhu R, Wan JM, Xu Y, Wu HR. Bcell translocation 1 gene inhibits cellular metastasisassociated behavior in breast cancer. Mol Med Rep 2014; 9: 2374–80. [DOI] [PubMed] [Google Scholar]
- Rivera-Gonzalez GC, Droop AP, Rippon HJ, Tiemann K, Pellacani D, Georgopoulos LJ, et al. Retinoic acid and androgen receptors combine to achieve tissue specific control of human prostatic transglutaminase expression: a novel regulatory network with broader significance. Nucleic Acids Res 2012; 40: 4825–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C, Unoki M, Nakamura Y. Identification and allelic frequencies of novel single-nucleotide polymorphisms in the DUSP1 and BTG1 genes. J Human Genet 2001; 46: 155–7. [DOI] [PubMed] [Google Scholar]
- Urzua U, Roby KF, Gangi LM, Cherry JM, Powell JI, Munroe DJ. Transcriptomic analysis of an in vitro murine model of ovarian carcinoma: functional similarity to the human disease and identification of prospective tumoral markers and targets. J Cell Physiol 2006; 206: 594–602. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shao C, Shi CH, Zhang L, Yue HH, Wang PF, et al. Change of the cell cycle after flutamide treatment in prostate cancer cells and its molecular mechanism. Asian J Androl 2005; 7: 375–80. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Gou WF, Chen S, Takano Y, Xiu YL, Zheng HC. BTG1 expression correlates with the pathogenesis and progression of ovarian carcinomas. Int J Mol Sci 2013; 14: 19670–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R, Li W, Xu Y, Wan J, Zhang Z. Upregulation of BTG1 enhances the radiation sensitivity of human breast cancer in vitro and in vivo. Oncol Rep 2015; 34: 3017–24. [DOI] [PubMed] [Google Scholar]
- Xiao F, Deng J, Guo Y, Niu Y, Yuan F, Yu J, et al. BTG1 ameliorates liver steatosis by decreasing stearoyl-CoA desaturase 1 (SCD1) abundance and altering hepatic lipid metabolism. Sci Signal 2016; 9: ra50. [DOI] [PubMed] [Google Scholar]
- Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res 2015; 75: 846–57. [DOI] [PubMed] [Google Scholar]
- Zhang T, Zhang J, Cui M, Liu F, You X, Du Y, et al. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia 2013; 15: 1282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Wang Y, Sun B, Xiao Z, Ye L, Zhang X. MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem Biophys Res Commun 2014; 444: 270–5. [DOI] [PubMed] [Google Scholar]
- Fassan M, Volinia S, Palatini J, Pizzi M, Baffa R, De Bernard M, et al. MicroRNA expression profiling in human Barrett's carcinogenesis. Int J Cancer 2011; 129: 1661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis 2012; 33: 1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Shi H, Li J, Yang Z, Fang R, Ye L, et al. MiR-30e suppresses proliferation of hepatoma cells via targeting prolyl 4-hydroxylase subunit alpha-1 (P4HA1) mRNA. Biochem Biophys Res Commun 2016; 472: 516–22. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Rouault J, Magaud J, Berthet C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett 2001; 497: 67–72. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ye LH, Zhang XD. A mutant of hepatitis B virus X protein (HBx Delta 127) enhances hepatoma cell migration via osteopontin involving 5-lipoxygenase. Acta Pharmacol Sin 2010; 31: 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YE, Wang Y, Chen FQ, Feng JY, Yang G, Feng GX, et al. Post-transcriptional modulation of protein phosphatase PPP2CA and tumor suppressor PTEN by endogenous siRNA cleaved from hairpin within PTEN mRNA 3′UTR in human liver cells. Acta Pharmacol Sin 2016; 37: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Micheli L, D'Andrea G, De Bardi M, Scheijen B, Ciotti M, et al. Altered cerebellum development and impaired motor coordination in mice lacking the Btg1 gene: Involvement of cyclin D1. Dev Biol 2015; 408: 109–25. [DOI] [PubMed] [Google Scholar]
- Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 2014; 26: 262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Tao T, Xu B, Lu K, Zhang L, Jiang L, et al. BTG1 potentiates apoptosis and suppresses proliferation in renal cell carcinoma by interacting with PRMT1. Oncol Lett 2015; 10: 619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun GG, Lu YF, Cheng YJ, Yang CR, Liu Q, Jing SW, et al. Expression of BTG1 in hepatocellular carcinoma and its correlation with cell cycles, cell apoptosis, and cell metastasis. Tumour Biol 2014; 35: 11771–9. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chong CC, Chen GG, Lai PB. A seven-microRNA expression signature predicts survival in hepatocellular carcinoma. PLoS One 2015; 10: e0128628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Dong W, Meng X, Liu H, Liao H, Liu S. MiR-511 inhibits growth and metastasis of human hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol 2015; 36: 4453–9. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol 2012; 18: 5442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical characteristics of HCC samples.