Abstract
IMPORTANCE
Dysfunction related to γ-aminobutyric acid (GABA)–ergic neurotransmission in the pathophysiology of major psychosis has been well established by the work of multiple groups across several decades, including the widely replicated downregulation of GAD1. Prior gene expression and network analyses within the human hippocampus implicate a broader network of genes, termed the GAD1 regulatory network, in regulation of GAD1 expression. Several genes within this GAD1 regulatory network show diagnosis- and sector-specific expression changes within the circuitry of the hippocampus, influencing abnormal GAD1 expression in schizophrenia and bipolar disorder.
OBJECTIVE
To investigate the hypothesis that aberrant DNA methylation contributes to circuit- and diagnosis-specific abnormal expression of GAD1 regulatory network genes in psychotic illness.
DESIGN, SETTING, AND PARTICIPANTS
This epigenetic association study targeting GAD1 regulatory network genes was conducted between July 1, 2012, and June 30, 2014. Postmortem human hippocampus tissue samples were obtained from 8patients with schizophrenia, 8 patients with bipolar disorder, and 8 healthy control participants matched for age, sex, postmortem interval, and other potential confounds from the Harvard Brain Tissue Resource Center, McLean Hospital, Belmont,Massachusetts. We extracted DNA from laser-microdissected stratum oriens tissue of cornu ammonis 2/3 (CA2/3) and CA1 postmortem human hippocampus, bisulfite modified it, and assessed it with the Infinium HumanMethylation450 BeadChip (Illumina, Inc). The subset of CpG loci associated with GAD1 regulatory network genes was analyzed in R version 3.1.0 software (R Foundation) using the minfi package. Findings were validated using bisulfite pyrosequencing.
MAIN OUTCOMES AND MEASURES
Methylation levels at 1308 GAD1 regulatory network–associated CpG loci were assessed both as individual sites to identify differentially methylated positions and by sharing information among colocalized probes to identify differentially methylated regions.
RESULTS
A total of 146 differentially methylated positions with a false detection rate lower than 0.05 were identified across all 6 groups (2 circuit locations in each of 3 diagnostic categories), and 54 differentially methylated regions with P < .01 were identified in single-group comparisons. Methylation changes were enriched in MSX1, CCND2, and DAXX at specific loci within the hippocampus of patients with schizophrenia and bipolar disorder.
CONCLUSIONS AND RELEVANCE
This work demonstrates diagnosis- and circuit-specific DNA methylation changes at a subset of GAD1 regulatory network genes in the human hippocampus in schizophrenia and bipolar disorder. These genes participate in chromatin regulation and cell cycle control, supporting the concept that the established GABAergic dysfunction in these disorders is related to disruption of GABAergic interneuron physiology at specific circuit locations within the human hippocampus.
One of the most widely replicated findings in the molecular analysis of schizophrenia (SZ) is downregulation of the glutamic acid decarboxylase 67 gene (GAD1),1–5 resulting in decreased expression of the glutamic acid decarboxylase 67-kDa enzyme and impaired γ-aminobutyric acid (GABA)–ergic neurotransmission in the brain. This decreased GABAergic tone in the brain of affected individuals is related to the cognitive dysfunction that is a core component of the disorder,6 and γ-band power, a physiological measure associated with GABAergic tone on electroencephalography, is abnormal in patients with SZ.7,8
While GAD1 has not been implicated in genome-wide association studies of psychotic disorders,9 GAD1 function is nevertheless robustly altered in these illnesses, suggesting that mechanisms beyond DNA sequence changes are active in the gene’s dysfunction. Prior microarray gene expression analyses in tissue microdissected from multiple sites within post-mortem human hippocampus of patients with SZ, patients with bipolar disorder (BD), and healthy control participants followed by network association analyses using the Ingenuity Pathway Analysis algorithm (Qiagen)10 have implicated a group of genes in the regulation of GAD1 expression.5,11 This group of genes, termed the GAD1 regulatory network, includes kain-ate receptor subunits, components of the Wnt and transforming growth factor β signaling pathways, cell cycle regulators, transcription factors, and chromatin-modifying enzymes. Multiple genes within the GAD1 regulatory network are differentially expressed in the hippocampus of patients with SZ, patients with BD, and control participants, and these altered expression patterns are distinct among diagnoses and locations within the circuitry of the hippocampus.5 These changes are most pronounced in the stratum oriens, where GABAergic interneurons are the sole neuronal phenotype,12 and are more prominent in subfields cornu ammonis 2 (CA2) and CA3 (together CA2/3) than in CA1.
An attractive candidate mechanism to produce disease-associated cell- and tissue-specific changes in gene activity such as these is modification of local chromatin structure through DNA methylation or histone modification.13–16 These epigenetic mechanisms act in a cell-specific manner and establish long-term and gene-specific expression levels in response to genetic and environmental cues in health and disease. Many of these cues may be homogeneous across all brain regions, such as an individual’s genetic background and environmental exposures like in utero maternal infection or malnutrition.17 Others, though, are specific to each subpopulation of neurons operating within the unique microenvironment of each distinct circuit of the brain, including patterns of synaptic input,18,19 extracellular matrix interactions,20 and exposure to neurotrophic factors.21 While there is rapidly growing interest in the mechanisms of chromatin modification across the full range of behavioral neuroscience, few studies to date have attempted to investigate differential effects among neuronal subpopulations beyond comparisons of cortical areas or large-scale structures in the human brain.
Herein, we offer insight into how chromatin dynamics, specifically DNA methylation, may be operative in generating distinct psychotic illness–associated GABAergic deficits in 2 closely related populations of neurons: GABAergic interneurons of the stratum oriens sampled from sectors CA2/3 vs CA1 of the human hippocampus. These locations are the seats of the second (CA2/3) and third (CA1) synapses within the trisynaptic pathway of the hippocampus, regions that contribute to distinct cognitive functions22,23 and symptoms in SZ.24
This study investigated the hypothesis that DNA methylation changes may play a role in sector-specific alteration of hippocampal gene expression within the GAD1 regulatory network in SZ and BD. The 27 members of this gene network (Table) described previously5 were chosen as genes of interest prior to study initiation. Methylation of DNA was measured using the Infinium HumanMethylation450 BeadChip (HM450; Illumina, Inc),25 a robust and widely used microarray platform for genome-wide measurement of DNA methylation. Data analysis was restricted to GAD1 regulatory network–associated probes and used traditional single-site analysis of covariance to identify differentially methylated positions (DMPs) as well as the more novel but well-established “bump hunting” strategy to identify differentially methylated regions (DMRs) by sharing information among spatially colocalized probes.26,27 The findings describe multiple sector- and diagnosis-specific alterations in DNA methylation that implicate this mechanism of chromatin remodeling in the dysregulation of a subset of these GABA-related genes in psychotic disorders.
Table.
Differentially Methylated Positions and Regions Within the GAD1 Regulatory Networka
| Gene | No. | P Value of Most Significant DMR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DMPs | Unique DMRsb | Total DMRsc | Unique DMR Probes | Total DMR Probes | Total Probes | DMPs/Probe | DMRs/Probe | Total DMR Probes/Probe | ||
| CCND1 | 7 | 3 | 3 | 13 | 13 | 80 | 0.088 | 0.038 | 0.163 | .002 |
| CCND2 | 9 | 6 | 10 | 18 | 29 | 80 | 0.113 | 0.075 | 0.363 | .001 |
| CTNNB1 | 0 | 2 | 2 | 3 | 3 | 21 | 0 | 0.095 | 0.143 | .008 |
| DAXX | 1 | 12 | 18 | 42 | 59 | 227 | 0.004 | 0.053 | 0.260 | <.001 |
| DLX2 | 2 | 3 | 3 | 12 | 12 | 63 | 0.032 | 0.048 | 0.190 | .002 |
| FOXG1 | 19 | 2 | 3 | 9 | 15 | 64 | 0.297 | 0.031 | 0.234 | .002 |
| GAD1 | 4 | 1 | 1 | 1 | 1 | 43 | 0.093 | 0.023 | 0.023 | .006 |
| GAD2 | 2 | 1 | 1 | 3 | 3 | 31 | 0.065 | 0.032 | 0.097 | .009 |
| GRIK1 | 1 | 0 | 0 | 0 | 0 | 15 | 0.067 | 0 | 0 | NA |
| GRIK2 | 6 | 2 | 2 | 10 | 10 | 54 | 0.111 | 0.037 | 0.185 | .005 |
| GRIK3 | 6 | 0 | 0 | 0 | 0 | 39 | 0.154 | 0 | 0 | NA |
| GRIK4 | 3 | 2 | 3 | 4 | 4 | 25 | 0.120 | 0.080 | 0.160 | .002 |
| GRIK5 | 5 | 0 | 0 | 0 | 0 | 51 | 0.098 | 0 | 0 | NA |
| GSK3B | 1 | 2 | 2 | 3 | 3 | 19 | 0.053 | 0.105 | 0.158 | .003 |
| HDAC1 | 2 | 2 | 2 | 11 | 11 | 31 | 0.065 | 0.065 | 0.355 | .005 |
| ID3 | 10 | 1 | 1 | 3 | 3 | 37 | 0.270 | 0.027 | 0.081 | .009 |
| IL1B | 1 | 0 | 0 | 0 | 0 | 12 | 0.083 | 0 | 0 | NA |
| LEF1 | 4 | 0 | 0 | 0 | 0 | 38 | 0.105 | 0 | 0 | NA |
| LHX2 | 5 | 0 | 0 | 0 | 0 | 34 | 0.147 | 0 | 0 | NA |
| MSX1 | 30 | 7 | 12 | 56 | 84 | 104 | 0.288 | 0.067 | 0.808 | <.001 |
| PAX5 | 3 | 0 | 0 | 0 | 0 | 49 | 0.061 | 0 | 0 | NA |
| RUNX2 | 5 | 1 | 1 | 5 | 5 | 61 | 0.082 | 0.016 | 0.082 | .007 |
| SMAD1 | 2 | 1 | 2 | 6 | 9 | 21 | 0.095 | 0.048 | 0.429 | .005 |
| SMURF1 | 6 | 4 | 6 | 11 | 15 | 50 | 0.120 | 0.080 | 0.300 | .002 |
| TGFB2 | 7 | 0 | 0 | 0 | 0 | 27 | 0.259 | 0 | 0 | NA |
| TGFBR1 | 3 | 1 | 2 | 1 | 2 | 10 | 0.300 | 0.100 | 0.200 | .008 |
| TLE1 | 2 | 1 | 2 | 3 | 5 | 22 | 0.091 | 0.045 | 0.227 | .006 |
| Total | 146 | 54 | 76 | 214 | 286 | 1308 | ||||
| Mean | 5.4 | 2.0 | 2.8 | 7.9 | 10.6 | 48.4 | ||||
Abbreviations: DMP, differentially methylated position; DMR, differentially methylated region; NA, not applicable.
The numbers of DMPs and DMRs and the total numbers of probes associated with each of the 27 genes within the GAD1 regulatory network are listed. As the detection of DMPs and DMRs is influenced by probe placement, the ratios of DMP- and DMR-associated probes to the total number of probes for each gene are listed.
Unique DMRs are the tally of DMRs associated with that gene, not double counting DMRs that appear in multiple comparisons.
Total DMRs are the total tally of DMRs across all 4 group comparisons (cornu ammonis 1 [CA1] for patients with schizophrenia vs control participants; CA2/3 for patients with schizophrenia vs control participants; CA1 for patients with bipolar disorder vs control participants; and CA2/3 for patients with bipolar disorder vs control participants).
Methods
Postmortem Human Hippocampus Samples
This epigenetic association study targeting GAD1 regulatory network genes was conducted between July 1, 2012, and June 30, 2014. Postmortem human hippocampus tissue samples from 8 patients with SZ, 8 patients with BD, and 8 healthy control participants matched for age, postmortem interval (PMI), sex, and pH were obtained from the Harvard Brain Tissue Resource Center. Demographic variables of the assembled cohort are provided in eTable 1 in the Supplement, and all cases were obtained by family referral; no cases were referred by a medical examiner’s office. Cases with documented history of illicit substance abuse were excluded. Institutional review board approval was obtained by the Harvard Brain Tissue Resource Center from McLean Hospital to collect, maintain, and distribute brain tissue along with deidentified clinical information to the investigators, and institutional review board approval was therefore not required for this specific study.
Fresh tissue was dissected as blocks cut at the level of the pulvinar thalami along the rostrocaudal axis of the hippocampus. Tissue was lightly fixed with ice-cold 0.1% formalin in 0.1M phosphate buffer (pH 7.2) for 90 minutes, followed by cryoprotection with 30% sucrose in phosphate buffer overnight prior to embedding in optimal cutting temperature compound (Sakura Finetek). Tissue samples were then stored at −80°C.
Tissue Processing
Frozen postmortem human hippocampus tissue samples were sectioned (30 μm) on a Microm HM560 cryostat at −20°C and mounted on polyethylene terephthalate frame slides (Leica). Slides were processed through a graded series of ice-cold acetone and ethanol, hydrated in phosphate-buffered saline, stained with cresyl violet, and dehydrated through ascending concentrations of ethanol. After drying, stratum oriens tissue was dissected from region CA2/3 or CA1 from approximately 60 sections per sample using a Leica LMD6500 laser microdissection system (eFigure 1 in the Supplement). We extracted DNA from collected tissue using the QIAamp DNA Micro Kit (Qiagen) and stored it at −80°C until further use.
Bead Arrays
We bisulfitemodified500 ng of genomic DNA with the EZ DNA Methylation Kit (Zymo Research) using modified incubation parameters as recommended by Illumina, Inc. Bisulfite-modified DNA was analyzed using the HM450 per the manufacturer’s protocol, with samples randomly distributed across 48 arrays on 4 slides to avoid batch effect.
Statistical Analysis
All statistical analysis was performed using R version 3.1.0 software (R Foundation). Raw intensity files (.idat files) were analyzed using the minfi package.28 Internal control probes were investigated and no outliers were identified among samples. Log ratios of methylation percentages, called M values,29 were extracted and preprocessed by stratified quantile normalization.27 The data set was then restricted to the 1308 probes interrogating sites within 10 kilobases (kb) of our predetermined 27 genes of interest. All GAD1 regulatory network genes are located on autosomes. Individual probes were assessed for differences across all groups by analysis of covariance (ANCOVA) with PMI, age, and sex as covariates, with significance reported as false detection rate (FDR) generated by Benjamini-Hochberg P value adjustment.30 The bump hunter function within minfi was used to identify DMRs between individual groups using permutation testing for significance based on 1000 permutations, again with PMI, age, and sex as covariates.
Correlation of DNA Methylation and Gene Expression
Our group has previously published microarray-generated gene expression data from equivalently microdissected tissue from a nonoverlapping cohort of postmortem human hippocampus.5 As the lack of overlap between the 2 cohorts (tissue from the earlier cohort has been expended) made correlation analysis at the level of single cases impossible, we assessed correlation between DNA methylation and gene expression at the level of averaged group differences. At each gene of interest we calculated the fold expression change between control and each patient group in each subfield (CA1 in patients with SZ vs control participants; CA2/3 in patients with SZ vs control participants; CA1 in patients with BD vs control participants; and CA2/3 in patients with BD vs control participants). At each assessed CpG site we calculated the DNA methylation change (ΔM value) in the same 4 comparisons. For each assessed CpG site we then used R software to calculate the Pearson product-moment correlation coefficient between the vectors representing the 4 DNA ΔM values at that probe and the 4 gene expression fold change values for the corresponding gene.
Bisulfite Pyrosequencing
Methylation measurements at 3 DMPs overlapping with DMRs were validated using bisulfite pyrosequencing.31 For each sample approximately 15 ng of unamplified bisulfite-modified DNA was used as template for polymerase chain reaction amplification of each site of interest using the primers (designed with MethPrimer32) and polymerase chain reaction conditions listed in eTable 2 in the Supplement. Polymerase chain reaction amplicons were sequenced with Pyromark MD96 (Qiagen) per the manufacturer’s protocol. Methylation measurements were assessed for significant differences among groups by ANCOVA with PMI, age, and sex as covariates.
Assessment of Potential Confounds
To investigate potential medication effects, bump hunting analysis was repeated in patient samples (SZ and BD together) from single subfields comparing cases with exposure to lithium carbonate, valproate sodium, or dibenzodiazepine-type antipsychotics vs those without such exposure (separate comparison for each medication) using the same parameters described earlier. Additionally, we assessed the relationship between tissue pH and methylation level by calculating the Pearson product-moment correlation coefficients and the associated level of significance at all 1308 GAD1 regulatory network–associated CpG sites. Finally, as copy number variation may possibly influence DNA methylation measurement by the HM450, we investigated copy number variation at all interrogated sites by comparing the total intensity of the methylated and unmethylated channels in each sample to that sum averaged across all samples.27
Results
Single-Site Analysis
Within the GAD1 regulatory network, 146 sites were differentially methylated among the 6 groups in our data set with FDR < 0.05. Sites of significant methylation change are not uniformly distributed throughout the GAD1 regulatory network but are enriched within particular genes, ranging from 0 (CTNNB1) to 30 (MSX1) DMPs (Table).
Bump Hunting
This hypothesis-driven study was underpowered to identify DMRs significant after correction for multiple comparisons (family wise error rate), so we proceeded to investigate DMRs with a nominal P < .01. This approach identified a total of 54 DMRs (27 hypermethylated, 27 hypomethylated), with an average of 2.0 DMRs per gene and a range of 0 (8 genes) to 12 (death-associated protein 6 gene [DAXX]) DMRs per gene (Table). The DMRs are highlighted in Figure 1 displaying the 3 genes with the 5 most significant DMRs, and the 3 most significant DMRs are depicted in greater detail in Figure 2.
Figure 1. Genomic Distribution of Differentially Methylated Regions (DMRs) and Differentially Methylated Positions at MSX1, DAXX, andCCND2.
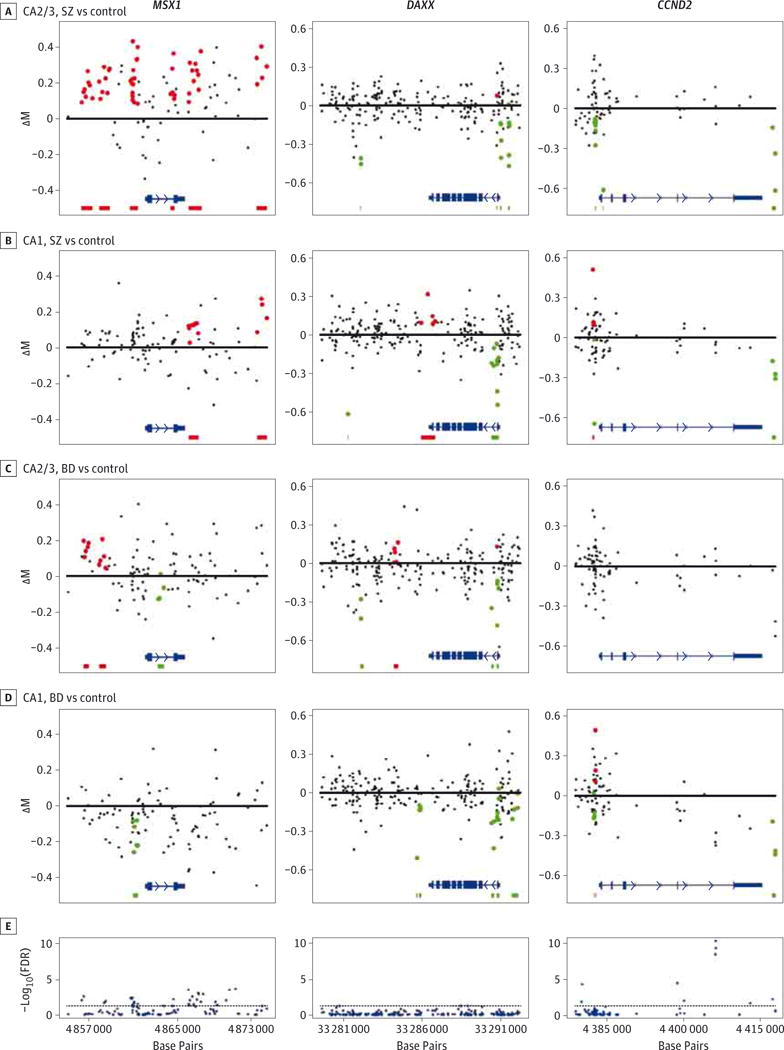
A–D, The DMRs between single sample groups, including cornu ammonis 2/3 (CA2/3) for patients with schizophrenia (SZ) vs control participants (A), CA1 for patients with SZ vs control participants (B), CA2/3 for patients with bipolar disorder (BD) vs control participants (C), and CA1 for patients with BD vs control participants (D). Each point represents the group difference between 8 patients and 8 control participants at a single CpG, with group methylation change (ΔM value) vs genomic position. Colored points are found within a DMR, with red DMRs hypermethylated and green DMRs hypomethylated. At the bottom of each plot, the location and structure of the gene are depicted in blue (arrowheads indicate direction of gene transcription); red and green horizontal bars indicate the footprints of hypermethylated and hypomethylated DMRs, respectively. E, Plots for −log10(false detection rate [FDR]) for each interrogated CpG along the length of the gene generated by analysis of covariance across all 6 sample groups. Dashed line indicates FDR = 0.05; points above this threshold are identified as differentially methylated positions.
Figure 2. The 3 Most Significant Differentially Methylated Regions (DMRs).
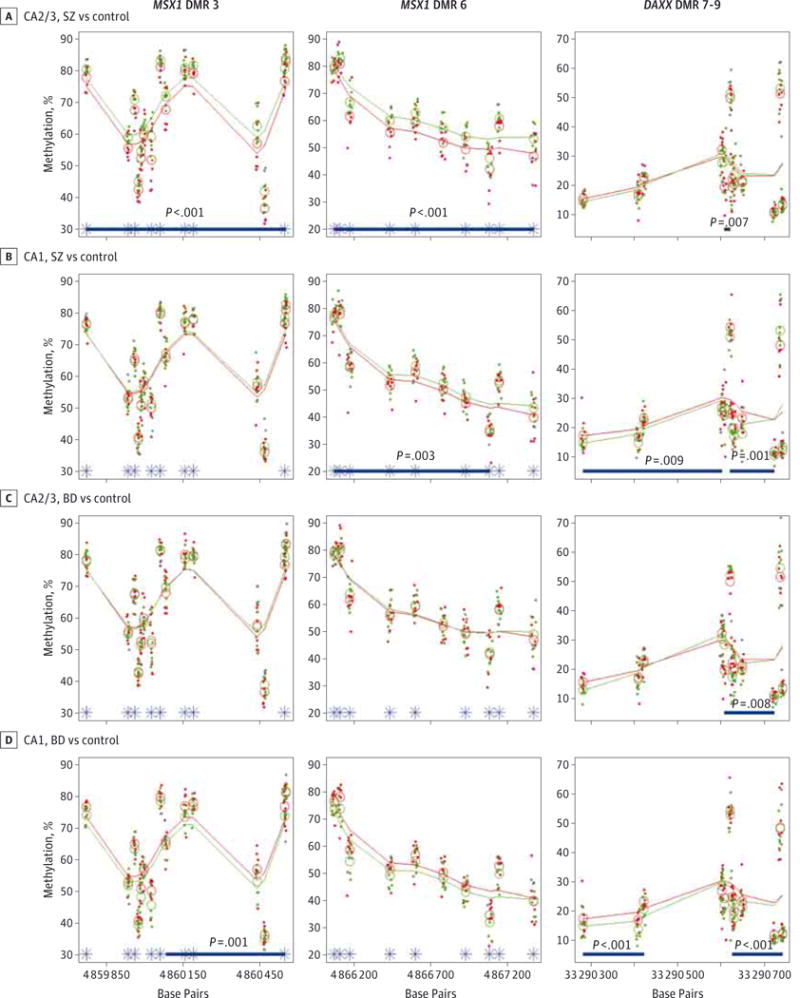
The 3 most significant DMRs are depicted, with each column representing a single genomic region. The DMRs are numbered from the p end to the q end of the genomic region associated with each gene (MSX1 DMR 3, MSX1 DMR 6, DAXX DMR 7–9). Group comparisons are shown for cornu ammonis 2/3 (CA2/3) for patients with schizophrenia (SZ) vs control participants (A), CA1 for patients with SZ vs control participants (B), CA2/3 for patients with bipolar disorder (BD) vs control participants (C), and CA1 for patients with BD vs control participants (D). Each point represents the DNA methylation level at the corresponding site in a single sample, with percentage of methylation vs genomic location. Open circles indicate group averages at each measured site; smoothed lines, running group averages; green, patients with SZ or BD; red, control participants; blue bars, footprint of an identified DMR for that comparison with the associated P value; and asterisks, location of differentially methylated positions.
DMR and DMP Distribution
There were greater numbers of DMRs and DMR-associated sites in SZ (44 and 181, respectively) than in BD (34 and 106, respectively) . While there were slightly more DMRs found in CA1 than in CA2/3 (40 vs 36, respectively), those within CA2/3 were markedly more significant, with 16 of the 20 most significant DMRs identified in CA2/3. A low amount of overlap was observed between DMPs and DMRs (17% of all DMR-associated sites are DMPs), although overlap is much greater in SZ than in BD (Figure 3). The 146 identified DMPs are slightly enriched at enhancer elements and normally distributed within CpG island shores and shelves, but they are significantly excluded from the overlapping categories of promoter regions and CpG islands . The DMRs are distributed largely as would be expected by chance but with slight enrichment at promoter sequences and CpG islands (Figure 3).
Figure 3. Functional Distribution of Differentially Methylated Positions (DMPs) and Differentially Methylated Regions (DMRs).
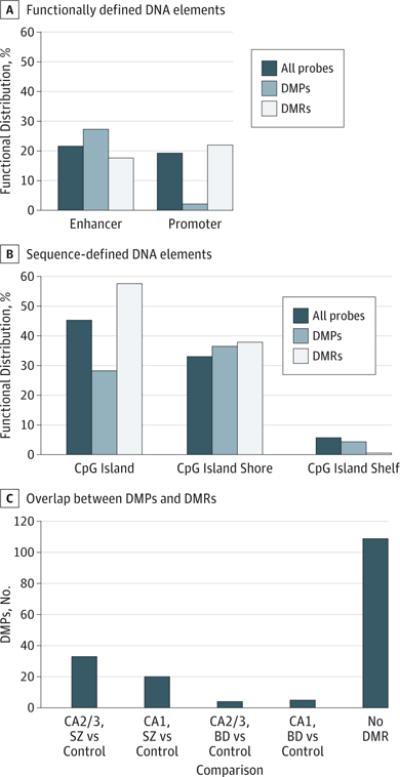
A and B, Distribution of DMPs and DMRs among functionally defined (A) and sequence-defined (B) genomic features. Percentages indicate total probe-associated CpG sites, DMPs, and DMR-associated sites found at the DNA elements relative to the total number of sites in each category. The DMPs are underrepresented within the spatially overlapping categories of promoter regions and CpG islands (functional elements, sequence elements, . C, Overlap of DMPs and DMRs within sample groups, showing the number of DMPs found within DMRs identified for any of the 4 comparisons (cornu ammonis 2/3 [CA2/3] for patients with schizophrenia [SZ] vs control participants; CA1 for patients with SZ vs control participants; CA2/3 for patients with bipolar disorder [BD] vs control participants; and CA1 for patients with BD vs control participants) as well as the number of DMPs that do not coincide with a DMR in any comparison.
Msh Homeobox1Gene
Across all 27 GAD1 regulatory network genes, 20% of all DMPs and 26% of all DMR-associated probes were found within 10 kb of the Msh homeobox 1 gene (MSX1). This gene contains the 2 most significant DMRs, one of which is also the only instance of direct overlap of a hypermethylated DMR in one group (SZ CA2/3; P < .001) with a hypomethylated DMR in another (BD CA1; P = .001). This DMR falls immediately upstream of the transcription start site (TSS), and methylation at all measured sites within this DMR is inversely correlated with MSX1 expression as would be expected for methylation of a promoter region (average r = −0.62) (Figure 4). Inverse correlation of methylation and expression is overrepresented at MSX1 with 3 positively and 27 inversely correlated DMPs and 1 positively and 11 inversely correlated DMRs.
Figure 4. Correlation Between DNA Methylation and Gene Expression.
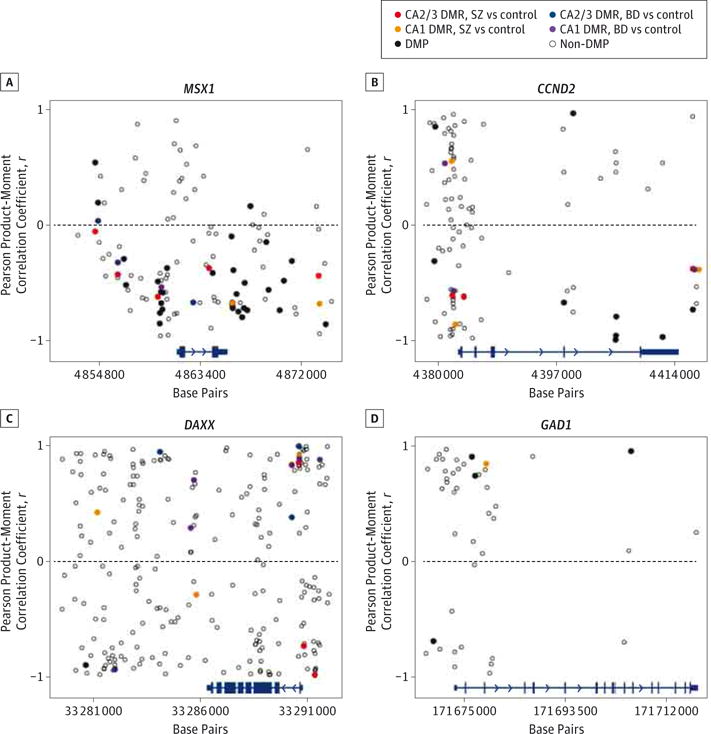
A–D, At each assessed CpG site for MSX1 (A), CCND2 (B), DAXX (C), and GAD1 (D), the Pearson product-moment correlation coefficient was calculated between group gene expression fold change and group methylation change for the 4 comparisons in this experiment (associated gene expression fold change correlated with methylation change [ΔM value] for the following: cornu ammonis 1 [CA1] for patients with schizophrenia [SZ] vs control participants; CA2/3 for patients with SZ vs control participants; CA1 for patients with bipolar disorder [BD] vs control participants; and CA2/3 for patients with BD vs control participants). Data are plotted with correlation vs genomic location. Gene location and structure are depicted at the bottom of each plot in blue (arrowheads indicate direction of gene transcription). Open circles indicate sites with nonsignificant methylation change; black circles, differentially methylated positions (DMPs); and colored circles, average methylation among all probes within single differentially methylated regions (DMRs).
Death-Associated Protein 6 Gene
While DAXX was associated with just 1 DMP, it was associated with the greatest number of DMRs of all GAD1 regulatory network genes and the third most significant DMR (P < .001). The single DAXX-associated DMP is strongly inversely correlated with gene expression (r = −0.90) (Figure 4). The DMRs at this gene are highly but variably correlated with DAXX expression, with 5 DMRs inversely correlated (average r = −0.78) and 13 positively correlated (average r = 0.75).
Cyclin D2 Gene
ThecyclinD2 gene (CCND2)containsthe most significant DMP across all groups, with FDR = 4 × 10−11, and this DMP is flanked by 2 DMPs both with FDR < 4 × 10−9.This region is hypomethylated but not identified as a significant DMR in the BD CA1 group (P = .01). These sites occur within the final intron of the gene, and methylation at these 3 sites was strongly inversely correlated with CCND2 expression (average r = −0.92) (Figure 4). This gene contains the fourth most significant DMR (P = .001). Two hypermethylated DMRs at the CCND2 promoter region were positively correlated with gene expression (average r = 0.74), and 4 hypomethylated DMRs were inversely correlated (average r = −0.86).
Glutamic Acid Decarboxylase 67 Gene
We observed 4 GAD1-associated DMPs, 3 of which are located within 4 kb of the TSS (FDR = 0.016−0.023) and another within the 11th intron of the gene (FDR = 1.8 × 10−5). A single DMR (P = .006) was identified in SZ CA1 immediately downstream of the third exon, colocalizing with a binding site for SUZ12 protein. At the GAD1 locus, methylation at all 4 DMPs was highly correlated with GAD1 expression, with the only DMP occurring upstream of the TSS inversely correlated (r = −0.70) (Figure 4) and the 3 DMPs within the gene body positively correlated with GAD1 expression (average r = 0.85). Methylation at the 1 DMR in this region was positively correlated with GAD1 expression (r = 0.84), as would be expected for gene-body methylation.33
Bisulfite Pyrosequencing Replication Analysis
The ANCOVAs of pyrosequencing methylation measurements of DMPs associated with MSX1, FOXG1, and RUNX2 yielded P = .001, P < .001, and P = .03, respectively. Box plots of pyrosequencing data are presented in eFigure 2 in the Supplement.
Assessment of Potential Confounds
Of the top 10 most significant DMRs from the original analysis, only 1 was found to be differentially methylated between groups separated by medication exposure (MSX1 DMR 6; original P < .001; analysis of lithium exposure, P = .007) (eFigure 3 in the Supplement). Testing within CA1, CA2/3, or all samples together identified no CpG sites where pH was correlated with methylation level with FDR < 0.05. Investigation of total probe intensities detected no distinct copy number variation at any CpG site associated with our genes of interest (eFigure 4 in the Supplement).
Discussion
Methylation changes in DNA are believed to be an important factor in the pathophysiology of psychotic disorders.34–37 Within this paradigm, this study is unique because it investigated a predetermined gene network relevant to these disorders at multiple sites within the neural circuitry of the human hippocampus. As is often the case in postmortem human brain tissue studies, this work was performed with a small sample size limited by the time- and labor-intensive methods used for sample preparation, and we must remain mindful of this fact while interpreting our findings. While the size of this study is similar to other recent work using the same or similar methods,27,38 replication of these findings and subsequent analysis of sites identified as DMPs or DMRs with additional methods are the next vital steps. Additionally, while our findings do not show methylation changes to be exclusive to the GAD1 regulatory network, these 27 genes were targeted based on prior gene expression studies also conducted in stratum oriens tissue containing only GABAergic interneurons. In conjunction with prior work, these data have identified regions within this preselected group of genes that hold promise for elucidating the pathophysiology of the well-documented GAD1 downregulation in psychotic illness.
In keeping with prior gene expression studies5 and current ideas about the cellular specificity of chromatin regulation,14,39 we found DNA methylation patterns to be distinct across diagnoses and across circuit locations within the trisynaptic pathway. Among all 1308 assessed CpG sites within the GAD1 regulatory network, 146 (11%) were identified as DMPs after correction for multiple comparisons. These data strongly suggest that DNA methylation is an active process in the dysregulation of GABAergic interneuronal function observed within the hippocampus in psychotic disorders, and the results identify genes and circuits warranting more targeted investigation.
The stratum oriens of CA2/3 and CA1 coincide with the second and third synapses within the trisynaptic pathway and as such represent phenotypically similar populations of neurons that perform distinct functional roles in distinct micro-circuits within the hippocampus. While some DMRs were similar between CA2/3 and CA1, most were unique to one region or the other. In fact, cluster analysis of the GAD1 regulatory network–restricted data set found that DNA methylation profiles differed more between CA2/3 and CA1 than between patients with SZ, patients with BD, and control participants. As with all postmortem human brain studies, medication exposure is a potential confound, but the fact that circuit location is more effective than diagnosis in determining methylation levels suggests that medication history is not a driving factor in our results. While our cohort was matched for postmortem interval and pH, an additional potential confound that must be considered in analysis of postmortem human tissue is autolysis, which may be associated with a prolonged agonal state at the time of death. Tissue pH is often used as an indicator of agonal state, and consistent with prior work showing no effect of pH on postmortem measurement of the highly stable epigenetic mark of cytosine methylation,40 pH and methylation measurements were not significantly correlated at any of the 1308 GAD1 regulatory network–associated probes.
While cluster analysis demonstrates that medications do not determine DMPs, we investigated further the effects of medication exposure on methylation within DMRs by comparing methylation levels in patients exposed to lithium, valproate, or dibenzodiazepine-type atypical antipsychotic medications vs those in patients without these exposures. Lithium has been shown to influence DNA methylation in human neuroblastoma cells in a gene-specific manner,41 valproate influences chromatin structure through its activity as a histone deacetylase inhibitor,42 and animal studies have found dibenzodiazepine compounds (including clozapine, olanzapine, and quetiapine fumarate) to influence DNA methylation patterns, while some other antipsychotic medications (specifically haloperidol and risperidone) do not.34 Of the top 10 most significant DMRs from the original analysis, none were differentially methylated in association with medication exposure except a single DMR at MSX1 in association with lithium (eFigure 3 in the Supplement). However, we do not believe that lithium exposure is causal to the original finding, as in the original analysis this region was significantly altered in the comparison between patients with SZ and control participants and not in the comparison between patients with BD and control participants, and within the SZ group there is only a single case with exposure to lithium (eTable 1 in the Supplement). This DMR remains significant on repeating the original analysis without including the lithium-exposed SZ case.
Our analysis used 2 approaches: (1) ANCOVA of single sites to identify DMPs across all 6 groups, and (2) the bump hunting strategy to identify DMRs between individual groups. In this study, DMPs indicate sites of differential DNA methylation across the entire data set, and cluster analysis indicates that most of this methylation difference is related to hippocampal sector rather than diagnostic category. On the other hand, DMRs are regions of methylation difference within individual comparisons (ie, CA2/3 in patients with SZ vs control participants). The DMPs but not DMRs appear to be excluded from promoter regions and CpG islands, the first being a functionally defined element and the second being a sequence-defined element with significant spatial overlap. For all other elements, the distribution of DMPs and DMRs is as would be expected by chance. While the interpretation of this finding is uncertain, it suggests as argued elsewhere26 that DMRs and bump hunting may be better suited to identify functionally significant changes in DNA methylation than is assessment of DMPs. As most methylation differences at DMPs are more related to circuitry than to diagnosis, most circuitry-dependent differential methylation appears excluded from these genomic features. Also, the greater overlap of DMPs with SZ DMRs than with BD DMRs suggests that SZ-associated methylation changes may be more circuitry specific than those associated with BD.
GAD1 displayed4 DMPsand1DMR associated with SZ CA1 at sites to our knowledge not previously investigated in human brain. The GAD1 promoter region has been investigated with various methods, including bisulfite sequencing of chromatin enriched for trimethylation of histone H3 lysine 4 (H3K4me3) or of histone H3 lysine 27 (H3K27me3).43 This approach assessed 12 CpG loci within 3 regions of the gene, and 1 of these loci is measured by an HM450 probe. While the prior study described SZ-associated methylation changes at the GAD1 promoter, both the prior study and our current data set found no methylation change at the overlapping CpG residue. Another study assessed postmortem human prefrontal cortex DNA enriched for regions of hypermethylation using restriction enzyme digestion interrogated using CpG island microarrays.37 This method did not find significant methylation change at GAD1, and the assessed promoter sequence has no overlap with any HM450 probes.
We identified 3 genes within the GAD1 regulatory network enriched for circuit- and disease-associated methylation changes containing the 5 most significant DMRs: MSX1, CCND2, and DAXX. Even among this subset of network genes, MSX1 stands out as it is associated with one-fourth of DMR-associated probes and one-fifth of DMPs identified across all 27 GAD1 regulatory network genes as well as the first, second, and fifth most significant DMRs. MSX1 encodes Msh homeobox 1, a regulator of early central nervous system and craniofacial development that continues to be expressed in adult brain; unique to the hippocampus, MSX1 is expressed at higher levels in the adult hippocampus than in the fetal hippocampus.44 It interacts with SUZ12, a component of the polycomb repressive complex 2, to direct polycomb repressive complex 2–mediated H3K27me3 to targeted genomic locations.45 Along with a striking number of the identified DMRs within the GAD1 regulatory network, SUZ12 binds a site approximately 1.5 kb downstream of the GAD1 TSS, and the H3K27me3 mark is increased at the GAD1 promoter region in the prefrontal cortex of patients with SZ.46 In addition, MSX1 functions in regulation of higher-order chromatin structures by recruiting H3K27me3-marked chromatin to the nuclear periphery.45 A 50-kb chromatin loop at the GAD1 locus functions in regulation of GAD1 expression, and this loop is disrupted in SZ.47
The most significant DMP and the fourth most significant DMR were associated with CCND2, which encodes cyclin D2, a highly conserved cyclin protein whose expression is tightly coupled to the cell cycle and in brain has been implicated in differentiation, maturation, and circuit integration of GABAergic interneurons.48 Mice deficient in CCND2 have been developed as a model of hippocampal hyperactivity displaying multiple cognitive deficits and molecular abnormalities observed in SZ.49 The third most significant DMR and the greatest number of DMRs were found at DAXX, which codes for death-associated protein 6, a heterochromatin-associated transcriptional regulator active in cell cycle control. In neurons, DAXX has been found to be a histone H3.3 chaperone sensitive to calcium influx and to mediate chromatin modification in response to neuronal activity.50
These 3 genes, MSX1, CCND2, and DAXX, are intriguing targets for future studies of how the 2 basic and fundamental cellular systems of chromatin dynamics and cell cycle regulation influence interneuronal physiology in SZ and BD. As discussed earlier, there is rapidly growing evidence for the importance of chromatin regulation in psychotic illness and neuroscience in general. Cell cycle reentry has been implicated in a number of neurodegenerative disorders, and a role for altered cell cycle–regulatory mechanisms has been suggested for nonneurodegenerative psychiatric disorders, including SZ and BD.51–53 The disrupted in schizophrenia 1 gene (DISC1) interacts with multiple signaling pathways, including Wnt and GABA signaling, to regulate adult neurogenesis in the hippocampus.54 Our data suggest multiple molecular targets that may offer insight into the mediators of cell cycle abnormalities in psychotic disorders as well as potential targets for intervention.
Conclusions
This work describes multiple gene-, circuit-, and diagnosis-specific DNA methylation changes relevant to GABAergic cell function in psychotic illness. Future studies will investigate the functional significance of changes observed at specific genomic loci and test their roles in perturbation of GABAergic interneuron function. Our data demonstrate the great benefit of more precise methods of sampling and analyzing postmortem human brain tissue and offer insight into the role of chromatin dynamics in the functional regulation of interneuronal subpopulations within a discrete microcircuit in the human hippocampus.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by grants MH077175 (Dr Benes) and MH/NS 077550 (Dr Benes) from the National Institutes of Health and by the William P. and Henry B. Test Endowment from Harvard University (Dr Benes). Dr Ruzicka is supported by a Dupont-Warren Fellowship from Harvard Medical School, the MD/PhD Fellowship from the American Psychiatric Association and Pfizer Pharmaceuticals, a NARSAD Young Investigator Award from the Brain and Behavior Research Foundation, and the Andrew P. Merrill Memorial Research Fellowship and Maria Lorenz Pope Fellowship from McLean Hospital.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Supplemental content at jamapsychiatry.com
Author Contributions: Dr Ruzicka had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Ruzicka, Benes.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: All authors.
Critical revision of the manuscript for important intellectual content: Ruzicka, Benes.
Statistical analysis: Ruzicka.
Obtained funding: Ruzicka, Benes.
Administrative, technical, or material support: Subburaju, Benes.
Study supervision: Subburaju, Benes.
Conflict of Interest Disclosures: None reported.
Contributor Information
W. Brad Ruzicka, Program in Structural and Molecular Neuroscience, McLean Hospital, Belmont, Massachusetts; Department of Psychiatry, Harvard Medical School, Boston, Massachusetts.
Sivan Subburaju, Program in Structural and Molecular Neuroscience, McLean Hospital, Belmont, Massachusetts; Department of Psychiatry, Harvard Medical School, Boston, Massachusetts.
Francine M. Benes, Program in Structural and Molecular Neuroscience, McLean Hospital, Belmont, Massachusetts; Department of Psychiatry, Harvard Medical School, Boston, Massachusetts; Program in Neuroscience, Harvard Medical School, Boston, Massachusetts.
References
- 1.Akbarian S, Kim JJ, Potkin SG, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52(4):258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- 2.Guidotti A, Auta J, Davis JM, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57(11):1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 3.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57(3):237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto T, Volk DW, Eggan SM, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23(15):6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104(24):10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahn RS, Keefe RS. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry. 2013;70(10):1107–1112. doi: 10.1001/jamapsychiatry.2013.155. [DOI] [PubMed] [Google Scholar]
- 7.Chen CM, Stanford AD, Mao X, et al. GABA level, gamma oscillation, and working memory performance in schizophrenia. Neuroimage Clin. 2014;4:531–539. doi: 10.1016/j.nicl.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Díez Á, Suazo V, Casado P, Martín-Loeches M, Molina V. Gamma power and cognition in patients with schizophrenia and their first-degree relatives. Neuropsychobiology. 2014;69(2):120–128. doi: 10.1159/000356970. [DOI] [PubMed] [Google Scholar]
- 9.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac Symp Biocomput. 2000:418–429. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 11.Benes FM. Regulation of cell cycle and DNA repair in post-mitotic GABA neurons in psychotic disorders. Neuropharmacology. 2011;60(7–8):1232–1242. doi: 10.1016/j.neuropharm.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Benes FM, Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25(1):1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 13.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465(7299):728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 2013;9(4):e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60(6):961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akbarian S, Huang HS. Epigenetic regulation in human brain: focus on histone lysine methylation. Biol Psychiatry. 2009;65(3):198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG. 2008;115(10):1243–1249. doi: 10.1111/j.1471-0528.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- 18.Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13(9):1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo JU, Ma DK, Mo H, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14(10):1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu R, Spencer VA, Bissell MJ. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. J Biol Chem. 2007;282(20):14992–14999. doi: 10.1074/jbc.M610316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu YZ, Chrivia JC, Latchman DS. Nerve growth factor up-regulates the transcriptional activity of CBP through activation of the p42/p44(MAPK) cascade. J Biol Chem. 1998;273(49):32400–32407. doi: 10.1074/jbc.273.49.32400. [DOI] [PubMed] [Google Scholar]
- 22.Kesner RP, Lee I, Gilbert P. A behavioral assessment of hippocampal function based on a subregional analysis. Rev Neurosci. 2004;15(5):333–351. doi: 10.1515/revneuro.2004.15.5.333. [DOI] [PubMed] [Google Scholar]
- 23.Goodrich-Hunsaker NJ, Hunsaker MR, Kesner RP. The interactions and dissociations of the dorsal hippocampus subregions: how the dentate gyrus, CA3, and CA1 process spatial information. Behav Neurosci. 2008;122(1):16–26. doi: 10.1037/0735-7044.122.1.16. [DOI] [PubMed] [Google Scholar]
- 24.Tamminga CA, Stan AD, Wagner AD. The hippocampal formation in schizophrenia. Am J Psychiatry. 2010;167(10):1178–1193. doi: 10.1176/appi.ajp.2010.09081187. [DOI] [PubMed] [Google Scholar]
- 25.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41(1):200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry. 2014;19(8):862–871. doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du P, Zhang X, Huang CC, et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 31.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2(9):2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 32.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 33.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 34.Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38(1):138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grayson DR, Jia X, Chen Y, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102(26):9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veldic M, Caruncho HJ, Liu WS, et al. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci U S A. 2004;101(1):348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wockner LF, Noble EP, Lawford BR, et al. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12(4):385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- 40.Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: in vitro and post-mortem brain studies. J Neurosci Methods. 2008;174(1):123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 41.Asai T, Bundo M, Sugawara H, et al. Effect of mood stabilizers on DNA methylation in human neuroblastoma cells. Int J Neuropsychopharmacol. 2013;16(10):2285–2294. doi: 10.1017/S1461145713000710. [DOI] [PubMed] [Google Scholar]
- 42.Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276(39):36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 43.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2(8):e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramos C, Martinez A, Robert B, Soriano E. Msx1 expression in the adult mouse brain: characterization of populations of beta-galactosidase-positive cells in the hippocampus and fimbria. Neuroscience. 2004;127(4):893–900. doi: 10.1016/j.neuroscience.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Kumar RM, Biggs VJ, et al. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev Cell. 2011;21(3):575–588. doi: 10.1016/j.devcel.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang HS, Matevossian A, Whittle C, et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27(42):11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bharadwaj R, Jiang Y, Mao W, et al. Conserved chromosome 2q31 conformations are associated with transcriptional regulation of GAD1 GABA synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. J Neurosci. 2013;33(29):11839–11851. doi: 10.1523/JNEUROSCI.1252-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leto K, Bartolini A, Di Gregorio A, et al. Modulation of cell-cycle dynamics is required to regulate the number of cerebellar GABAergic interneurons and their rhythm of maturation. Development. 2011;138(16):3463–3472. doi: 10.1242/dev.064378. [DOI] [PubMed] [Google Scholar]
- 49.Gilani AI, Chohan MO, Inan M, et al. Interneuron precursor transplants in adult hippocampus reverse psychosis-relevant features in a mouse model of hippocampal disinhibition. Proc Natl Acad Sci U S A. 2014;111(20):7450–7455. doi: 10.1073/pnas.1316488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michod D, Bartesaghi S, Khelifi A, et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 2012;74(1):122–135. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benes FM, Lim B, Subburaju S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc Natl Acad Sci U S A. 2009;106(28):11731–11736. doi: 10.1073/pnas.0903066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benes FM. Relationship of GAD(67) regulation to cell cycle and DNA repair in GABA neurons in the adult hippocampus: bipolar disorder versus schizophrenia. Cell Cycle. 2010;9(4):625–627. doi: 10.4161/cc.9.4.10820. [DOI] [PubMed] [Google Scholar]
- 53.Fan Y, Abrahamsen G, McGrath JJ, Mackay-Sim A. Altered cell cycle dynamics in schizophrenia. Biol Psychiatry. 2012;71(2):129–135. doi: 10.1016/j.biopsych.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Wu Q, Li Y, Xiao B. DISC1-related signaling pathways in adult neurogenesis of the hippocampus. Gene. 2013;518(2):223–230. doi: 10.1016/j.gene.2013.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


