Significance
We describe the interaction between an intrinsically disordered protein (IDP), NUPR1, and the well-folded C-terminal region of RING1B protein of the Polycomb complex, involving residues Ala33 and Thr68 of the IDP. These observations are significant because they raise the possibility that NUPR1 aids the function of the members of the Polycomb complex, playing an active role in carcinogenesis.
Keywords: IDP, NUPR1, Polycomb group, RING1B
Abstract
Intrinsically disordered proteins (IDPs) are ubiquitous in eukaryotes, and they are often associated with diseases in humans. The protein NUPR1 is a multifunctional IDP involved in chromatin remodeling and in the development and progression of pancreatic cancer; however, the details of such functions are unknown. Polycomb proteins are involved in specific transcriptional cascades and gene silencing. One of the proteins of the Polycomb complex is the Ring finger protein 1 (RING1). RING1 is related to aggressive tumor features in multiple cancer types. In this work we characterized the interaction between NUPR1 and the paralogue RING1B in vitro, in silico, and in cellulo. The interaction occurred through the C-terminal region of RING1B (C-RING1B), with an affinity in the low micromolar range (∼10 μM). The binding region of NUPR1, mapped by NMR, was a hydrophobic polypeptide patch at the 30s region of its sequence, as pinpointed by computational results and site-directed mutagenesis at Ala33. The association between C-RING1B and wild-type NUPR1 also occurred in cellulo as tested by protein ligation assays; this interaction is inhibited by trifluoperazine, a drug known to hamper binding of wild-type NUPR1 with other proteins. Furthermore, the Thr68Gln and Ala33Gln/Thr68Gln mutants had a reduction in the binding toward C-RING1B as shown by in vitro, in silico, and in cellulo studies. This is an example of a well-folded partner of NUPR1, because its other interacting proteins are also unfolded. We hypothesize that NUPR1 plays an active role in chromatin remodeling and carcinogenesis, together with Polycomb proteins.
Gene expression control occurs through the combined activities of transcription factors and chromatin regulators, considered as effectors of epigenetic mechanisms. Posttranslational modifications of nucleosomal histones, DNA methylation, local compaction, and long-range interactions determine a variety of chromatin structures that can affect the transcriptional output.
A group of chromatin regulators are the Polycomb Repressive complexes (PRCs) (1, 2). These Polycomb group (PcG) proteins are encoded by genes initially discovered over 60 years ago as repressors of the Hox genes in Drosophila (3). The PcG proteins form two main silencing heteromeric complexes called PRC1 and PRC2; both are highly conserved in eukaryotes and either in isolation or synergistically can silence genes (4, 5). The catalytic functions of both complexes include ubiquitin-ligase (H3K119ub1) and methyltransferase activity (H3K27Me3), respectively. In mammals, the PRC2 core is formed by, at least, four heteromeric units, one of which is EZH2 (or its paralog, EZH1), the methyltransferase that catalyzes the trimethylation of histone H3 at Lys27 (5). This modification can act as an anchoring site of PRC1 complexes containing chromobox (CBX) proteins. These CBX proteins recruit PRC1 complex onto PRC2-enriched chromatin, facilitating the monoubiquitylation. The PRC1-dependent monoubiquitylation at Lys119 of histone H2A correlates with transcriptional repression (6–9). This process is carried out by a heterodimeric RING finger E3 ligase, whose subunit binding the E2 ligase is RING1B, or its paralog RING1A. The other member of the heterodimer is one out of a group of six Polycomb Ring finger proteins (whose best-known member is perhaps BMI1), thought to act as a positive cofactor (10, 11). It has been shown that RING1B is involved in several human cancers (12–15), especially in human hepatocellular carcinomas and pancreatic cancer (16). However, its exact function during the development of such cancers is yet unknown.
We have described the conformational properties of the C-terminal domain of RING1B (C-RING1B), encompassing the residues 227–334 (17), and shown that it is a dimer of well-formed monomers. The X-ray structure of the monomers of C-RING1B resembles that of a ubiquitin module, and it serves to interact with CBX proteins (18). Furthermore, we have also shown that C-RING1B is capable of interacting with RING1 and YY1 binding protein (RYBP) (19, 20), which is a highly basic, oligomeric, intrinsically disordered protein (IDP) that interacts with DNA (19) and other proteins involved in apoptosis (21–23). The CBX proteins binding to C-RING1B undergo a tightening in their structures (24), and their presence hampers RYBP binding (25). Taken together, these results suggest that C-RING1B could be involved in binding to different partners, acting as a modulator of several protein cascades.
NUPR1 is an 82-residue-long (8-kDa), highly basic, monomeric IDP that is overexpressed during the acute phase of pancreatitis (26). As happens with many IDPs (27–29), NUPR1 does not have stable secondary and tertiary structure in any region of its sequence. Similarly to other chromatin proteins, NUPR1 binds to DNA (30); it can be regulated by cell signaling cascades to transduce gene regulatory, morphogenetic signals from cell membrane to the nucleus. Although NUPR1 is considered to function as a scaffold protein in transcription, and as an essential element for the stress cell response and in cell-cycle regulation, the distinct function mediated by this protein is currently debated (31, 32). Indeed, NUPR1 participates in the regulation of apoptosis by forming a complex with another IDP, prothymosin α (33), as well as being involved in DNA repair (34). In addition, NUPR1 plays key roles in pancreatic tumorigenesis, acting downstream of the KrasG12D oncogenes, which are critical for pancreatic carcinogenesis (35). Therefore, given the similarities in the physicochemical properties of RYBP and NUPR1 (namely, high isoelectric point, DNA-binding features, and an intrinsic disordered state), and the involvement of C-RING1B in some types of cancer, we hypothesize that this domain might also bind to NUPR1.
To test this hypothesis we carried out the in vitro characterization of the binding between both proteins by using spectroscopic (namely, fluorescence, NMR, and CD) and biochemical (proteolysis degradation) techniques. Furthermore, in cellulo assays were carried out by means of the protein ligation assay (PLA) and NUPR1 knock-out cells by CRISPR/Cas9n-mediated genome editing to assess the interaction specificity. Our hypothesis-driven experiments show that there was binding between these proteins both in cellulo and in vitro, with an affinity of ∼10 μM. The binding region of NUPR1 involves a hydrophobic patch at the 30s region of the protein, but the protein remained disordered upon binding. Blind in silico studies carried out by using the X-ray structure (18) of C-RING1B in interaction with all possible polypeptide patches of NUPR1 show that the binding region involves Ala33 of NUPR1 and its hydrophobic surroundings, as well as another hydrophobic patch around Thr68. We also tested the importance of these regions in binding to C-RING1B by generating the single, Thr68Gln, and the double, Ala33Gln/Thr68Gln, mutants (the single mutant Ala33Gln could not be expressed in Escherichia coli). Although the interaction with C-RING1B was not fully abolished in vitro, both the single and double mutants had larger dissociation constants (i.e., less affinity toward C-RING1B) than wild-type NUPR1, as measured by fluorescence and NMR. Thus, both positions (Ala33 and Thr68) are important for the binding of NUPR1 to C-RING1B, and Ala33 seems to be particularly critical to achieve local hydrophobic clustering. PLA was also performed in cellulo with the three NUPR1 mutants, and the results showed a complete disappearance of the interaction with C-RING1B. Interestingly enough, in contrast to the previously characterized and identified protein partners of NUPR1 [prothymosin α and male-specific lethal (MSL) proteins], C-RING1B is a well-folded protein. Our findings suggest that NUPR1 may play a C-RING1B–associated function within the Polycomb pathway, which may help to explain both its gene regulatory and oncogenic functions.
Results
C-RING1B and NUPR1 Interact in Vitro.
To test whether C-RING1B and NUPR1 interacted in vitro we followed a four-part approach. First, we tried isothermal titration calorimetry (ITC) experiments at several pH values above 7.0 between wild-type NUPR1 and C-RING1B; in all conditions explored we did not observe a binding-associated heat signal. Because we observed binding by using other techniques (discussed below), the absence of thermogram in ITC could be due to (i) a low affinity due to a high flexibility in the complex (36, 37) or (ii) a very small amount of heat exchange with the environment. Based on our results (discussed below), we suggest that the most plausible explanation is the latter.
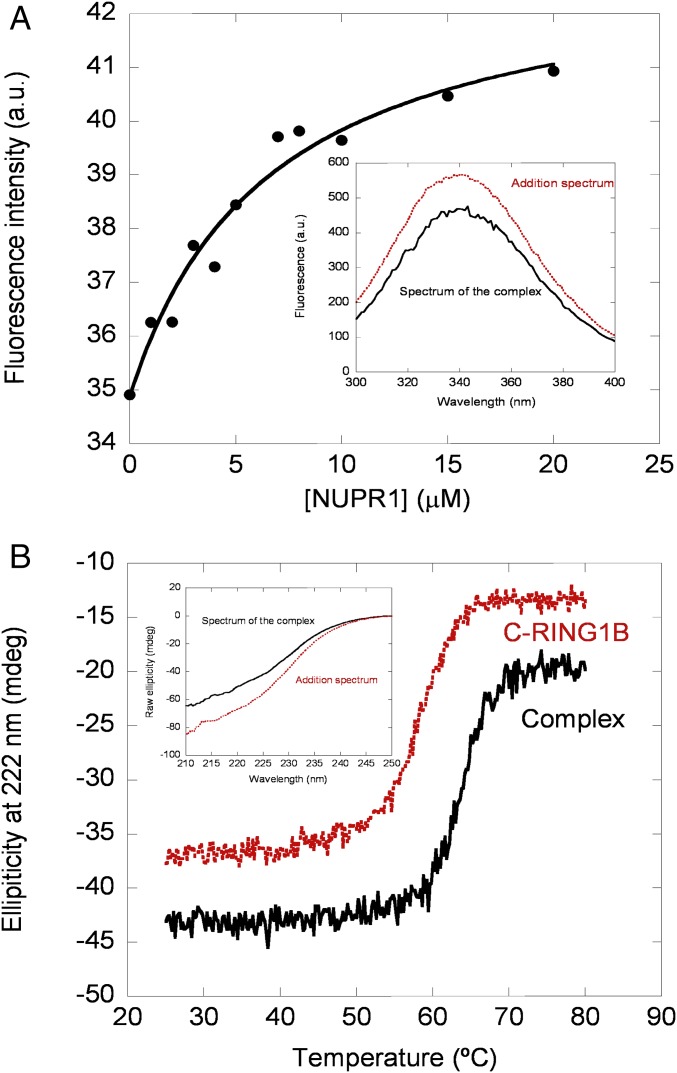
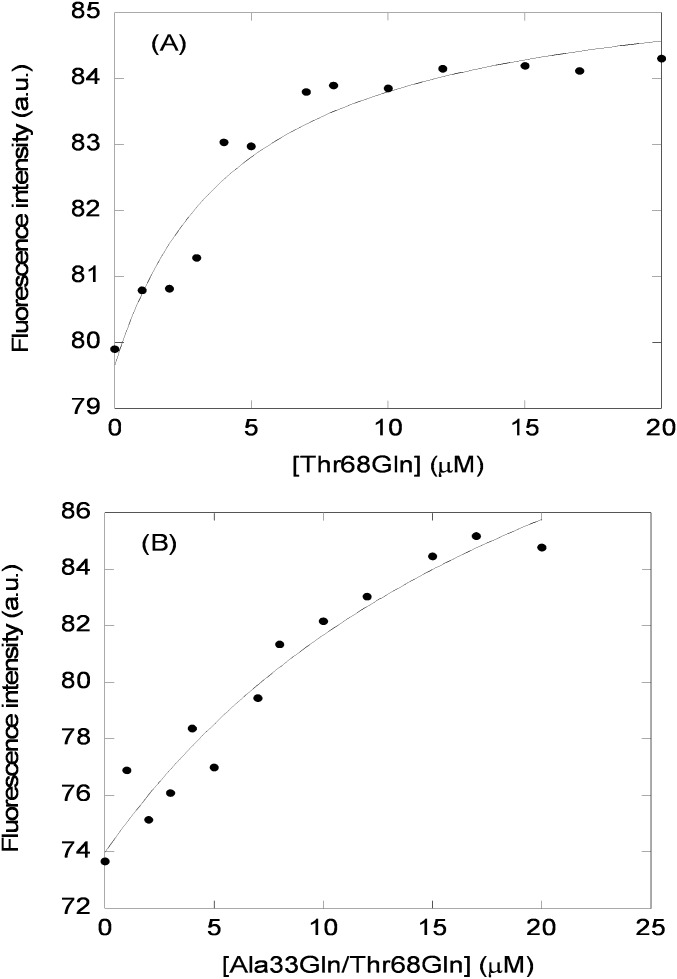
Second, we used fluorescence spectroscopy to determine whether there was either a change in the maximum wavelength or in the intensity when the spectrum of the complex was compared with that obtained from the addition of each one of the isolated proteins. A variation in fluorescence intensity was observed when the complex formed (Fig. 1A, Inset), but there were no changes in the maximum wavelength of the spectrum. We can conclude that the two proteins bind, and binding alters mainly the environment of the sole tryptophan of C-RING1B. Therefore, we used fluorescence to determine the affinity constant (Fig. 1A), leading to a Kd of 6 ± 2 μM, which results in a free energy of binding of −7.1 ± 0.3 kcal/mol. We also measured the binding of both mutants, Thr68Gln and Ala33Gln/Thr68Gln, to C-RING1B. Both mutants were designed to hamper the interaction based on our previous in silico results of the whole intact NUPR1 (38). The titration curves were always noisier than those of wild type (Fig. S1), yielding Kds of 9 ± 2 μM and 14 ± 5 μM for Thr68Gln and Ala33Gln/Thr68Gln, respectively, which are slightly larger than that measured in the wild type. These results suggest that the affinity of both mutants is not as large as that of the wild-type protein in vitro.
Fig. 1.
Spectroscopic characterization of the binding between C-RING1B and wild-type NUPR1. (A) Changes in the intensity at 330 nm with increasing amounts of wild-type NUPR1 after excitation at 295 nm. The line through the data is the fitting to Eq. 1, yielding a Kd of 6 ± 2 μM. (Inset) Fluorescence spectra of the complex between wild-type NUPR1 and C-RING1B (continuous black line) and that obtained by the addition of the spectra of each protein (dotted red line). (B) Thermal denaturations followed by the changes in ellipticity at 222 nm of the complex of the two proteins and that of isolated C-RING1B. (Inset) Far-UV CD spectra of the complex (continuous black line) and that obtained by the addition of the spectra of each protein (dotted red line). a.u., arbitrary units; mdeg, millidegrees.
Fig. S1.
Fluorescence characterization of the binding between C-RING1B and NUPR1 mutants. (A) Changes in the intensity at 330 nm with increasing amounts of Thr68Gln after excitation at 295 nm. The line through the data is the fitting to Eq. 1, yielding a Kd of 9 ± 2 μM. (B) Changes in the intensity at 330 nm with increasing amounts of Ala33Gln/Thr68Gln after excitation at 295 nm. The line through the data is the fitting to Eq. 1, yielding a Kd of 14 ± 5 μM. a.u., arbitrary units.
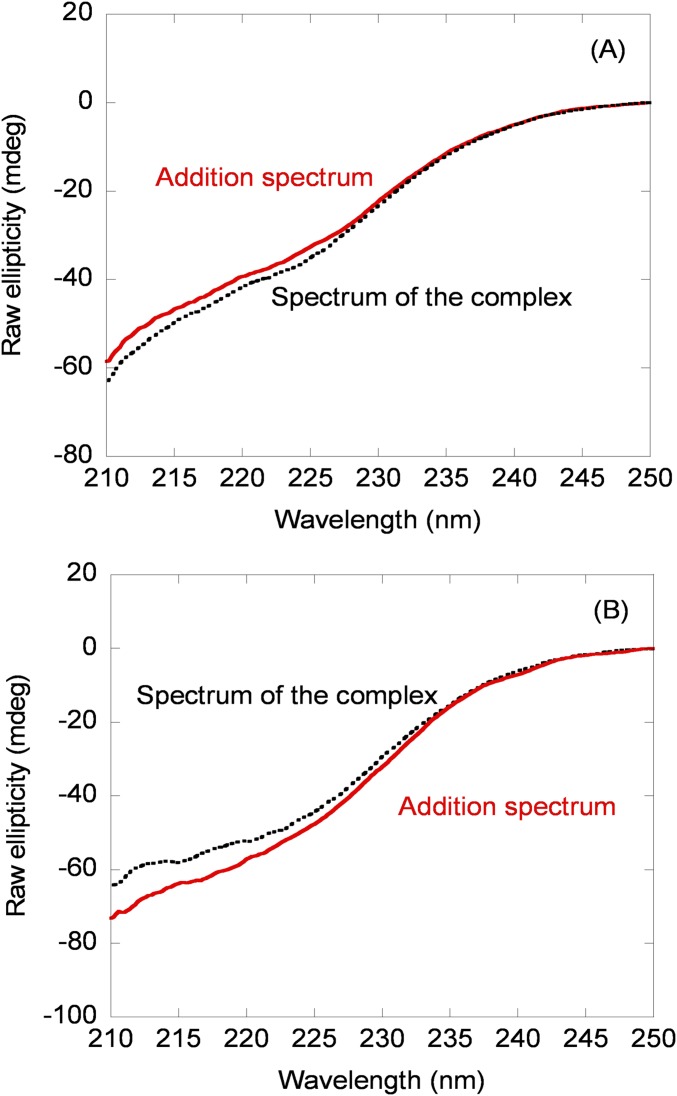
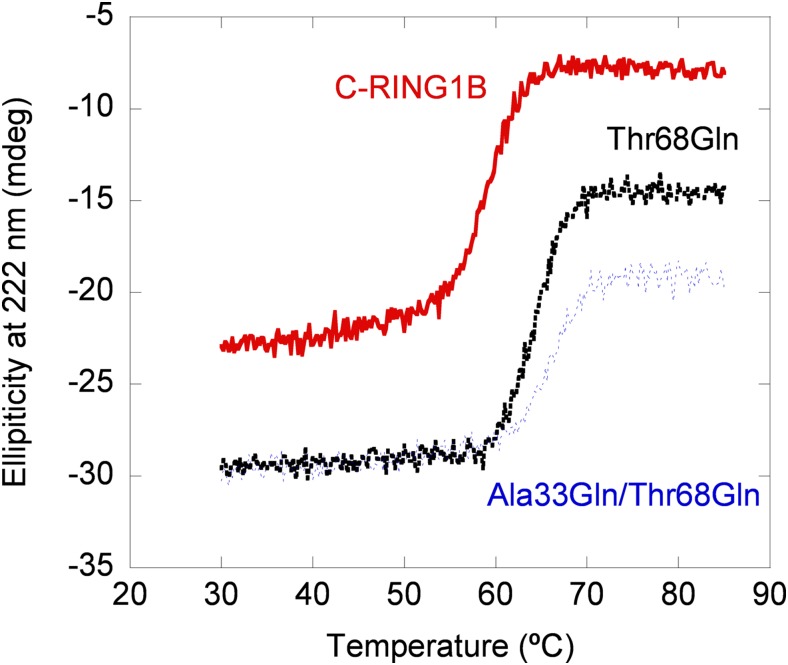
In the third step of our approach we carried out far-UV CD experiments. As in fluorescence, the addition spectrum (obtained from the sum of the isolated spectra of both proteins) for the wild-type NUPR1 was different from that of the complex (Fig. 1B, Inset). In both mutants there were differences between the addition spectrum of both C-RING1B and the corresponding mutant and that of the matching complex, but the changes were not as large as those of the wild-type protein (Fig. S2), suggesting a decreased affinity. Next, we carried out thermal denaturations of the corresponding complex and isolated C-RING1B: The interaction with wild-type or NUPR1 mutants should increase the thermal stability of C-RING1B if the complex dissociates before the rate-limiting step of denaturation. As expected, the presence of wild-type NUPR1 increased the apparent thermal denaturation midpoint (Fig. 1B). The presence of any of the mutants in solution with C-RING1B did also increase, as much as with the wild-type NUPR1, the apparent thermal midpoint for C-RING1B (Fig. S3). We did not determine Kd from changes in the denaturation midpoints due to the irreversibility of C-RING1B thermal denaturations (17) (with all NUPR1 species we observed precipitation). Thus, we conclude that we could detect binding by far-UV CD of wild-type NUPR1 and C-RING1B, leading to changes in the secondary structure of, at least, one of the proteins. We could also detect binding of C-RING1B to NUPR1 mutants, but in both cases the changes in the secondary structure were smaller compared with those observed in the binding to wild-type NUPR1.
Fig. S2.
Far-UV CD characterization of the binding between C-RING1B and NUPR1 mutants by using steady-state spectra. (A) Far-UV CD spectra of the complex (black line) and that obtained by the addition of the spectra of each protein (red line) in the mutant Thr68Gln. (B) Far-UV CD spectra of the complex (black line) and that obtained by the addition of the spectra of each protein (red line) in the mutant Ala33Gln/Thr68Gln. mdeg, millidegrees.
Fig. S3.
Far-UV CD characterization of the binding between C-RING1B and NUPR1 mutants by using thermal denaturations. Thermal denaturations followed by the changes in ellipticity at 222 nm of the complex with C-RING1B of the two NUPR1 mutants and that of isolated C-RING1B. To allow for a comparison the traces of the thermal denaturations have been scaled up, and then the y axis does not represent the measured ellipticity. mdeg, millidegrees.
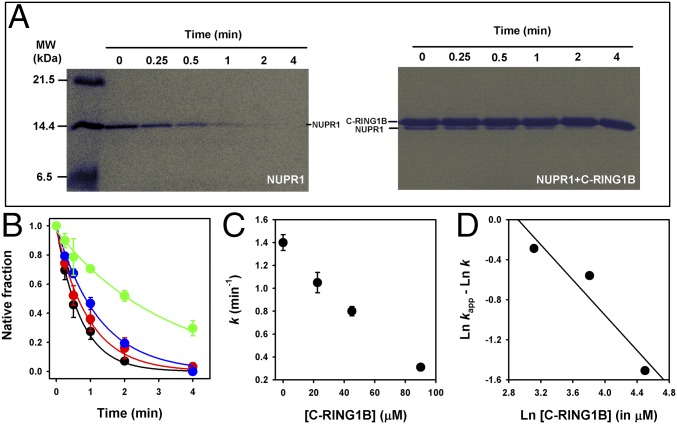
Finally, we tried to characterize the binding by means of proteolysis degradation (39). In Fig. 2A we show representative gels for the degradation of wild-type NUPR1 by thermolysin either in the absence or in the presence of C-RING1B. Even with a high proportion of acrylamide in the Tris-Tricine gels, some overlap occurred between the two bands. However, it did not preclude us from carrying out independent densitometric analyses of the bands. In Fig. 2 B and C we show the protection against degradation of wild-type NUPR1 by C-RING1B in a concentration-dependent manner. The analyses, using a simple model (SI Materials and Methods), are shown in Fig. 2D. The slope of this plot is −0.9 ± 0.2, consistent with a 1:1 stoichiometry (note that this slope should be equal to −1), and supports the adequacy of the proposed model. The y axis intercept is 2.6 ± 1.1 [= ln(Kd) in micromolar], corresponding to a Kd = 12 ± 5 μM. This value is similar, within the error, to that measured by fluorescence and leads to a free energy of −6.6 ± 0.7 kcal/mol.
Fig. 2.
Protection of wild-type NUPR1 toward degradation by thermolysin in the presence of C-RING1B. (A) Representative gels of the degradation kinetics of wild-type NUPR1 (10 μM) in the absence or presence of 45 μM (in protomer units) of C-RING1B. C-RING1B was resistant to proteolysis under our conditions (10 nM thermolysin). (B) Kinetics of degradation of wild-type NUPR1 in the absence (black symbols) or in the presence of C-RING1B [22.5 μM, red symbols; 45 μM, blue symbols, 90 μM, green symbols (all indicated in protomer units)]. Data are shown as means, and errors are SDs as obtained from three independent experiments. (C) First-order rate constants for wild-type NUPR1 degradation at different C-RING1B concentrations. (D) Plot based on a simple kinetic model to estimate the Kd value of the interaction between NUPR1 and C-RING1B. MW, molecular weight.
Thus, to sum up, the three techniques used show that C-RING1B and wild-type NUPR1 interact in vitro with a low affinity constant in the range of 10 μM. This interaction seems to be decreased in the two NUPR1 mutants.
The C-RING1B Binding Maps to a Hydrophobic Region Located at the 30s of NUPR1 Sequence.
To characterize the molecular bases behind the formation of the C-RING1B/NUPR1 complex we sought to determine the NUPR1 regions involved in binding. Because we have previously reported the NMR assignment of all residues of wild-type NUPR1 (34) (Biological Magnetic Resonance Data Bank no. 19364), we used 1H-15N heteronuclear single quantum correlation (HSQC) spectra of wild-type NUPR1 to monitor changes upon C-RING1B addition.
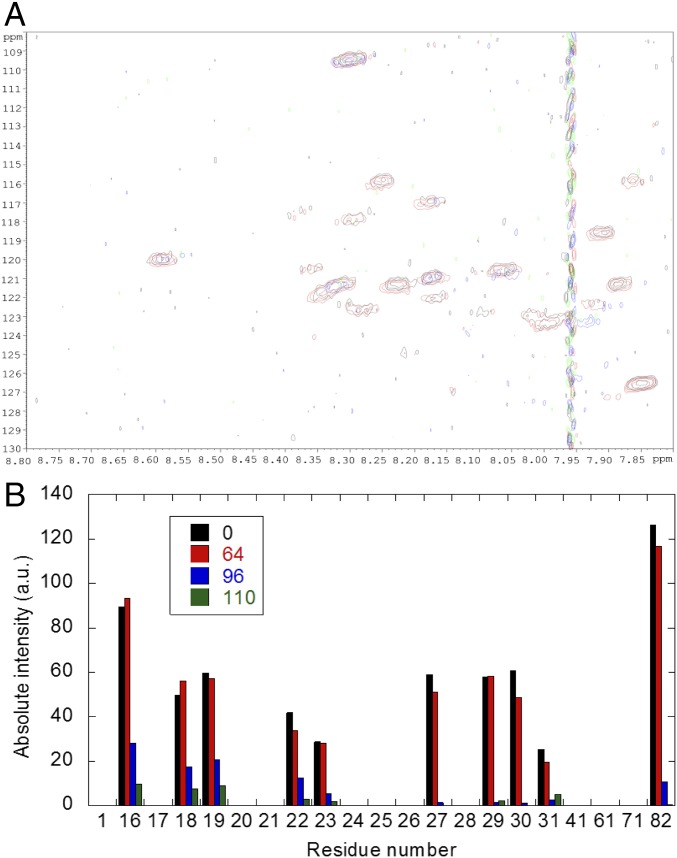
We carried out the experiments at pH 7.0, where C-RING1B was stable (17). At this pH, the most protected residues from exchange of wild-type NUPR1 that could be unambiguously assigned were (Fig. 3A) Gly16, Glu18, Asp19, Ser22 (Ser10), Ser23 (Asn53), Ser27, Leu29, Tyr30, Ser31, and Arg82 (the residues within parentheses are those where signal overlapping occurs). Most of these residues were close to the hydrophobic 30s region (38) or, alternatively, they were partly sequestered from the solvent by local structure or hydrogen bonding (40, 41). Upon addition of C-RING1B, the cross-peaks began to disappear (Fig. 3A), and those whose signal intensities decreased faster were Ser22 (Ser10), Ser23 (Asn53), Ser27, Leu29, Tyr30, and Ser31; this decrease suggests a closer proximity to the Polycomb protein. However, the residues whose cross-peak intensities were the largest at the highest wild-type [NUPR1] were Gly16, Glu18, and Asp19 (i.e., their cross-peak intensities decreased more slowly) (Fig. 3B); this result suggests that these residues are less close to C-RING1B. However, the decrease in intensities did not follow a hyperbolic behavior for any of the residues (Fig. 3B). Moreover, there was no variation in the chemical shifts of any of the residues, and then, given the exquisite sensitivity of chemical shifts, it seems that NUPR1 remained disordered upon binding. Furthermore, because there were changes only in the intensities, the equilibrium exchange between the free and bound wild-type NUPR1 must be intermediate to slow within the NMR time scale. It is important to note here that we observed larger intensity values for some NUPR1 residues at larger C-RING1B concentrations [see, for instance, Fig. 3A, residues Gly16, Glu18, and Leu29 at 0 and 64 μM (in protomer units) of C-RING1B, black and red bars, respectively]. We think that those variations are due to error of the measurements in the spectra, even though we corrected the measured values by the figure obtained when setting the receiving gain of the spectrometer. It is also well known that the measurement of intensities in 2D HSQC NMR spectra is plagued with several difficulties (42), and even the viscosity of the medium (which is altered by the presence of other molecules) can affect the value measured (43), even though at the total protein concentrations used in this work (∼500 μM) we have not observed broadening in the spectra of isolated NUPR1 (34) or C-RING1B (17). The facts that (i) broadening is only observed when titration is carried out (i.e., when the corresponding protein partner is added) and (ii) broadening of the signals of C-RING1B in the HSQC spectra follows a different pattern when the mutants are titrating (with molecular weight similar to that of the wild type, discussed below) suggest that the binding between the two proteins is mainly responsible for the broadening of signals. We also observed a large change in the intensities of residues ongoing from 64 to 96 μM (in protomer units) of C-RING1B; the reasons for this change are not fully understood, but they could be due to the increase of the dimeric population of C-RING1B.
Fig. 3.
Interaction of wild-type NUPR1 and C-RING1B mapped by HSQC spectra of NUPR1. (A) Overlay of wild-type NUPR1 spectra in the presence of different amounts of C-RING1B: 0 (black), 64 (red), 96 (blue), and 110 (green) μM (all indicated in protomer units). (B) Intensity decrease of cross-peaks (corrected by the value of the receiver gain of the spectrometer in each titrating point) in the 1H-15N HSQC spectra of wild-type NUPR1 as [C-RING1B] was increased (color code is the same as above). The signals appearing between 8.15 and 8.30 (1H dimension) and between 123 and 121 ppm (the 15N one) could not be unambiguously assigned. a.u., arbitrary units.
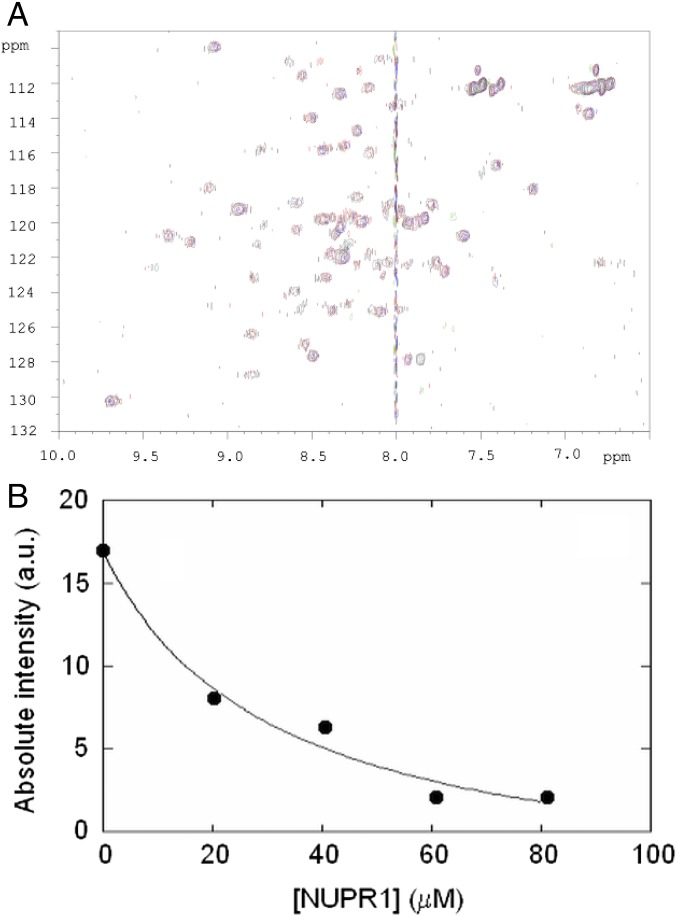
We also tested complex formation by using 15N-labeled C-RING1B, to which increasing amounts of wild-type NUPR1 were added (Fig. 4A). As happened with the 1H-15N HSQC spectra of wild-type NUPR1, the cross-peak intensities of the spectra of C-RING1B decreased upon wild-type NUPR1 addition. We did not observe changes in the chemical shifts of any cross-peak, suggesting that (i) binding is intermediate to slow within the NMR time scale and (ii) the structure of C-RING1B remained basically unaltered. In this case, however, the intensities for all its cross-peaks decreased at the same rate in a hyperbolic manner. Fitting of intensity decrease of the cross-peak appearing at 9.40 ppm in the spectrum of C-RING1B to Eq. 1 led to a Kd of 31 ± 16 μM (Fig. 4B), a slightly higher value than those measured by fluorescence and proteolysis. As a control, we also added to a 15N-labeled C-RING1B sample increasing amounts of unlabeled, intact RYBP (Fig. S4). As with wild-type NUPR1, we also observed a decrease of cross-peak intensity as [RYBP] was raised.
Fig. 4.
Interaction of wild-type NUPR1 and C-RING1B mapped by HSQC spectra of C-RING1B. (A) Overlay of C-RING1B selected spectra in the presence of different amounts of NUPR1: 0 (black), 20 (red), and 61 (blue) μM. (B) Fitting of the absolute intensity (corrected by the value of the receiver gain of the spectrometer in each titrating point) of the cross-peak of C-RING1B, appearing at 9.40 ppm to Eq. 1; the Kd was 31 ± 16 μM. a.u., arbitrary units.
Fig. S4.
Interaction of RYBP and C-RING1B mapped by HSQC spectra of C-RING1B. Overlay of spectra of C-RING1B in the presence of different amounts of RYBP at 0 (black), 20 (red), 40 (blue), and 60 (green) μM (all in protomer units).
We also tried to determine the binding affinity toward C-RING1B of the Thr68Gln and Ala33Gln/Thr68Gln mutants by means of HSQC NMR spectra of 15N-labeled C-RING1B. As it happens with fluorescence, the affinity of the mutants was substantially decreased compared with that of the wild-type NUPR1, with values (Kds) of 65 ± 15 μM and 115 ± 85 μM for the single and double mutants, respectively (Fig. S5). Thus, all of the techniques suggest that the interaction with C-RING1B is decreased by the two mutations, and it is lower in the double mutant. Our results with the mutants also pinpoint the 30s region of NUPR1 as a key polypeptide patch to attain binding.
Fig. S5.
Fitting of the absolute intensity (corrected by the value of the receiver gain in each titrating point) of the cross-peak of C-RING1B, appearing at 9.40 ppm to Eq. 1 for the Thr68Gln (A) and Ala33Gln/Thr68Gln (B) mutants. The Kd for Thr68Q was 65 ± 15 μM and 115 ± 85 μM for the double mutant. a.u., arbitrary units.
In conclusion, the NMR experiments further confirm that there is binding between the two proteins, involving residues around the 30s region of NUPR1 sequence.
In Silico Calculations Show That the NUPR1 Patch Involved in the Binding Is Also the 30s Region.
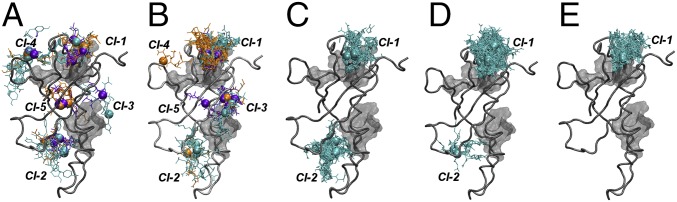
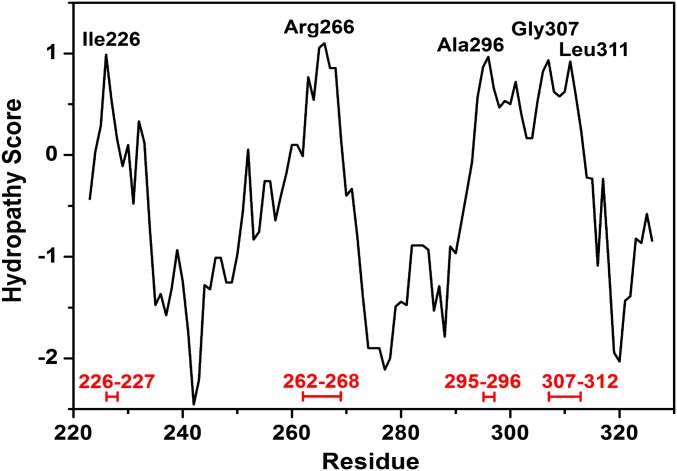
Fig. 5 shows docking poses obtained for representative portions of the wild-type NUPR1 sequence in our molecular docking calculations. The NUPR1 region around Ala33 shows the most favorable docking scores, with predicted binding energies up to –7.8 kcal/mol for the capped seven-residue fragment. This value is similar to the free energy experimentally obtained by fluorescence and proteolysis. The interaction sites of this fragment with C-RING1B, as well as those obtained with other shorter (five-residue and three-residue) fragments centered on Ala33, were observed to group in five well-defined locations, hereafter named clusters Cl-1 to Cl-5 (Fig. 5A). All these polypeptide patches comprise highly hydrophobic regions in C-RING1B, according to the Kyte–Doolittle hydropathy scale (44) (Fig. S6). Cluster Cl-1 is in between the two C-RING1B regions 226–227 and 307–312, both clusters Cl-2 and Cl-3 involve the region 262–268, cluster Cl-4 Ring1B comprises the polypeptide patch 295–296, and, finally, cluster Cl-5 is close to amino acids 307–312.
Fig. 5.
Docking poses of selected portions of wild-type NUPR1 sequence to C-RING1B. Fragments of NUPR1 are centered on (A) Ala33 and (B) Thr68. Fragment length for wild-type NUPR1 is either seven (cyan), five (orange), or three (purple) residues, with Cα atoms of the central amino acid represented as spheres; clusters Cl-1 to Cl-5 are also labeled. Regions with high hydrophobicity in C-RING1B are represented as transparent gray surfaces. The structures (C–E) report docked structures of seven-residue fragments centered around Lys47, Lys76, and Asp19, respectively. Figures were produced with VMD.
Fig. S6.
Hydropathy plot of C-RING1B. Selected protein residues are labeled, and the most hydrophobic segments are highlighted in red. The hydrophobicity scale of Kyte–Doolittle was used, by considering a window size of nine consecutive residues.
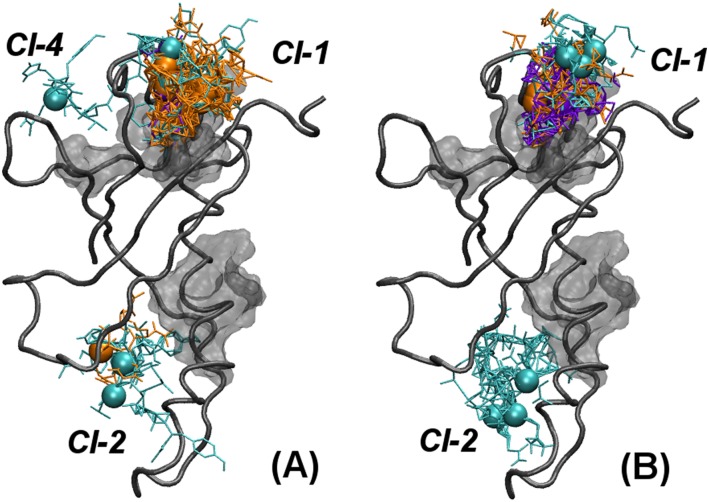
Both the binding affinity and the number of recurring locations, identified through the molecular docking procedure, decreased for protein fragments centered on other residues along the NUPR1 sequence. The region around the other key amino acid Thr68, which together with Ala33 constitutes the most hydrophobic regions of its sequence (38) and is another anchoring site to NUPR1 molecular partners, showed docking scores of about –6.0 kcal/mol. However, Thr68 has a slightly higher preference for binding to cluster Cl-1 of C-RING1B (Fig. 5B), compared with the preferred locations for Ala33. The number of clusters reduced to two (Cl-1 and Cl-2) for the most hydrophilic segments of the NUPR1 sequence, such as the highly charged basic regions around Lys47 and Lys76 (Fig. 5 C and D). Moreover, it reduced to only Cl-1 for the highly acidic region centered on Asp19 (Fig. 5E), which includes all of the residues whose cross-peak intensities decreased more slowly in our NMR experiments. Thus, the two clusters Cl-1 and Cl-2 represent C-RING1B–interacting regions with low specificity, which could assist the binding preferably through electrostatic interactions.
Our simulations reveal additional details about residues of C-RING1B involved in the binding of NUPR1, which were obscured in our experiments in vitro. In fact, NUPR1 binds close to Leu311 and Glu312 in Cl-1, and it interacts with most of the residues 262–268 region in Cl-2, and preferentially with residues Tyr262 and Val265 in Cl-3. In the cluster Cl-4, both residues Ala33 and Thr68 of NUPR1 bind to region 295–296 of C-RING1B, with some of their adjacent amino acids interacting more closely with the nearby region 307–312. Finally, Ala33 and Thr68, in Cl-5, interact with the region 307–312, and in particular with residues Gly307 and Ser308.
Although the two regions around Ala33 and Thr68 have in common the possibility of anchoring to several C-RING1B polypeptide patches, they also show some differences in their respective binding features. In fact, the affinity obtained for the seven-residue fragment centered on Ala33 decreased by up to 2 kcal/mol, when the length of the sequence portion was reduced to five or three residues. Conversely, the predicted binding energy for the region around Thr68 was not length-dependent on the NUPR1 fragment considered. These features may be explained in terms of a high specificity in the interaction of the side chain of Thr68 with the surface of C-RING1B within the clusters. In contrast, for Ala33 the binding was due to less-localized interactions that depended on a larger portion of the NUPR1 sequence, and it also included significant contributions by other adjacent protein residues.
We also carried out docking simulations of NUPR1 fragments including both Ala33Gln and Thr68Gln mutations. Both were observed to significantly alter the binding with C-RING1B, leading to interaction energies less favorable by ∼2 kcal/mol and to a reduced number of binding clusters (Fig. S7) compared with the wild-type protein. In the case of the Thr68Gln mutant, the binding affinity of the region around residue 68 toward C-RING1B becomes comparable to that of highly hydrophilic NUPR1 regions, such as the charged segments around Asp19, Lys47, and Lys76 (discussed above). For the Ala33Gln mutant, the binding properties were still not completely equivalent to the ones observed for the most hydrophilic regions of NUPR1, although they seemed to be drastically reduced.
Fig. S7.
Docking poses of selected portions of mutated NUPR1 sequence to C-RING1B. Fragments of NUPR1 mutants Ala33Gln and Thr68Gln centered on (A) Gln33 and (B) Gln68. Fragment length for NUPR1 is either seven (cyan), five (orange), or three (purple) residues, with Cα atoms of the central amino acid represented as spheres; clusters Cl-1, Cl-2, and Cl-4 are also labeled. Regions with high hydrophobicity in C-RING1B are represented as transparent gray surfaces. Figures were produced with VMD (66).
Then, our in silico results suggest that NUPR1 and C-RING1B interact through the same hydrophobic patch of the former protein mapped by NMR experiments, and that the mutations at Ala33 and Thr68 decreased the binding toward C-RING1B to a remarkable extent.
Evidence for the Interaction of RING1B with NUPR1 in an Intracellular Environment.
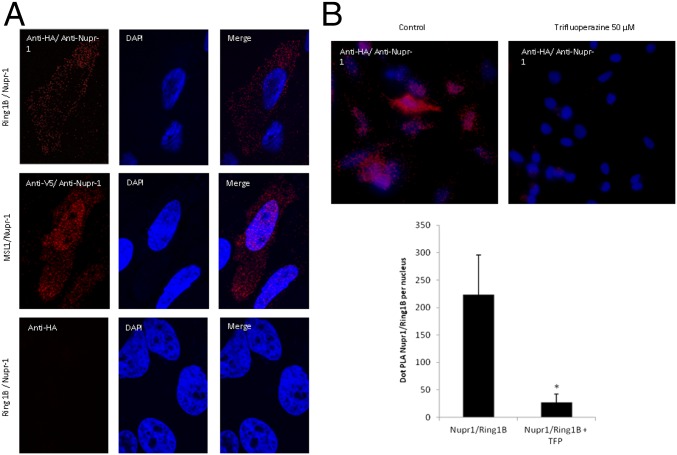
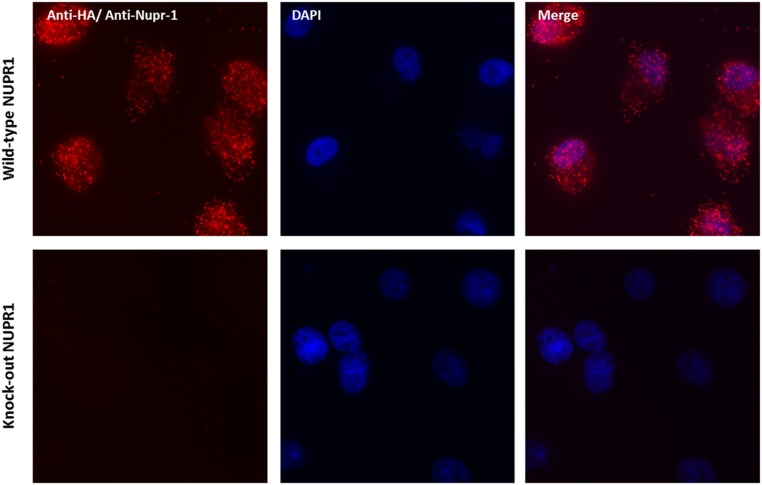
To test whether interaction between C-RING 1B and wild-type NUPR1 occurred within cells we used HeLa cells transfected with NUPR1-FLAG and RING1B-HA. Subsequently, we sought to detect their interaction using the Duolink in situ assay, which resolves the binding of proteins that occurs at distances less than 16 Å. Our results show that RING1B efficiently interacted with wild-type NUPR1 (Fig. 6A), showing red fluorescent spots, corresponding to the PLA signals. We also showed the specificity of wild-type NUPR1 for C-RING1B with the CRISPR/Cas9n method, by simultaneously developing control and NUPR1 knockout cell lines (Fig. S8). In this experiment, only PLA-positive signals were observed in control cells, and they were abolished in NUPR1 knockout cells, demonstrating that the interaction between both proteins is specific.
Fig. 6.
In situ PLA of C-RING1B and wild-type NUPR1. (A) Mouse anti-HA and rabbit anti-human wild-type NUPR1 were used to reveal the interaction between the proteins in HeLa cells. (B) PLA was carried out as described (34); cells were treated beforehand with TFP 50 μM for 24 h. ImageJ was used to count the number of red dots. Data are the means of 10 fields each containing not fewer than 150 nuclei counted (*P ≤ 0.001). (Magnification: A, 60×; B, 20×.)
Fig. S8.
Specific interaction between C-RING1B and wild-type NUPR1. In situ PLA in CRISPR/Cas9n control cells (with wild-type NUPR1) and CRISPR/Cas9n NUPR1 knockout cells. Mouse anti-HA and rabbit anti-NUPR1 were used to detect the protein interaction by PLA. DAPI staining was used to detect nuclei and combined with the PLA fluorescence in the merged panel. (Magnification: 40×.)
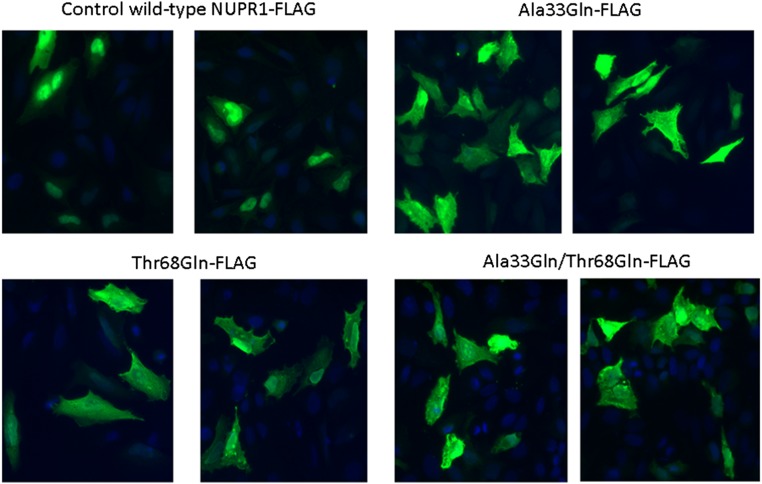
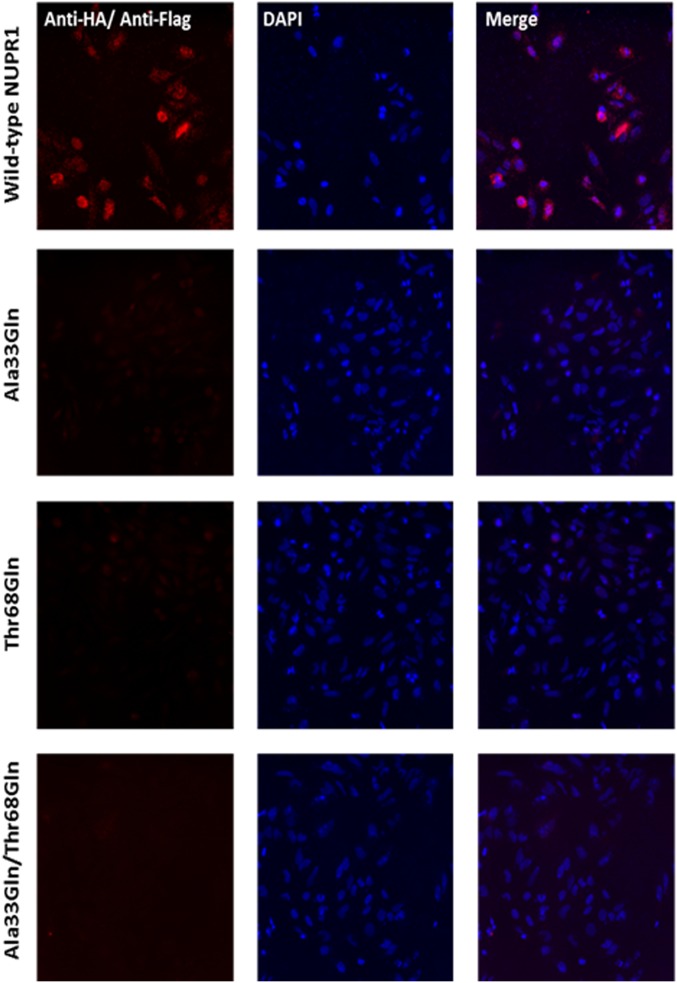
We carried out experiments with expression of the proteins with the FLAG label for both single (Ala33Gln and Thr68Gln) and double (Ala33Gln/Thr68Gln) mutants. In all cases (Fig. S9) we observed protein expression, but in wild-type NUPR1 transfected cells the fluorescence signal appeared more concentrated in the nucleus than when the NUPR1 mutants were used. That is, in the mutants the signals appeared also in the cytoplasm of the cells, as if a complete translocation of the protein was hampered. Finally, we performed the PLAs to test the interaction between C-RING1B and the three NUPR1 mutants in cellulo (Fig. S10). Under our conditions no positive PLA signals were observed in NUPR1 single mutants or in the double one, indicating that the two regions around Ala33 and Thr68 are crucial for the interaction between C-RING1B and NUPR1 also in an intracellular environment.
Fig. S9.
Immunofluorescence in HeLa cells transfected with wild-type and NUPR1 mutants. Mouse anti-FLAG primary antibody and Alexa 488-labeled goat anti-mouse secondary antibody were used to reveal the localization of the protein. DAPI staining was used to detect nuclei and it was combined with the Alexa 488 fluorescence in the merged panel. (Magnification: 20×.)
Fig. S10.
In situ PLA between C-RING1B and NUPR1 mutants. Mouse anti-FLAG and rabbit anti-HA were used to reveal the interaction between the proteins. Nuclei staining was performed with DAPI and combined with the PLA fluorescence in the merged images. (Magnification: 10×.)
We have previously shown (38) that the interaction of wild-type NUPR1 with MSL1 can be hampered by the addition of a drug (trifluoperazine, TFP) competing for the same binding site of the protein. Therefore, we wondered whether the interaction between RING1B and wild-type NUPR1 could be also inhibited by TFP. To this end, we incubated the HeLa transfected cells, as shown above, with TFP at a concentration of 50 μM. In this case, the positive red fluorescent signal in the cells was decreased, indicating that TFP blocked the interaction of the two proteins (Fig. 6B).
Thus, taken together, these results demonstrate that a specific C-RING1B/NUPR1 complex forms within cultured cells, where residues Ala33 and Thr68 of NUPR1 seem to intervene in the interacting region. These findings confirm the biological relevance of our simulations and the biophysical and biochemical in vitro experiments.
SI Materials and Methods
Materials.
Deuterium oxide was obtained from Apollo Scientific. Sodium trimethylsilyl [2,2,3,3-2H4] propionate (TSP), imidazole, Trizma base, nickel resin, and thermolysin from Bacillus thermoproteolyticus rokko were from Sigma-Aldrich. Dialysis tubing, with a molecular weight cutoff of 3,500 Da, was from Spectrapor (Spectrum Laboratories). Amicon centrifugal devices with a cutoff molecular weight of 3,000 Da were from Millipore. Standard suppliers were used for all other chemicals. Water was deionized and purified on a Millipore system.
Protein Expression and Purification.
Wild-type NUPR1 and C-RING1B were produced and purified in LB media as described (17, 30) in C41 cells. For 15N-labeled NUPR1 and C-RING1B, the same protocol as in LB was used, containing 15NH4Cl as the sole source of nitrogen. RYBP was expressed in LB as described (19).
We chose to mutate the positions, namely Ala33 and Thr68, based on our previous MD results of the isolated wild-type protein (38), where those residues are involved in hydrophobic clusters, acting as small “tongs” ready to interact with other regions (and/or molecules). We decided to mutate both residues to Gln to disrupt any possible contact with the rest of the polypeptide chain. The three mutants (a single at each position and a double one) were obtained by using the Quick Site Directed Mutagenesis kit (Stratagene), with the following pair of primers for Ala33Gln mutation: forward: ctgacctctatagcctgcagcattcctacctcggagg and reverse: cctccgaggtaggaatgctgcaggctatagaggtcag. The following primers were used for the Thr68Gln mutation: forward: ggcacgagaggaaactggtgcagaagctgcagaattcagagag and reverse: ctctctgaattctgcagcttctgcaccagtttcctctcgtgcc. Mutations were double-checked by DNA sequencing at GATC Biotech.
Expression of the three mutants was carried out initially under the same conditions as for the wild type. However, we observed that their expression was basically different from that of the wild type. The Thr68Gln was expressed nearly at the same amount as the wild-type NUPR1 (8 mg/L of culture versus 10–12 mg/L culture); however, the expression of Ala33Gln/Thr68Gln was substantially decreased (2–3 mg/mL of culture) and we did not obtain any expression of Ala33Gln. In the latter mutant, we tried several strains (C41, C43, and BL21) with different temperatures (25, 30, and 37 °C) after induction with several final amounts of isopropyl β-d-1-thiogalactopyranoside (0.5 and 1 mM); in all cases, we were unsuccessful. A possible explanation is that Ala33 maintains a local compactness of the protein that favors its expression; loss of this local compactness in the Ala33Gln mutant is partially (but not fully) prevented by the concomitant Thr68Gln mutation. Although indirectly, this result further supports the presence of long-range interactions between the two regions, which has been suggested by in silico results (38) and biophysical characterization of the electrostatics of the wild-type NUPR1 (41). Given the poor expression of the mutants in rich media we did not attempt to express them in minimal one. The mutants were purified in rich media following the same protocol as in the wild type (30).
CRISPR/Cas9n Clone Development.
Cells were seeded in 12-well plates and transfected with 1 µg of NUPR1 double nickase plasmid or double control nickase plasmid (Santa Cruz Biotechnology), delivered with 2 μL of Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) per well, following the manufacturer’s protocol. Transfected cell selection was performed with puromycin and after 72 h and isolation of single-cell colonies was done. Complete allelic knockouts were confirmed by Sanger sequencing.
CRISPR/Cas9n clones were developed to test whether the interaction between the two proteins is specific, by using a PLA assay. Control clones, with wild-type NUPR1 expression, and knockout clones, with a complete lack of expression of the protein NUPR1, have been used for this purpose. In these experiments, red fluorescence corresponds to the PLA-positive signal, and it indicates that the two molecules belong to the same protein complex. The objective of the assay was to demonstrate that the positive interaction that we showed in the PLA experiments in wild-type cells (red fluorescence) corresponds exclusively to the interaction between NUPR1 and RING1B, and it is not due to any possible interaction with other unstructured proteins with similar amino acid composition or to an uncommon unspecific binding of the primary antibodies. For this reason, we used wild-type and knockout clones, whose sole difference is the presence or the absence of one of the proteins of the complex under study (in this case, NUPR1). If the interaction between the proteins is specific, no positive signal should be observed in the knockout clones. However, if the interaction observed in the wild-type cells corresponded to an unspecific binding with other proteins, red fluorescence would be detected.
Basis of the PLA Approach.
Proximity ligation assay is a technology that allows us to visualize protein interactions with an easy-to-execute immunodetection protocol that is similar to traditional immunofluorescence. Briefly, the fixed samples are incubated with two primary antibodies from two different species and each one will bind one of the proteins of the complex under study. Then, secondary antibodies conjugated with oligonucleotides (named PLA probe MINUS and PLA probe PLUS) are added to the reaction and incubated. After that, the ligation solution, consisting of two oligonucleotides and ligase enzyme, is added to the samples. The oligonucleotides will hybridize to the two PLA probes and join to form a closed circle if they are in close proximity. Finally, the amplification solution, consisting of nucleotides and fluorescently labeled oligonucleotides, is added together with polymerase enzyme. The oligonucleotide arm of one of the PLA probes acts as a primer for a rolling-circle amplification (RCA) reaction using the ligated circle as a template, generating a repeated sequence product. At the microscope, we are able to visualize red fluorescent spots because fluorescently labeled oligonucleotides will hybridize to the RCA product.
We have chosen this technique because it combines the specificity of two primary antibodies (already validated in one of our laboratories by immunofluorescence) and two secondary antibodies, with the sensitivity afforded by RCA to detect protein interaction in cells. The pair of oligonucleotide-labeled secondary antibodies generates an amplified signal only when the probes are in close proximity (<40 Å).
Moreover, the PLA approach allows us to test in cellulo the results obtained in vitro, providing valuable information about the interaction of wild-type or mutant proteins on a more realistic model. Therefore, it is possible to demonstrate whether these proteins interact within a physiological context, in the presence of many more factors compared with the in vitro assays (where only the two proteins are present in solution).
Immunofluorescence.
HeLa cells were seeded in six-well plates on coverslips and transfected as previously described with NUPR1-FLAG and the different mutants. After fixation, cells were incubated with a mouse anti-FLAG (Sigma-Aldrich) primary antibody. After washing out the first antibody, cells were incubated with Alexa Fluor 488-labeled secondary antibody. Image acquisition of Alexa Fluor 488-derived fluorescence and DAPI staining was performed using a Nikon Eclipse 90i fluorescence microscope.
Proteolysis by Thermolysin.
Thermolysin was dissolved in Tris (50 mM, pH 7) containing NaCl (150 mM) and CaCl2 (100 mM), and stored at −80 °C; its concentration was measured by using ε280nm= 66,086 M−1·cm−1. Proteolysis was performed at 25 °C in Tris 50 mM, NaCl 150 mM, pH 7, using 10 μM of wild-type NUPR1 and 0–90 μM of C-RING1B (protomer units); the reaction was initiated by the addition of thermolysin (at a final concentration of 10 nM and 10 mM CaCl2) in a final volume of 200 μL. At different times, 20 μL of the reaction mixture were withdrawn and proteolysis was quenched by addition of 5 μL of EDTA, 100 mM, pH 8, mixed with Laemmli’s buffer and denatured at 95 °C for 5 min. Samples were resolved in 16% acrylamide Tris-Tricine PAGE gels, stained with Coomassie Blue, and densitometered using ImageJ (https://imagej.nih.gov/ij/). After normalization of the intensities by using that of the control sample in the absence of thermolysin, the intensity (I) versus time (t) curves were analyzed using a single exponential function:
| [S1] |
where k is the proteolysis rate constant and I0 the initial intensity.
To estimate the Kd for the interaction between the wild-type NUPR1 and C-RING1B we considered first the initial cleavage of NUPR1 (where N is the intact NUPR1 and F is the proteolyzed products) by thermolysin to follow a one-step model with first-order kinetics (k is the rate constant): , for which the rate law is
| [S2] |
In the presence of C-RING1B, the cleavage of wild-type NUPR1 is considered to follow first-order kinetics, with a rate constant kapp (whose value depends on [C-RING1B]): , where NL is the 1:1 C-RING1B/NUPR1 complex. In this case, the rate law is
| [S3] |
Assuming that wild-type NUPR1 in complex with C-RING1B is largely protected toward proteolysis, we can use the following model: , where the rate law is
| [S4] |
Because , we can substitute [N] into Eq. S4, and combine it with Eq. S3 to yield
| [S5] |
This leads to
| [S6] |
Therefore, a plot of ln(kapp) − ln(k) versus ln([C-RING1B]) yields a slope of −1 (if the stoichiometry of the interaction is 1:1) and a y axis intercept equals to ln(Kd).
Discussion
An IDP Partner for PcG Proteins.
NUPR1 is not the sole IDP capable of interacting with C-RING1B, because also RYBP protein binds to it (19, 20, 24, 25). The main physicochemical difference between RYBP and NUPR1 is that the former self-associates (both are IDPs, DNA-binding proteins with a high isoelectric point). We know that the C-terminal domain of RYBP becomes ordered upon binding to C-RING1B (25), but from the NMR spectra of NUPR1 (Fig. 4A), NUPR1 remained disordered. We have several pieces of evidence to support this hypothesis. First, we did not observe large changes in the steady-state far-UV CD spectra of the complex and that obtained from the sum of the isolated proteins. Second, we did not observe two transitions, or alternatively a very broad one, in the far-UV CD thermal denaturations (which could indicate the presence of two well-folded proteins that are dissociating). Finally, we could argue that the flexibility of the formed complex, where NUPR1 is not folded, was responsible for the absence of thermogram in ITC (37). The persistently disordered conformation of NUPR1 seems to be a general feature (33, 34, 38): Whatever molecule (synthetic, DNA, or protein) is bound to it, it is not capable of altering its disordered nature. However, binding to C-RING1B had an intermediate-to-slow exchange rate within the NMR time scale (either for C-RING1B or NUPR1). This time-scale regime was also kept in binding of C-RING1B to RYBP (Fig. S4), and it could be a feature derived from the disordered nature of both proteins. This finding means that the small variations in ellipticity at 222 nm observed in the far-UV CD upon binding (Fig. 1B) did not involve formation of a rigid structure, or at least not in NUPR1.
The interaction between the two proteins was also specific, as shown by two pieces of evidence: (i) the results in cellulo of the PLAs in NUPR1 knockout cells by CRISPR/Cas9n methods and (ii) the results obtained with the Thr68Gln and Ala33Gln/Thr68Gln variants of NUPR1 in vitro and in cellulo, whose mutations at those two specific residues decreased the interaction between NUPR1 and C-RING1B. These experimental results seemed to be confirmed by the docking in silico of the complexes of C-RING1B with NUPR1 mutants.
The monomeric CBX domains are also partially disordered in the absence of C-RING1B (24), but they seem to fold upon binding (25). The fact that the chemical shifts in the spectra of C-RING1B were not altered upon addition of NUPR1 (Fig. 4) species suggests that the basic scaffold of the protein remained unaltered (as happens upon addition of RYBP; Fig. S4).
Structural Determinants of the C-RING1B/NUPR1 Interaction in NUPR1.
The recognition region of NUPR1 involved residues around the 30s, as suggested by the NMR results with the wild type and the findings with the mutant species (Fig. 3), and further confirmed by molecular dynamics (MD) simulations (Fig. 5 and Fig. S7). The aromatic residues in this region also intervene in the binding to prothymosin α (33); furthermore, previous MD simulations suggest they also play a role in the binding to DNA (45) and in the binding of drugs against NUPR1 (38). In fact, in this work, we further show that one of those drugs (TFP) also hindered the binding between C-RING1B and wild-type NUPR1. Thus, we suggest that the same “hot-spot” region is used for binding any of the natural NUPR1 partners (DNA, MSL1, prothymosin α, and C-RING1B) and synthetic drugs.
The functional plasticity of NUPR1 is behind its high promiscuity; in multipartite binding, IDPs use small linearly arranged sequences of residues to mediate binding, the so-called short lineal motifs (SLiMs). These SLiMs tend to range from 3 to 13 residues in length, and therefore a single IDP can accommodate several SLiMs, which facilitates multivalent interactions with other molecules (46). However, a single IDP can have multiple SLiMs capable of recognizing distinct binding partners, acting as a scaffold for a high-order biomolecular complex formation (47). Our results suggest that NUPR1 has several SLiMs facilitating multivalent interactions, and these SLiMs are critical to allow the binding toward its different biomolecular partners. In our previous studies we have also shown that Thr68 is involved in the binding of NUPR1 to other macromolecules (38). The region around Thr68, together with the polypeptide patch around the 30s, is the most hydrophobic one on the Kyte–Doolitle scale (Fig. S6). In this work, because NMR experiments were carried out at pH 7.0 [where the hydrogen exchange with the solvent is faster than at pH 4.5 (40)], most of the residues of NUPR1 were not observed (because they are not hydrogen-bonded for long periods of time or, alternatively, they are not buried within a well-folded, compact structure). Thus, we did not have any experimental NMR evidence that this other region of NUPR1 was involved in binding to C-RING1B; however, the MD simulations suggest that Thr68 intervened in binding to NUPR1 (Fig. 5), similarly to the 30s region, and our mutagenesis studies also indicate that that amino acid intervenes in binding. It is interesting to note that some SLiMs in other IDPs have also been shown to function as high promiscuous binders; for instance, the transactivation domain of p53 is known to interact with multiple proteins by using the same binding motif with structurally distinct binding modes upon engagement of its several partners (48). Our results suggest that in NUPR1, Ala33, and Thr68 are also promiscuous SLiMs.
The measured Kd of NUPR1 for C-RING1B is similar to that observed for MSL1 (∼3 μM) (34), for prothymosin α (∼6 μM) (33), and for different drugs capable of binding to NUPR1 and competing with the binding between NUPR1 and MSL1 (38). Such affinity is relatively small, but we believe that this low value is necessary to obtain a proper regulation of the several pathways where NUPR1 intervenes (32), achieving a high specificity despite a low affinity. Affinities with a similar order of magnitude (i.e., in the range 1–10 μM) have also been described in the formation of “fuzzy” complexes, involving at least an IDP, which remains disordered upon binding (49, 50).
Finally, although the observation of how a sole mutation of a single residue in an IDP causing disruption of the interactions with molecular partners could seem unusual, there are a few examples in the literature. For instance, in the N-terminal membrane-binding site of α-synuclein, two familial mutations (Ala30Pro and Glu46Lys) have been hypothesized to alter membrane binding by changing the population of its conformational ensemble (51, 52).
Structural Determinants of the C-RING1B/NUPR1 Interaction in C-RING1B.
The X-ray structures of C-RING1B with CBX [3GS2 (25)] and the C-terminal region of RYBP [3IXS (25)] show that any of the latter proteins pack against the side chains of Val229, Pro234, Tyr247, Ile248, Thr250, Ala254, His258, Leu259, Tyr262, Val265, and Pro324, together with electrostatic interactions involving Glu227, Arg247, Lys249, and Arg266 (25) of C-RING1B. Many of these residues were observed to interact with NUPR1 in our in silico models (discussed above). Interestingly enough, from our fluorescence titration experiments (Fig. 1) the sole tryptophan (Trp319), or its surroundings (such as Pro324), of C-RING1B were also affected by NUPR1, although, probably, the environment of some of the seven tyrosines was also affected by the presence of the IDP. As we observed in our MD simulations, the sole Trp319 is close to cluster Cl-1, and then we had direct evidence (fluorescence results) of the validity of the MD findings. Therefore, as with NUPR1, the “hot-spot” region of C-RING1B in the interaction with any molecular partner is the same.
Although the binding regions identified for C-RING1B are fairly exposed to the solvent, the two patches accessible to any residue of NUPR1 (Cl-1 and Cl-2, Fig. 5) have a relatively large surface area, whereas only Ala33 and Thr68 can dock into smaller and more specific binding locations (Cl-3, Cl-4, and Cl-5). These two residues are not very far away in the ensemble conformation obtained in our MD simulations of the whole protein (38), and they can contribute to hold nearby binding partners in a sort of “weak hydrophobic clamp.” In such a mechanistic model these two residues would act as gentle tongs ready to close when another hydrophobic region (from a biomolecule or from a drug) is in the surroundings, as those in Cl-3, Cl-4, and Cl-5. When one end of the tong (for instance, Ala33) could be altered (by mutation), the binding could be severely crippled, although interaction with the other end of the tong (Thr68) would still be present and could be sufficient to maintain the binding. We can conclude, based on the hydrophobicity plots (Fig. S6) and our own mutagenesis studies, that hydrophobicity has a central role for both proteins in determining their interactions; however, additional and significant contributions include local geometric features and other particular characteristics of the specific binding residues involved. Examples include the possibility of hydrogen bonding for the side chain of Thr68 in NUPR1, or electrostatic interactions for both proteins.
Conversely to what happens with NUPR1, the affinities of C-RING1B for CBX or the C-terminal region of RYBP are much larger [in the nanomolar range (25)]. This result implies that in the simultaneous presence of the three proteins (C-RING1B, NUPR1, and either CBX or RYBP), unless there is a kinetic effect C-RING1B will first bind to CBX or RYBP, due to their larger affinity.
C-RING1B is a dimeric protein (its self-association constant is ∼200 μM) (17). Except for the NMR titration experiments, the rest of the experiments were carried out under conditions where the dominant C-RING1B species was the monomer. These findings suggest that the “competent” species in binding to NUPR1 is the C-RING1B monomer. The other biomolecules known to interact with NUPR1 are either monomeric (prothymosin α) (33) or oligomeric (MSL1) (34), but in the latter case we do not know the exact self-association state of the protein. The fact that the titration of 15N-labeled NUPR1 did not lead to a reliable estimation of Kd, conversely to what we observed in the titration of 15N-labeled C-RING1B (where the concentration was held constant at 125 μM, corresponding to a 30% population of dimer) with any of the NUPR1 species, further suggests that the monomeric species of C-RING1B is the “competent” one. A similar behavior, when involving self-associated species, which is the titrating molecule in the titration experiment, has been observed in fluorescence experiments of protein–DNA interactions (53).
Possible Biological Implications.
At this point, it is important to stress that the discovery of this interaction in cellulo between the two proteins was prompted by a comparison with the physicochemical properties of other already-known partners of C-RING1B with the features of NUPR1. Thus, it should be interesting to test whether similar hypothesis-driven studies can be carried out with other partners of both proteins to identify possible protein cascades.
In light of these findings, and on the basis of the known functions of both proteins NUPR1 and C-RING1B, it becomes important to speculate on the potential implications of their interactions for cancer development. Members of the Polycomb complexes of epigenomic regulators are well-characterized cell fate regulators, through their ability to silence entire gene expression networks that give rise to distinct phenotypes (6, 54). In addition, PRCs are important elements in stem-cell renewal and differentiation (54, 55). These proteins are among the best-characterized “epigenomic oncogenes,” playing significant roles in a large variety of solid tumors, as well as lymphomas and leukemias. Therefore, due to their ability to regulate gene expression processes that determine cellular patterns of morphogenesis, Polycomb complexes are key regulators of cell development, maintain tissue homeostasis, and, when altered, give rise to cancer. NUPR1 plays a major role in pancreatic ductal adenocarcinoma (PDAC), because the oncogenic KrasG12D expression in the mouse pancreas is unable to promote precancerous lesions in the absence of NUPR1 expression (31, 32, 35). In addition, RING1B is weakly expressed in pancreatic intraepithelial neoplasias but strongly expressed in PDAC [as well as in hepatocellular carcinoma (14)]. Therefore, we speculate that NUPR1 and C-RING1B might intervene in regulating some of the pathways involved in chromatin-mediated processes, gene regulation, and/or carcinogenesis, where the binding between both proteins can be shifted by the presence of other proteins such as RYBP and CBX (due to the higher value of their affinity constants). Alternatively, it could be suggested that under conditions of low expression of RYBP, NUPR1 could help in carrying out its same functions. The expression of RYBP has been reported to be reduced in lung, cervical, prostate, and liver cancers; in addition, RYBP has an apoptosis-inducing capacity, inhibiting cancer growth, metastasis, and chemoresistence in vitro and in vivo (56). Because NUPR1 in complex with prothymosin α can regulate the cell’s apoptotic response (31, 33), we hypothesize that in the absence of RYBP (as in some types of cancer) the cell expresses larger amounts of NUPR1 to compensate for that loss, taking over the functions of RYBP in a sort of self-defense mechanism. In this sense, it is interesting to note that in noncanonical complexes of PRC1 other DNA-binding proteins are heterodimeric transcription factors, such as the Myc-associated factor X (ref. 57 and references therein). Therefore, in any of the possible scenarios NUPR1 could be a modulator of the activity of PcG proteins by binding to RING1B and controlling its binding to other macromolecules.
Our work provides structural and functional evidence describing the interaction of NUPR1 with RING1B, through its C-terminal region. Thus, it becomes relevant to discuss the potential roles of this interaction. In this regard, we underscore the fact that RING1B is a ubiquitin E3 ligase, whose catalytic activity is activated by interaction with other proteins. Noteworthy, however, it has been previously shown that RING1B can also interact with other DNA-binding proteins, which are not part of the PcG (57). These interactions are thought to direct the E3 ligase activity on other chromatin proteins that do not belong to the Pc complex. Furthermore, RING1B not only self-ubiquitinates but also interacts with other proteins, modulating their degradation (57, 58). Thus, it seems that RING1B has both Pc-dependent and Pc-independent functions by associating with other chromatin proteins. Our data reveal that RING1B binds to another IDP chromatin protein, and they pave the way for future investigation as to whether this complex plays a role in either Pc-dependent or -independent processes.
Materials and Methods
Fluorescence.
Fluorescence spectra were collected on a Cary Varian spectrofluorimeter (Agilent) interfaced with a Peltier, at 25 °C. Experiments were carried out at pH 6.8 (20 mM phosphate buffer), where C-RING1B has a well-folded conformation (17). The samples were prepared the day before and left overnight at 5 °C. A 1-cm-pathlength quartz cell (Hellma) was used.
Steady-state spectra.
Protein samples were excited at 280 and at 295 nm to characterize a possible different behavior of tryptophan or tyrosine residues in C-RING1B (59). There are only two tyrosine residues in NUPR1 and mutants; C-RING1B has a single tryptophan and seven tyrosines. Sample concentration was 28 μM of C-RING1B (in protomer units) and 14 μM of wild-type NUPR1. The slit widths were 5 nm for both the excitation and emission light. The experiments were recorded between 300 and 400 nm. The signal was acquired for 1 s and the increment of wavelength was set to 1 nm. Blank corrections were made in all spectra.
Binding experiments.
Increasing amounts of wild-type NUPR1 or mutants, in the range 0–20 μM, were added to a solution with a fixed concentration of C-RING1B (5 μM of protomer). Under these conditions, C-RING1B is mainly in its monomeric form [because its self-association constant is ∼200 μM (17)]. The experimental set was that described above. The titrations were repeated three times. The fluorescence values of a blank solution containing only wild-type NUPR1 or its mutants were subtracted for each point. The dissociation constant of each complex, Kd, was calculated by fitting the plot of the observed fluorescence change of C-RING1B versus added NUPR1 proteins to the following equation (53, 60):
| [1] |
where Fmeas is the measured fluorescence at any particular concentration of NUPR1 species after subtraction of the blank; ΔFmax is the difference in the fluorescence of C-RING1B when all of NUPR1 species is forming the complex with the fluorescence of isolated C-RING1B; F is the fluorescence intensity when no NUPR1 species has been added; [C-RING1B]T is the constant, total protein concentration of C-RING1B (5 μM in protomer units); and [NUPR1]T is the total concentration of NUPR1 species, which is varied during the titration. The Kd was determined by following the fluorescence at 330 nm. Each titration was repeated three times. At all used concentrations the absorbance of NUPR1 species was kept lower than 0.2 units of absorbance (at 280 nm) to avoid inner filter effects during fluorescence excitation (61).
CD.
CD spectra were collected on a Jasco J810 or J815 spectropolarimeters (Jasco) fitted with thermostated cell holders and interfaced with Peltiers. The instruments were periodically calibrated with (+) 10-camphorsulphonic acid.
Far-UV steady-state spectra.
Isothermal wavelength spectra of either the isolated proteins or the complex of C-RING1B (31 μM protomer units) and wild-type NUPR1 (at 20 μM) at pH 6.8 (20 mM phosphate buffer) were acquired at a scan speed of 50 nm/min with a response time of 2 s and averaged over four scans at 25 °C. For the experiments with the mutants the concentrations of any of the two species were the same as for the assays with the wild-type NUPR1. Measurements were performed in 0.1-cm-pathlength quartz cells. All spectra were corrected by subtracting the baseline. The samples were prepared the day before and left overnight at 5 °C.
Thermal denaturation experiments.
Thermal denaturation experiments of isolated C-RING1B (31 μM protomer units) and of the complex [formed by 31 μM (protomer units) of C-RING1B and 20 μM of NUPR1 species] were performed at heating rates of 60 °C/h and a response time of 8 s at pH 6.8 (20 mM phosphate buffer). Thermal scans were collected at 222 nm from 25 to 80 °C in 0.1-cm-pathlength cells. The possibility of drifting of the CD spectropolarimeter was tested by running two samples containing only buffer, before and after the thermal experiments; no difference was observed between the scans. It is important to note that isolated wild-type NUPR1 does not show any sigmoidal transition because it is an IDP (30). However, thermal denaturations of C-RING1B are irreversible (17); in fact, precipitation of C-RING1B was always observed either in the presence or the absence of any of the NUPR1 species.
NMR Spectroscopy.
The NMR experiments were acquired on a Bruker Avance DRX-500 spectrometer equipped with a triple resonance probe and z-pulse field gradients. Experiments were acquired at 25 °C and pH 7.2 (Tris, 50 mM); probe temperature was calibrated with a methanol NMR standard (62). The peaks in the 2D 1H-15N HSQC NMR spectra (63) of NUPR1 were identified by using previously determined assignments at pH 4.5 (34). Spectra were acquired in echo–antiecho mode, with the carrier frequency at 8.00 ppm. For experiments using 15N-labeled NUPR1, a [NUPR1] of 150 μM was used, and increasing concentrations of unlabeled C-RING1B (64, 96, and 110 μM, in protomer units) were added. At each step, the final volume of the resulting sample was reduced to 500 μL by using Amicon centrifugal devices. For experiments with 15N-labeled C-RING1B, a [C-RING1B] of 125 μM (in protomer units) was used in all experiments, and increasing concentrations of unlabeled wild-type NUPR1 (20, 40, 60, and 80 μM) were added. Similar concentrations were used for Thr68Gln, but higher ones were used for the double mutant (Fig. S5). Spectra for the mutants were acquired in the TPPI mode (62). The final volume of the resultant sample (after addition of the corresponding titrating protein) was reduced to 500 μL by using Amicon centrifugal devices at each step.
The spectra were typically acquired with 2 K complex points in the 1H dimension, 60 complex points in the 15N dimension, with 32 or 64 scans. Typical spectral widths were 6,000 (1H) and 1,500 (15N) Hz. The resulting matrix of each experiment was zero-filled to double the number of original points in all dimensions and shifted squared sine-bell apodization functions were applied, before Fourier transformation. NMR data were processed and analyzed using Topsin 1.3 (Bruker). Signal intensities in the NMR titration experiments were measured by using the same program, and in each spectrum intensities were corrected by the corresponding value of the receiver gain at each particular concentration of the proteins that was being added during the titration. Spectra were calibrated with external TSP for 1H and for the indirect dimensions as described (64).
PLA.
HeLa cells were seeded in six-well plates on coverslips and transfected with 2 µg of DNA (NUPR1-FLAG and RING1B-HA) RING1B-HA and the mutants NUPR1-Ala33Gln-FLAG NUPR1-Thr68Gln-FLAG and NUPR1-Ala33Gln/Thr68Gln-FLAG) and 6 μL of Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) per well. Cells were assayed 24 h posttransfection. Cells were washed twice in PBS, fixed, washed twice again, permeabilized in PBS/0.1% Triton X-100, and saturated with blocking solution for 30 min before immune-staining with Duolink by using PLA Technology (Sigma-Aldrich) following the manufacturer’s protocol. Slides were processed for in situ PLA by using sequentially the Duolink In Situ Detection Reagents Red, Duolink In Situ PLA Probe Anti-Mouse MINUS, and Duolink In Situ PLA Probe Anti-Rabbit PLUS. The following antibodies were used: mouse monoclonal anti-HA (12CA5; Santa Cruz Biotechnology), mouse monoclonal antibody anti-FLAG M2 (Sigma-Aldrich), rabbit monoclonal antibody anti HA (Cell Signaling Technology), and rabbit anti-human NUPR1 antibody chemically synthesized (Neosystem) following previously described methods (65). In these experiments, red fluorescence corresponds to the PLA-positive signal, and it indicates that the two molecules belong to the same protein complex. Blue fluorescence corresponds to nuclei (so-called DAPI staining). To check the specificity of the PLA signal, negative control experiments omitting one of the primary antibodies and positive control with NUPR1 and MSL1 (MSL1-V5 plasmid) interaction (34) were performed. Unspecific binding was optimized by using the proper antibodies and optimizing the transfection times. Protein overexpression was used to obtain a clearer and better signal.
Preparations were mounted using Prolong Gold antifade reagent (Invitrogen) and image acquisition was carried out on an LSM 510 META confocal microscope (Zeiss) and on a Nikon Eclipse 90i fluorescence microscope. Statistical analysis was performed by using a Student’s t test and the measurement was expressed as mean ± SD.
In Silico Methods.
Model of the C-RING1B structure.
The structure was built by using as a starting template the X-ray structure of unbound C-RING1B [3H8H (18)], which includes protein residues 224–330. Missing side-chain atoms in five solvent-exposed Glu/Lys residues were built in silico by using VMD (66). The N-terminal portion and two missing loops (residues 240–242 and 273–284) were reconstructed through the I-TASSER prediction server (67), with the rest of the protein as a guiding template.
The complete structure was refined through MD simulations, to obtain a representative ensemble. The protein was equilibrated first in a short NVT run (10 ps) performed in the presence of restraints on the positions of nonhydrogen atoms determined in crystallography, and then in a constraint-free run carried out for 1 ns at room temperature and pressure. The simulation package GROMACS (68) was used with the AMBER ff99SB force field (69). Other simulation conditions that included thermostat and barostat reference values and coupling times, treatment of electrostatics and van der Waals interactions, use of restraints and time step in the integration of the equations of motions, and treatment of periodic boundary conditions were as described (70, 71).
Molecular docking.
The binding affinity of NUPR1 (wild-type or mutated species) and C-RING1B was evaluated by performing a systematic molecular docking. Portions of the NUPR1 sequence containing seven, five, or three consecutive protein residues were considered. The rest of the backbone was truncated and (when necessary) each fragment was capped with an acetyl or amide group at the N or C terminus, respectively. We used this procedure, instead of considering the entire polypeptide chain, to reduce the exceedingly large number of possible degrees of freedom in the calculation. This allows for an efficient sampling of all local regions of NUPR1 bound to C-RING1B, whereas the rest of the protein is assumed to interact mainly with the solvent.
The graphical interface AutoDock Tools 1.5.6 (72) was used to delete apolar hydrogen atoms and select permissible dihedral angle torsions for the fragments; molecular docking was performed by using AutoDock Vina (73). The docking search included the whole surface of C-RING1B, and it was performed with high exhaustiveness: The number of internal independent runs, and consequently the simulation time, was higher than the default value by a factor 12.5 (a linear increase of the simulation time corresponds to an exponential increase in the probability of finding proper energetic minima). For each simulation, the best 20 docking conformations were obtained. This number was reduced by eliminating the binding modes with a difference in the affinity score <0.5 kcal/mol with respect to the most favorable conformation.
Data Availability.
All of the materials (plasmid constructs) and data are available from the authors upon request.
Acknowledgments
We thank the two anonymous reviewers for helpful suggestions and discussions. J.L.N. thanks May García, María del Carmen Fuster, and Javier Casanova for excellent technical assistance. This work was supported by Spanish Ministry of Economy and Competitiveness Grants CTQ 2015-64445-R (to J.L.N.) and BIO2015-66426-R (to Á.L.P.); Spanish regional government Junta de Andalucía Grant P11-CTS-07187 (to Á.L.P.); La Ligue Contre le Cancer, INCa, Canceropole Provence-Alpes-Côtes d’Azur, Direction Générale de l’Offre de Soins (Sites de Recherche Intégrée sur le Cancer), and INSERM (J.L.I.); and Fundación Alfonso Martín-Escudero (P.S.-C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619932114/-/DCSupplemental.
References
- 1.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 2.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 4.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaustov L, et al. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem. 2011;286:521–529. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grau DJ, Antao JM, Kingston RE. Functional dissection of Polycomb repressive complex 1 reveals the importance of a charged domain. Cold Spring Harb Symp Quant Biol. 2010;75:61–70. doi: 10.1101/sqb.2010.75.056. [DOI] [PubMed] [Google Scholar]
- 10.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto Y, Abe A, Emi N. Clarifying the impact of polycomb complex component disruption in human cancers. Mol Cancer Res. 2014;12:479–484. doi: 10.1158/1541-7786.MCR-13-0596. [DOI] [PubMed] [Google Scholar]
- 13.Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y, Hu B, Wei L, Jiang D, Zhu M. Upregulated expression of polycomb protein Ring1 contributes to poor prognosis and accelerated proliferation in human hepatocellular carcinoma. Tumour Biol. 2015;36:9579–9588. doi: 10.1007/s13277-015-3721-7. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, et al. Polycomb group oncogene RING1 is over-expressed in non-small cell lung cancer. Pathol Oncol Res. 2014;20:549–556. doi: 10.1007/s12253-013-9727-9. [DOI] [PubMed] [Google Scholar]
- 16.Grzenda A, Ordog T, Urrutia R. Polycomb and the emerging epigenetics of pancreatic cancer. J Gastrointest Cancer. 2011;42:100–111. doi: 10.1007/s12029-011-9262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czypionka A, et al. The isolated C-terminal domain of Ring1B is a dimer made of stable, well-structured monomers. Biochemistry. 2007;46:12764–12776. doi: 10.1021/bi701343q. [DOI] [PubMed] [Google Scholar]
- 18.Bezsonova I, et al. Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins. Biochemistry. 2009;48:10542–10548. doi: 10.1021/bi901131u. [DOI] [PubMed] [Google Scholar]
- 19.Neira JL, et al. The transcriptional repressor RYBP is a natively unfolded protein which folds upon binding to DNA. Biochemistry. 2009;48:1348–1360. doi: 10.1021/bi801933c. [DOI] [PubMed] [Google Scholar]
- 20.García E, Marcos-Gutiérrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng L, Schickling O, Peter ME, Lenardo MJ. The death effector domain-associated factor plays distinct regulatory roles in the nucleus and cytoplasm. J Biol Chem. 2001;276:31945–31952. doi: 10.1074/jbc.M102799200. [DOI] [PubMed] [Google Scholar]
- 22.Danen-van Oorschot AA, et al. Human death effector domain-associated factor interacts with the viral apoptosis agonist Apoptin and exerts tumor-preferential cell killing. Cell Death Differ. 2004;11:564–573. doi: 10.1038/sj.cdd.4401391. [DOI] [PubMed] [Google Scholar]
- 23.Gervais FG, et al. Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel partner Hippi. Nat Cell Biol. 2002;4:95–105. doi: 10.1038/ncb735. [DOI] [PubMed] [Google Scholar]
- 24.Wang R, et al. Structural transitions of the RING1B C-terminal region upon binding the polycomb cbox domain. Biochemistry. 2008;47:8007–8015. doi: 10.1021/bi800857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure. 2010;18:966–975. doi: 10.1016/j.str.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallo GV, et al. Cloning and expression of the rat p8 cDNA, a new gene activated in pancreas during the acute phase of pancreatitis, pancreatic development, and regeneration, and which promotes cellular growth. J Biol Chem. 1997;272:32360–32369. doi: 10.1074/jbc.272.51.32360. [DOI] [PubMed] [Google Scholar]
- 27.Uversky VN. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tompa P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Encinar JA, et al. Human p8 is a HMG-I/Y-like protein with DNA binding activity enhanced by phosphorylation. J Biol Chem. 2001;276:2742–2751. doi: 10.1074/jbc.M008594200. [DOI] [PubMed] [Google Scholar]
- 31.Cano CE, Hamidi T, Sandi MJ, Iovanna JL. Nupr1: The Swiss-knife of cancer. J Cell Physiol. 2011;226:1439–1443. doi: 10.1002/jcp.22324. [DOI] [PubMed] [Google Scholar]
- 32.Goruppi S, Iovanna JL. Stress-inducible protein p8 is involved in several physiological and pathological processes. J Biol Chem. 2010;285:1577–1581. doi: 10.1074/jbc.R109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malicet C, et al. Regulation of apoptosis by the p8/prothymosin alpha complex. Proc Natl Acad Sci USA. 2006;103:2671–2676. doi: 10.1073/pnas.0508955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguado-Llera D, et al. Deciphering the binding between Nupr1 and MSL1 and their DNA-repairing activity. PLoS One. 2013;8:e78101. doi: 10.1371/journal.pone.0078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamidi T, et al. Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J Clin Invest. 2012;122:2092–2103. doi: 10.1172/JCI60144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linkuvienė V, Krainer G, Chen WY, Matulis D. Isothermal titration calorimetry for drug design: Precision of the enthalpy and binding constant measurements and comparison of the instruments. Anal Biochem. 2016;515:61–64. doi: 10.1016/j.ab.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Chodera JD, Mobley DL. Entropy-enthalpy compensation: Role and ramifications in biomolecular ligand recognition and design. Annu Rev Biophys. 2013;42:121–142. doi: 10.1146/annurev-biophys-083012-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neira JL, et al. Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Sci Rep. 2017;7:39732. doi: 10.1038/srep39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park C, Marqusee S. Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 40.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 41.Neira JL, Rizzuti B, Iovanna JL. Determinants of the pKa values of ionizable residues in an intrinsically disordered protein. Arch Biochem Biophys. 2016;598:18–27. doi: 10.1016/j.abb.2016.03.034. [DOI] [PubMed] [Google Scholar]
- 42.Rai RK, Tripathi P, Sinha N. Quantification of metabolites from two-dimensional nuclear magnetic resonance spectroscopy: Application to human urine samples. Anal Chem. 2009;81:10232–10238. doi: 10.1021/ac902405z. [DOI] [PubMed] [Google Scholar]
- 43.Pastore A, Temussi PA. The emperor’s new clothes: Myths and truths of in-cell NMR. Arch Biochem Biophys. 2017 doi: 10.1016/j.abb.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 45.Urrutia R, et al. Evidence supporting the existence of a NUPR1-like family of helix-loop-helix chromatin proteins related to, yet distinct from, AT hook-containing HMG proteins. J Mol Model. 2014;20:e2357. doi: 10.1007/s00894-014-2357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittag T, et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 48.Berlow RB, Dyson HJ, Wright PE. Functional advantages of dynamic protein disorder. FEBS Lett. 2015;589:2433–2440. doi: 10.1016/j.febslet.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan H, et al. Fuzzy complex formation between the intrinsically disordered prothymosin α and the Kelch domain of Keap1 involved in the oxidative stress response. J Mol Biol. 2013;425:1011–1027. doi: 10.1016/j.jmb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Miskei M, Antal C, Fuxreiter M. FuzDB: Database of fuzzy complexes, a tool to develop stochastic structure-function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res. 2017;45:D228–D235. doi: 10.1093/nar/gkw1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fusco G, et al. Direct observation of the three regions in α-synuclein that determine its membrane-bound behaviour. Nat Commun. 2014;5:3827. doi: 10.1038/ncomms4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekhar A, Kay LE. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc Natl Acad Sci USA. 2013;110:12867–12874. doi: 10.1073/pnas.1305688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beckett D. Measurement and analysis of equilibrium binding titrations: A beginner’s guide. Methods Enzymol. 2011;488:1–16. doi: 10.1016/B978-0-12-381268-1.00001-X. [DOI] [PubMed] [Google Scholar]
- 54.Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 55.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W, et al. RYBP expression is associated with better survival of patients with hepatocellular carcinoma (HCC) and responsiveness to chemotherapy of HCC cells in vitro and in vivo. Oncotarget. 2014;5:11604–11619. doi: 10.18632/oncotarget.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vidal M, Starowicz K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp Hematol. 2017;48:12–31. doi: 10.1016/j.exphem.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Zaaroor-Regev D, et al. Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc Natl Acad Sci USA. 2010;107:6788–6793. doi: 10.1073/pnas.1003108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmid FX. Optical spectrosocopy to characterize protein conformation and conformational changes. In: Creighton TE, editor. Protein Structure. 2nd Ed. Oxford Univ Press; Oxford: 1997. pp. 261–297. [Google Scholar]
- 60.Royer CA, Scarlata SF. Fluorescence approaches to quantifying biomolecular interactions. Methods Enzymol. 2008;450:79–106. doi: 10.1016/S0076-6879(08)03405-8. [DOI] [PubMed] [Google Scholar]
- 61.Birdsall B, et al. Correction for light absorption in fluorescence studies of protein-ligand interactions. Anal Biochem. 1983;132:353–361. doi: 10.1016/0003-2697(83)90020-9. [DOI] [PubMed] [Google Scholar]
- 62.Cavanagh JF, Wayne J, Palmer AG, III, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Academic; San Diego: 1996. [Google Scholar]
- 63.Bodenhausen G, Ruben D. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 64.Wishart DS, et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 65.Vasseur S, et al. Cloning and expression of the human p8, a nuclear protein with mitogenic activity. Eur J Biochem. 1999;259:670–675. doi: 10.1046/j.1432-1327.1999.00092.x. [DOI] [PubMed] [Google Scholar]
- 66.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 69.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guzzi R, Rizzuti B, Bartucci R. Dynamics and binding affinity of spin-labeled stearic acids in β-lactoglobulin: Evidences from EPR spectroscopy and molecular dynamics simulation. J Phys Chem B. 2012;116:11608–11615. doi: 10.1021/jp3074392. [DOI] [PubMed] [Google Scholar]
- 71.Pantusa M, Bartucci R, Rizzuti B. Stability of trans-resveratrol associated with transport proteins. J Agric Food Chem. 2014;62:4384–4391. doi: 10.1021/jf405584a. [DOI] [PubMed] [Google Scholar]
- 72.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the materials (plasmid constructs) and data are available from the authors upon request.