Significance
The search for anticancer drugs continues to be greatly pursued. The nucleoside analog gemcitabine, which targets ribonucleotide reductase (RR) as a diphosphate and DNA polymerases as a triphosphate, is the standard first-line treatment in patients with pancreatic cancer. However, its cytotoxicity to normal dividing tissues leads to unwanted side effects. Here, we have discovered a nonnucleoside RR inhibitor, naphthyl salicylic acyl hydrazone (NSAH), that has efficacy similar to gemcitabine and the potential to be modified to provide safer and more effective cancer therapies.
Keywords: ribonucleotide reductase, cancer chemotherapy, small molecule, drug discovery, enzyme regulation
Abstract
Human ribonucleotide reductase (hRR) is crucial for DNA replication and maintenance of a balanced dNTP pool, and is an established cancer target. Nucleoside analogs such as gemcitabine diphosphate and clofarabine nucleotides target the large subunit (hRRM1) of hRR. These drugs have a poor therapeutic index due to toxicity caused by additional effects, including DNA chain termination. The discovery of nonnucleoside, reversible, small-molecule inhibitors with greater specificity against hRRM1 is a key step in the development of more effective treatments for cancer. Here, we report the identification and characterization of a unique nonnucleoside small-molecule hRR inhibitor, naphthyl salicylic acyl hydrazone (NSAH), using virtual screening, binding affinity, inhibition, and cell toxicity assays. NSAH binds to hRRM1 with an apparent dissociation constant of 37 µM, and steady-state kinetics reveal a competitive mode of inhibition. A 2.66-Å resolution crystal structure of NSAH in complex with hRRM1 demonstrates that NSAH functions by binding at the catalytic site (C-site) where it makes both common and unique contacts with the enzyme compared with NDP substrates. Importantly, the IC50 for NSAH is within twofold of gemcitabine for growth inhibition of multiple cancer cell lines, while demonstrating little cytotoxicity against normal mobilized peripheral blood progenitor cells. NSAH depresses dGTP and dATP levels in the dNTP pool causing S-phase arrest, providing evidence for RR inhibition in cells. This report of a nonnucleoside reversible inhibitor binding at the catalytic site of hRRM1 provides a starting point for the design of a unique class of hRR inhibitors.
Ribonucleotide reductase (RR) is a ubiquitous multisubunit enzyme that catalyzes the rate-determining step of dNTP synthesis, and its regulation is essential for maintaining a balanced dNTP pool (1). RR is essential for cell growth and genomic stability, as nucleotide imbalances lead to replication stress and severe effects on cell growth and survival (2). Therefore, the activity of RR is tightly regulated at various levels, such as transcription (3), allostery (1), protein inhibition in Saccharomyces cerevisiae by sml1(4) and cellular localization (5). RR consists of two subunits: (i) the catalytic subunit called RR1 (α) contains the catalytic or C-site and two allosteric sites, the specificity or S-site and the activity or A site (Fig. 1A), and (ii) the small subunit called RR2 (β) that houses a free radical essential for catalysis (6). The catalytic site contains three essential cysteine residues that conduct thiol-based redox chemistry to reduce the ribose substrate to 2′-deoxyribose (7) (Fig. 1A). In the presence of effectors, α exists as a dimer which in the presence of the allosteric effectors dATP and ATP form α6β2 and α6βn (n = 2, 4, 6) multimers, respectively (8–12). Current interest in RR research is increasing our understanding of higher-order oligomerization, enzyme turnover, and the mechanism of allosteric regulation that governs substrate specificity, catalysis, and inhibition of this critical enzyme (8, 9, 11–13).
Fig. 1.
Structural organization of the regulatory subunit hRRM1 and steady-state kinetic studies of NSAH–hRR. (A) A structure of the hRRM1 dimer showing the nonnucleoside small-molecule (magenta) NSAH binding at the C-site. The nucleotide effector TTP is shown as ball and stick (cyan) bound at the S-site, which controls the specificity of the C-site. Active-site cysteine residues are shown in yellow. (B) The specific activity of non–drug-treated hR1 (blue, 162.3 nmol⋅min−1⋅mg−1) is comparable to that of the diluted solution of hR1 in the presence of 50 µM NSAH (green, 152.3 nmol⋅min−1⋅mg−1). However, an undiluted solution of hR1 in the presence of 50 µM NSAH showed no activity (red). This indicates that NSAH inhibits reversibly. (C) Plot of velocity versus [14C-ADP] for hRRM1 in the presence of 0, 1, 5, 10, 25, and 50 µM NSAH. [14C-ADP] ranged from 0.5 to 5.0 mM.
Human RR (hRR holocomplex) is an important target for cancer chemotherapy. The nucleotide analogs of chemotherapeutics such as fludarabine, clofarabine, and cladribine target all three sites of hRRM1 for inhibition (11, 14, 15) (Fig. 1A). In contrast, hydroxyurea (HU) blocks catalysis by targeting the di-iron cluster of RR2 (16, 17). Gemcitabine diphosphate, clofarabine, and cladribine nucleotides inhibit hRRM1 by stabilizing a form of the α6 complex (10, 11, 13, 15). Such nucleotide-analog inhibitors of hRR, especially gemcitabine, are effective anticancer agents and can sensitize cancer cells to ionizing radiation and to DNA-damaging drugs (18). Indeed, gemcitabine is a mainstay of the cancer chemotherapy toolbox and is combined with radiation and/or another chemotherapeutic agent, such as cisplatin, in clinical protocols for multiple types of cancer (19).
In addition to its direct anticancer effects, hRR inhibition also aids the ability of nucleotide analogs to be incorporated into DNA. Inhibition of hRR lowers dNTPs, which enhances the ability of gemcitabine triphosphate to be incorporated into growing DNA strands by DNA polymerase. However, gemcitabine causes serious side effects due to its cytotoxicity to normal cells caused by DNA chain termination (20, 21), irreversible inhibition of hRR (22), and inhibition of numerous other enzymes that recognize phosphate moieties, including deoxycytidine deaminase (23), thymidylate synthase (24), CTP-synthase (25, 26), and topoisomerase 1 (27, 28).
We hypothesize that competitive, reversible, nonnucleoside hRR inhibitors should be less toxic to normal cells, and provide unique leads for medicinal chemistry and structure-based drug design to advance identification of unique cancer chemotherapeutics. Here, we describe the identification and characterization of a hydrazone compound, naphthyl salicylic acyl hydrazone (NSAH), that inhibits hRR reversibly with micromolar affinity in vitro. The crystal structure of the NSAH complex with hRR together with steady-state kinetic data demonstrate that it binds in the C-site of hRRM1. Importantly, the IC50 for NSAH is within twofold of that of gemcitabine for growth inhibition of multiple cancer cell lines. NSAH was shown to inhibit hRR in cells, as demonstrated by depressed dGTP and dATP levels, which preceded early S-phase cell cycle arrest. However, NSAH demonstrated little measurable cytotoxicity against normal mobilized peripheral blood progenitor cells. Thus, these data identify NSAH as a nonnucleoside competitive inhibitor of hRRM1 and reveal its improved selectivity for tumor compared with normal cells relative to existing therapeutics, providing a starting point for rational fragment-based drug design of a unique class of hRR inhibitors.
Results
NSAH Acts as a Reversible Competitive Inhibitor of hRR.
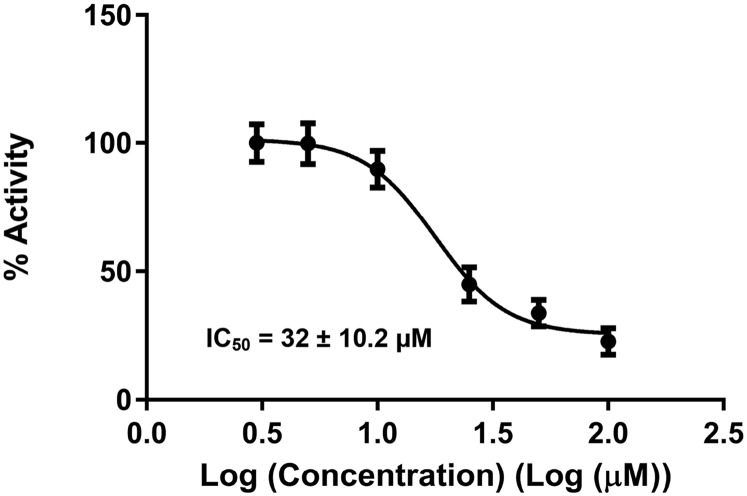
NSAH was discovered as a potential hRRM1 inhibitor by an in silico screening strategy previously used, which is briefly described in Materials and Methods (29). Docking experiments were conducted targeting the catalytic site of the hRRM1 dimer using the program Schrödinger (30, 31). The majority of hits were filtered out through a cancer cell growth inhibition assay, as described in Materials and Methods. The experimentally confirmed top two hits were both hydrazone derivatives, NSAH and a related naphthyl acyl hydrazone (NAH). NSAH had a lower cell-free IC50 (32 µM versus 40 µM) and cell-based IC50 (∼250 nM versus 600 nM), which established NSAH as our lead compound. NSAH is fully compliant with the Lipinski Rule of Five (Table S1). Binding of NSAH to hRRM1 was validated using fluorescence quenching, and in vitro multiple-turnover RR kinetic analyses were used to analyze the mode of inhibition, as in a similar study (29) (Fig. 1 B and C). hRR enzymatic IC50 values were determined using the method described in ref. 29. Six inhibitor concentrations were used ranging from 5 to 100 µM, which yielded an IC50 of 32 ± 10 µM (Fig. S1). The direct binding of NSAH to hRRM1 was assessed using a fluorescence quenching assay (29), which gave a KD of 37 ± 4 µM (Fig. 2 A and B).
Table S1.
Lipinski properties for NSAH and other hits
 |
Fig. S1.
Sigmoidal dose–response curve for NSAH inhibition of hRRM1.
Fig. 2.
NSAH binding and structural studies with hRRM1. (A) Quenching of tryptophan fluorescence of hRRM1 by ligand NSAH. Tryptophan fluorescence spectra of hRRM1 (0.2 mg/mL in buffer A) at the indicated concentration in µM of NSAH. (B) Variation of the extent of fluorescence quenching [(F0 − F)/F0, where F0 and F are the fluorescence intensities at 340 nm in the absence and in the presence of NSAH of 0.2 mg/mL of hRRM1]. (C) 2Fo−Fc electron density (blue) of NSAH contoured at 1σ, compound NSAH shown in magenta. Active-site cysteines and interacting residues are shown in green. (D) Comparison of binding of NSAH and substrate at the catalytic site of hRRM1. NSAH is shown in magenta, and GDP is in green. (E) Interactions of NSAH at the hRRM1 catalytic site. Hydrogen bonds are shown in black dashed lines.
The mechanism of inhibition was analyzed using a series of multiple-turnover kinetic experiments. Importantly, under steady-state reaction conditions, we observe linear reaction kinetics in both the presence and absence of inhibitor consistent with a rapid equilibrium mode of binding. Nonetheless, hydrazones are prone to act as electrophiles, especially when the azomethine carbon of the C=N bond is unsubstituted; therefore, we tested whether NSAH functions as an irreversible inhibitor of hRR, perhaps by forming covalent bonds with the enzyme by nucleophilic attack of the C=N group. We used a preincubation–dilution experiment to test whether NSAH inhibition of hRR was time dependent or sensitive to dilution of preformed enzyme–inhibitor complexes. As shown in Fig. 1B, addition of inhibitor and substrate simultaneously or preincubation of RR with 50 µM NSAH showed complete inactivation of enzyme activity consistent with rapid inhibitor binding. If dissociation of the inhibitor is slow, then enzyme–inhibitor complexes formed at high concentrations will persist after dilution and inhibition will be maintained. Conversely, if inhibition is rapidly reversible, then dilution to a lower concentration of inhibitor below the IC50 should result in restoration of enzyme activity. The diluted sample retains essentially full activity and is consistent with rapid dissociation of the compound from the enzyme without significant rebinding, consistent with a reversible mechanism.
Next, we further characterized the mechanism of inhibition using steady-state inhibition kinetics. The kinetics of untreated (wild-type) hRRM1 at varying substrate concentrations and hRRM1 in the presence of 1, 5, 10, 25, and 50 µM NSAH were determined, and the data were globally fitted to a general rate equation as described in Materials and Methods (Fig. 1C). In this mechanism, the binding of the inhibitor is described by dissociation constant Ki and binding to the ES complex by αKi. A value of α ≫ 1 provided a better fit to the data, consistent with very weak binding to the ES complex and therefore a competitive mechanism. A double-reciprocal plot shows that all datasets are observed to converge upon a common y intercept near the origin. This is a clear indication that, although Km is dependent upon [I], Vmax remains constant consistent for all [I], as expected for a competitive inhibition model. Based on global fitting to the model, the Ki value of NSAH = 5.0 ± 1.5 µM, Km = 198 ± 55 µM, and kcat = 13 ± 1 s−1.
An X-Ray Structure of hRRM1 Shows NSAH Bound at the Catalytic Site.
The 3D structure in the dimeric form of hRRM1 bound to NSAH was determined to 2.66-Å resolution (Table S2). The structure was refined to acceptable R and Rfree values with good geometrical parameters (Table S2). The Fourier 2|Fo|–|Fc| electron density map and |Fo|–|Fc| omit map clearly shows NSAH bound at the C-site (Fig. 2C and Fig. S2). There is unambiguous density for NSAH bound to monomer B, whereas monomer A has partial density. An effector TTP binds to the S-site in a manner identical to the previously described TTP–GDP-bound hRRM1 structure (9). NSAH binds to the C-site close to the natural substrate GDP by adopting a U shape (Fig. 2 C and D). For example, compared with GDP binding, the naphthyl moiety binds in the diphosphate binding region, whereas the hydrazone backbone binds within 1 Å of the ribose, and the benzene moiety partially overlaps with the edge of the ribose and the substrate base (Fig. 2D). The C7 atom of the hydrazone backbone is an isosteric center that adopts the transisomer configuration (Fig. 2C). The compound is observed to form three hydrogen bonds with the enzyme and makes 64 contacts with the binding site that are within 4-Å distance (Fig. 2E, Fig. S3, and Table S3). Among the catalytic residues, such as Cys-218, Asn-426, and Glu-430, only Cys-218 and Glu-430 interact with the inhibitor. Compared with the hRRM1–TTP–GDP structure, the three catalytic residues of hRRM1–TTP–NSAH did not show any significant conformational changes upon ligand binding. However, the loop 2 (residues 285–295) conformation appears to be identical to the hRRM1–TTP (3HNC) rather than the hRRM1–TTP–GDP (3HND) structure (Fig. S4).
Table S2.
Data collection and refinement statistics for X-ray crystal structures of hRRM1 bound with NSAH
| Data statistics | NSAH |
| Wavelength, Å | 0.98 |
| No. of unique reflections | 50,742 |
| Resolution range, Å | 110–2.66 (2.80–2.66) |
| Monomer per asymmetric unit | 2 |
| Completeness, % | 99.1 (95.8) |
| Rmerge, %* | 14.4 (58.4) |
| I/σ(I) | 9.0 (1.1) |
| Space group | P212121 |
| Unit cell parameter | a = 69.52 b = 114.3 c = 220.4 α = β = γ = 90 |
| Redundancy | 5.4 (5.1) |
| Refinement statistics | |
| Resolution range, Å | 110.18–2.66 |
| No. of reflections | 48,650 |
| No. of reflections used for Rfree | 1,998 (3.94% of the overall reflections) |
| Rwork/Rfree, %† | 19.91/24.97 |
| Total no. of atoms | 11,714 |
| Protein | 11,598 |
| Ligand | 83 |
| Water | 33 |
| Average B value, Å2 | |
| Protein | 56 |
| Ligand (NSAH) | 86 |
| TTP | 58 |
| Mg2+ | 46 |
| Ramachandran plot, residues in, % | |
| Core regions | 89.1 |
| Allowed regions | 9.6 |
| Generously allowed regions | 1.1 |
Rmerge = Σ|I – <I>|/Σ<I>, where I and <I> are the measured and averaged intensities of multiple measurements of the same reflection, respectively. The summation is over all of the observed reflections.
R = Σ|Fo – Fc|/Σ|Fo| calculated for all observed data. Rfree = Σ|Fo – Fc|/Σ|Fo| calculated for a specified number of randomly chosen reflections that were excluded from the refinement. Values in parenthesis are used for the highest-resolution shell.
Fig. S2.
Fo−Fc simulated annealing omit map contoured at 2.5σ. Interacting residues are shown in green sticks.
Fig. S3.
Ligplot of NSAH (purple in color) interactions with human RR1. Polar interactions are shown in green dotted line. Other interacting residues are shown in red lines. Carbon atoms are shown in black, oxygen atoms are shown in red, and nitrogen atoms are shown in blue color.
Table S3.
Total 64 contacts were found between the human RR1 and the ligand at 4 Å using CCP4 program suite
| Source atoms | Target atoms | Distance |
| (NSAH)/O1[O]: | 217(SER)./OG[O]: | 2.48 |
| 446(LEU)./CD1[C]: | 3.91 | |
| 218(CYS)./SG[S]: | 3.76 | |
| 217(SER)./CB[C]: | 3.74 | |
| (NSAH)/C6[C]: | 217(SER)./OG[O]: | 3.64 |
| 218(CYS)./SG[S]: | 3.76 | |
| (NSAH)/C1[C]: | 218(CYS)./SG[S]: | 3.69 |
| (NSAH)/C[C]: | 218(CYS)./SG[S]: | 3.37 |
| (NSAH)/O[O]: | 217(SER)./OG[O]: | 3.09 |
| 218(CYS)./SG[S]: | 2.92 | |
| 217(SER)./CB[C]: | 3.85 | |
| 217(SER)./CA[C]: | 3.72 | |
| 217(SER)./C[C]: | 3.18 | |
| 217(SER)./O[O]: | 3.12 | |
| 218(CYS)./N[N]: | 3.68 | |
| (NSAH)/C5[C]: | 247(GLY)./N[N]: | 3.72 |
| 247(GLY)./CA[C]: | 3.75 | |
| (NSAH)/C4[C]: | 428(LEU)./CD2[C]: | 3.63 |
| (NSAH)/C3[C]: | 428(LEU)./CD2[C]: | 3.89 |
| (NSAH)/C2[C]: | 429(CYS)./SG[S]: | 3.82 |
| (NSAH)/N[N]: | 603(PRO)./CD[C]: | 3.99 |
| 429(CYS)./SG[S]: | 3.67 | |
| (NSAH)/N1[N]: | 602(MET)./CB[C]: | 3.93 |
| 603(PRO)./CD[C]: | 3.37 | |
| (NSAH)/C7[C]: | 604(THR)./N[N]: | 3.72 |
| 602(MET)./CB[C]: | 3.70 | |
| 602(MET)./CG[C]: | 3.71 | |
| 603(PRO)./N[N]: | 4.00 | |
| 603(PRO)./CD[C]: | 3.67 | |
| (NSAH)/C8[C]: | 604(THR)./N[N]: | 3.68 |
| 604(THR)./CA[C]: | 3.85 | |
| (NSAH)/C9[C]: | 605(ALA)./N[N]: | 3.77 |
| (NSAH)/O2[O]: | 605(ALA)./N[N]: | 3.96 |
| 605(ALA)./CB[C]: | 3.98 | |
| (NSAH)/C10[C]: | 606(SER)./N[N]: | 3.56 |
| 606(SER)./CB[C]: | 3.96 | |
| 606(SER)./OG[O]: | 3.04 | |
| 605(ALA)./N[N]: | 3.83 | |
| (NSAH)/C11[C]: | 606(SER)./N[N]: | 3.67 |
| 606(SER)./CB[C]: | 3.70 | |
| 606(SER)./OG[O]: | 3.21 | |
| 607(THR)./N[N]: | 3.84 | |
| 202(SER)./OG[O]: | 3.36 | |
| (NSAH)/C12[C]: | 607(THR)./OG1[O]: | 3.62 |
| (NSAH)/C13[C]: | 604(THR)./CA[C]: | 3.83 |
| (NSAH)/C17[C]: | 607(THR)./CB[C]: | 3.43 |
| 607(THR)./CG2[C]: | 3.02 | |
| 607(THR)./OG1[O]: | 2.82 | |
| 202(SER)./N[N]: | 3.68 | |
| 202(SER)./OG[O]: | 3.97 | |
| 201(ALA)./CA[C]: | 3.67 | |
| (NSAH)/C16[C]: | 607(THR)./CB[C]: | 3.77 |
| 607(THR)./CG2[C]: | 3.49 | |
| 607(THR)./OG1[O]: | 2.95 | |
| 200(HIS)./O[O]: | 3.84 | |
| 201(ALA)./N[N]: | 3.78 | |
| 201(ALA)./CA[C]: | 3.25 | |
| 201(ALA)./CB[C]: | 3.98 | |
| (NSAH)/C15[C]: | 607(THR)./OG1[O]: | 3.86 |
| 201(ALA)./CA[C]: | 3.72 | |
| 201(ALA)./CB[C]: | 3.95 | |
| (NSAH)/C14[C]: | 602(MET)./CG[C]: | 3.69 |
| 602(MET)./SD[S]: | 3.95 | |
| 602(MET)./CE[C]: | 3.65 |
Fig. S4.
Superposition of three structures shown in stereo, hRRM1–TTP–NSAH (magenta), hRRM1–TTP (3HNC; cyan), and hRRM1–TTP–GDP structure (3HND; green). Arg-293 in 3HND is depicted in a ball-and-stick model, which sits on loop 2.
The naphthalene ring is buried within a pocket containing a mixture of hydrophobic and hydrophilic residues. Usually, when nucleoside diphosphate substrates are bound, this pocket will form multiple hydrogen bonds with the phosphate groups, primarily through the backbone atoms of Ser-202, Ser-606, and Ser-610 (9, 32). The naphthalene ring interacts with four hydrophobic residues: Met-602, Ala-201, Ala-605, and Leu-428; and three hydrophilic residues: Cys-218, Ser-606, and Thr-607 (Table S3). These interactions consist of six carbon–carbon and five carbon–polar atom contacts. The polar hydroxyl group on the C2 position of the naphthyl ring makes a contact with the backbone amide atom of Ala-605 (Fig. 2E). The hydrazone backbone is found near the region usually occupied by the ribose ring of the natural substrate (Fig. 2D). The oxygen atom of the carbonyl group accepts a hydrogen bond from the hydroxyl group of Ser-217 (Fig. 2E). The hydroxyl group on carbon 2 of the benzene ring is observed to form a hydrogen bond with both the hydroxyl group of Ser-217 at 2.6 Å and the carbonyl oxygen atom of Ser-217 at 2.9 Å. The hydroxyl group also contacts the sulfur atom of Cys-218 at 3.0 Å. The carbon atoms of the benzene ring form van der Waals contacts with Leu-428 and Cys-429 (Fig. 2E and Fig. S3). The cognate substrates are known to make contact with a 9-aa–containing loop 2 that is involved in substrate recognition in RR (32–34) (Fig. S4). Residues 292–293 are disordered in our structure. This is possibly because the benzene moiety does not reach far enough to make contact with loop 2. Nevertheless, future generations of analogs of NSAH may be designed to take advantage of interacting with loop 2. Based on the structure, most of the binding energies derive from three hydrogen bonds and 64 van der Waals contacts (Table S3). The inhibitor has a total surface accessible area of 490 Å2, of which 401 Å2 is buried upon binding to the protein.
NSAH Shows Tumor Cell-Selective Cytotoxicity.
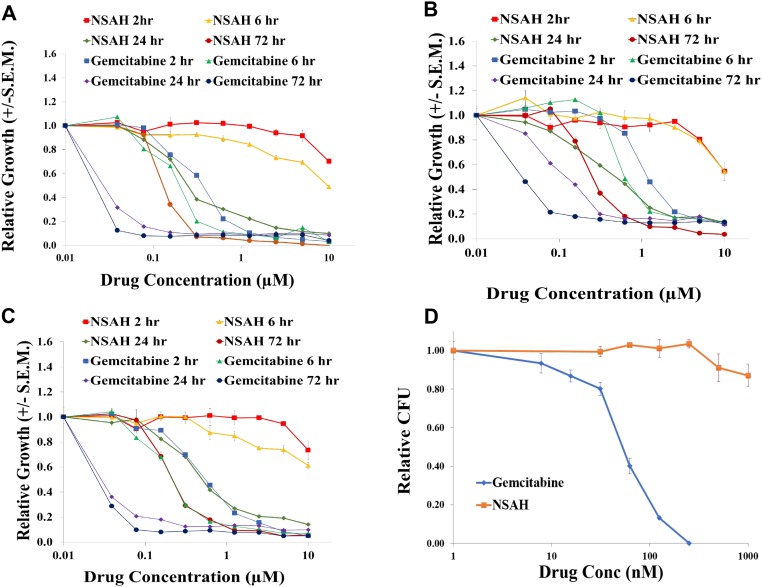
To determine the time dependence of the growth inhibitory activity of NSAH and gemcitabine, three established cancer cell lines were exposed to a full concentration range of either drug for 2, 6, 24, or 72 h (Fig. S5 A–C). In groups with less than 72-h exposure, the drugs were removed, and drug-free medium was added for the remaining time. Continuous 72-h exposure demonstrated growth inhibition in all three cell lines, with IC50 values for gemcitabine ranging from 30 to 100 nM. NSAH treatment resulted in IC50 values ranging from 220 to 500 nM. Both drugs showed a significant decrease in activity with shorter exposures; however, the time dependence of the growth inhibition was greater with NSAH than with gemcitabine. Gemcitabine exposures of 2 h were 15- to 20-fold less effective than continuous exposure, whereas NSAH showed less than 60% growth inhibition at the highest dose tested (10 µM), which was 20- to 50-fold higher than the IC50 values for continuous exposure (Table S4).
Fig. S5.
Cell growth-inhibitory activity of NSAH. A–C show time dependence of the growth-inhibitory activity of NSAH and gemcitabine. D shows the relative cytotoxicity of NSAH and gemcitabine in normal human blood progenitor cells.
Table S4.
Time dependence of the growth-inhibitory activity of NSAH and gemcitabine
| Cell line | Drug exposure, h | Drug | IC50, µM |
| MDA-231 | 2 | NSAH | >10.0 |
| MDA-231 | 6 | NSAH | >10.0 |
| MDA-231 | 24 | NSAH | 0.658 |
| MDA-231 | 72 | NSAH | 0.498 |
| MDA-231 | 2 | GEM | 1.930 |
| MDA-231 | 6 | GEM | 1.540 |
| MDA-231 | 24 | GEM | 0.274 |
| MDA-231 | 72 | GEM | 0.100 |
| HCT116 | 2 | NSAH | >10.0 |
| HCT116 | 6 | NSAH | >10.0 |
| HCT116 | 24 | NSAH | 0.572 |
| HCT116 | 72 | NSAH | 0.221 |
| HCT116 | 2 | GEM | 0.487 |
| HCT116 | 6 | GEM | 0.343 |
| HCT116 | 24 | GEM | 0.058 |
| HCT116 | 72 | GEM | 0.030 |
| Panc1 | 2 | NSAH | >10.0 |
| Panc1 | 6 | NSAH | >10.0 |
| Panc1 | 24 | NSAH | 0.898 |
| Panc1 | 72 | NSAH | 0.442 |
| Panc1 | 2 | GEM | 0.680 |
| Panc1 | 6 | GEM | 0.406 |
| Panc1 | 24 | GEM | 0.084 |
| Panc1 | 72 | GEM | 0.046 |
To examine the relative cytotoxicity of NSAH and gemcitabine in normal human blood progenitor cells, mobilized peripheral blood mononuclear cells were tested in a 14-d colony-forming unit (CFU) assay. Gemcitabine was chosen as a reference, due to its established activity as an RR inhibitor and clinically used anticancer agent. The IC50 for gemcitabine against the normal blood progenitor cells was similar to that for the cancer cell lines at 35 nM. However, in stark contrast to gemcitabine, NSAH demonstrated little measurable cytotoxicity against normal mobilized peripheral blood progenitor cells at the maximum concentration tested (1 µM). This illustrated a superior therapeutic index using mobilized blood progenitor cells as a surrogate for normal bone marrow (the most common site of dose-limiting toxicities for gemcitabine) and suggests that NSAH will demonstrate improved therapeutic indices in animal model studies and subsequent clinical trials compared with gemcitabine (Fig. S5D).
dNTP Pool and Cell Cycle Analysis.
To evaluate the role of RR as the cytotoxic target for NSAH, we compared its effects on dNTPs to that of HU at their respective IC90 values. As expected for inhibitors of RR in solid tumor cells, both drugs decreased dATP and dGTP by >60% within 24 h (35) (Table S5). Whereas HU produced a greater decrease in dATP and dGTP at 4 h, the duration of decrease was longer with NSAH (through 24 h). Both drugs produced little or no decrease in dCTP and TTP at 4 h, whereas by 24 h HU increased both dCTP and TTP by more than 200%.
Table S5.
dNTP pool and cell cycle analysis with NSAH and HU treatment HCT116 cells
| pmol/106 cells | Cell cycle | |||||||
| Treatment | Time (h) | dCTP | dTTP | dATP | dGTP | S, % | G1, % | G2, % |
| Control | 4 | 7.17 | 42.04 | 6.28 | 0.88 | 35.9 | 32.3 | 31.7 |
| NSAH | 4 | 9.73 | 45.50 | 2.45 | 1.36 | 36.2 | 36.8 | 26.9 |
| 24 | 9.09 | 32.40 | 2.12 | nd | 44.1 | 42.6 | 13.4 | |
| Hydroxyurea | 4 | 11.23 | 48.29 | nd | nd | 56.9 | 35.2 | 7.9 |
| 24 | 29.14 | 112.03 | 0.63 | 5.47 | 74.5 | 15.3 | 10.3 | |
nd, not detected.
The changes in dATP and dGTP levels preceded an increase in the proportion of cells in S phase and a decrease in cells in G2/M phase after NSAH treatment (Table S5). At 4 h after treatment, there was no significant change in cell cycle distribution. Twenty-four hours after drug addition, an increase in S phase (44.1%, compared with 35.9% in controls) and a decrease in G2/M phase (13.4%, compared with 31.7% in controls) were observed. The changes after HU were more rapid and dramatic, with accumulation of cells in S phase and loss of cells in G2/M phase observed after 4 h, and these changes were maintained at 24 h. The block in S-phase progression corresponds well to the extent and timing of the decreases in dATP and dGTP observed with NSAH and HU.
Discussion
Decades of clinical use of HU to treat patients with cancer have validated hRR as an important target for cancer chemotherapy. However, clinical use of HU has been hampered by its lack of potency. Improved potency has been attained by several nucleotide analogs that are used clinically as drugs to target hRRM1 (10, 13, 22). Although the role of hRR inhibition is considered to be secondary to DNA chain termination as the primary mechanism of cytotoxicity by nucleoside drugs where the triphosphate form of the drug is incorporated (22), work reported by the Stubbe laboratory highlighted the importance of hRR inhibition by nucleotide analogs (10, 11). The three analogs that are best characterized biochemically are gemcitabine (10), clofarabine (11, 13), and cladribine (15). Gemcitabine diphosphate is a substoichiometric mechanism-based inhibitor that targets hRRM1’s catalytic site, resulting in the formation of a tight α6β6 holocomplex (10, 36). Clofarabine is a reversible inhibitor of hRRM1 that forms persistent hexamers α6 (13) by binding both at the catalytic (C-site) and allosteric sites. Both drugs cause severe side effects due to their cytotoxicity on normal cells (37, 38). Nevertheless, these reports highlight the need to develop novel classes of inhibitors against hRR with greater selectivity for tumor cells.
One of our in silico screens, focusing on the catalytic site of hRRM1, yielded a nonnucleoside small molecule that we named NSAH. Biochemical, structural, and cellular characterization of NSAH reveals that it is a competitive, reversible inhibitor, binding at the catalytic site of hRRM1. Here, we report a reversible, nonnucleoside small molecule binding at the catalytic site of hRRM1 and provide a template for the development of highly specific, noncovalent hRR inhibitors with a far superior therapeutic index to the existing hRRM1 drugs.
The IC50 of NSAH against cell-free hRR enzymatic activity was determined to be 32 ± 10 µM, and it binds the hRRM1 catalytic subunit with a KD of 37 ± 4 µM (Figs. 1B and 2 A and B). Steady-state kinetics revealed that NSAH is a competitive inhibitor of hRRM1 with a Ki of 5.0 µM, as demonstrated by a globally fitted double-reciprocal plot of hRRM1 in the presence of 0, 1, 5, 10, 25, and 50 µM NSAH (Fig. 1C). The crystal structure reveals that NSAH binds at the catalytic site, partially overlapping with the natural substrate, GDP.
The reversibility of NSAH was determined by enzymatic preincubation–dilution experiments and cell culture experiments. In the preincubation–dilution experiments (24), the enzyme recovers full activity upon dilution, demonstrating that NSAH is a reversible inhibitor (Fig. 1B). In cell culture experiments, a head-to-head comparison was made between gemcitabine and NSAH. Cell viability was assessed in a 3-d growth assay with variable drug exposure times, where drugs were removed at various times and cells were allowed to grow for a total of 72 h, regardless of the length of drug exposure. In these experiments, NSAH showed little toxicity at concentrations up to 10 µM (maximum concentration administered) when administered for only 2 or 6 h. However, when drug exposure was extended to 24 or 72 h, significantly lower IC50 values were observed. Specifically, the IC50 values for the three cell lines tested (HCT116, Panc1, and MDA-MB-231) increased 23.3×, 22.6×, and >100×, respectively, when the exposure times were reduced from 24 to 6 h, whereas the same reduction in exposure times with gemcitabine lead to only 6.0×, 4.8×, and 5.6× increases in IC50 values, respectively. This analysis demonstrates that, in comparison with NSAH, gemcitabine cytotoxicity is less time dependent and more consistent with covalent inhibition.
Currently, NSAH is an initial hit against hRR, and it has not been fully optimized; like similar analogs, it may inhibit other targets (39) and potentially coordinate metal ions (39). Because metal chelation would interfere with the RR enzyme activity assay, NSAH was tested for Fe2+ and Mg2+ binding by UV spectroscopy, but no such binding was found. The potential off-target effects may contribute to the disparity between the enzymatic IC50 and the cell growth inhibition IC50 at 72 h, further underscoring the importance of improving the target specificity of NSAH toward hRRM1 through knowledge-based design. Nonetheless, its activity against several cancer cell lines and relative lack of cytotoxicity against normal blood progenitor cells, in contrast to the widely used drug gemcitabine, suggests a promising future for this class of compounds (Fig. S5 and Table S4). Mobilized human blood progenitor cells, isolated from a normal donor, were assayed for sensitivity to gemcitabine and NSAH. In a 14-d colony-forming survival assay, gemcitabine demonstrated toxicity against normal blood progenitor cells that was similar to that observed against cancer cells (CFU IC50 of 35 nM). In contrast, NSAH demonstrated very moderate toxicity at 1 µM (87% survival); this dose is twofold to fivefold greater than the IC50 observed in cancer cells. These data demonstrate a significantly greater therapeutic ratio for NSAH compared with gemcitabine. Although NSAH is not currently suitable for clinical testing, it offers a promising starting point for a unique class of chemotypes that will be improved by using knowledge-based design cycles.
To establish whether NSAH inhibits hRR in cells, dNTP pools and cell cycle distributions were assessed after drug treatment. The hallmark of a hRR drug in solid tumor cells is a depression of dATP and dGTP levels in the dNTP pool preceding cell cycle arrest at the S phase. Table S5 summarizes the results where NSAH was compared with HU. The finding that NSAH and HU, at equitoxic concentrations in HCT116 cells, produced similar effects on dNTPs with a block in S-phase progression, supports that RR is inhibited in cells. The relatively lesser effect on cell cycle progression observed with NSAH compared with HU could be due to the longer-lasting effects of NSAH on dNTP pools. The extended decrease in dNTPs with NSAH could cause a block in cell cycle progression at a point in early S phase that is indistinguishable from G1 phase in this analysis, resulting in an underestimation of the cell cycle perturbation caused by NSAH.
An effective drug against hRR must necessarily satisfy two criteria, safety and efficacy. Several factors are likely to affect the latter, including target specificity and potency. Most nucleoside drugs, such as gemcitabine, derive their efficacy by inhibiting multiple targets, such as hRR, DNA polymerase, thymidylate synthase, and CTP synthase (22–24, 26). Designing a highly specific, reversible small-molecule hRR inhibitor lacking effects of additional cytotoxic targets is likely to pay a penalty in efficacy. The key to overcoming this deficiency will be improvement of the potency against hRRM1, which is achievable, because the crystal structure of NSAH can guide design efforts.
Although NSAH provides a promising starting point for rational drug design, several modifications will need to be made to address its current unsuitability as a clinical drug. First, the hydrazone backbone chain is highly labile under mildly acidic conditions. Incorporating this region of the molecule within a heterocycle will improve the stability of the compound overall. Additionally, structure–activity relationship studies focused on the naphthalene and benzene ring systems will provide insight into variations that could improve the compound’s potency toward hRR. Finally, growing the compound with fragments reaching toward the loop 2 region of the C-site will improve additional binding interactions with hRRM1 and potentially lead to improved inhibitory potency (9, 33, 34, 40) (Fig. S4). NSAH’s inability to interact with loop 2 leads the hRRM1–TTP–NSAH structure to have an almost identical conformation to the hRRM1–TTP (3HNC) structure, which is different from the hRRM1–TTP–GDP structure (3HND, Fig. S4).
Interactions with loop 2 are expected to reduce the mobility of the loop and stabilize it, enhancing binding energy as observed with the recently determined Escherichia coli complexes 5. This is based on previous studies where Arg-293 and Gln-288 (hRR numbering), or their equivalent residues in S. cerevisiae, E. coli, and Thermotoga maritima RRs, play an important role in substrate recognition (32–34, 41). In particular, in T. maritima and E. coli RR1 structures, arginine forms a salt bridge with the β-phosphate of the substrate, which is a crucial interaction. Hence, engaging these residues by a potential inhibitor would be highly advantageous.
Materials and Methods
NSAH was purchased from ChemDiv (catalog number 2066-0112).
In Silico Screening of NSAH.
The University of Cincinnati drug library, containing ∼350,000 compounds, was screened in silico against hRRM1 (PDB ID code 3HND) using the Glide docking module of the Schrӧdinger 9.3 modeling software suite. See Supporting Information for more details.
Protein Expression and Purification of hRRM1.
The hRRM1 protein was expressed in E. coli BL21 DE3 (RIL) and purified using peptide affinity chromatography, as described previously (9). The homogeneous protein was pooled and concentrated to 20–25 mg/mL, as quantified by UV absorbance spectroscopy, as described previously.
Establishing Reversible Inhibition of NSAH of hRR.
In assay buffer, 50 µM NSAH was incubated on ice with 2.5 µmol of hRRM1 for 30 min. The assay sample was then diluted by a factor of 5, and enzyme activity was assayed in triplicate (29). As a control, the assay was also performed for non–drug-treated hRRM1 and for hRRM1 with 50 µM NSAH without dilution.
Crystallization and Data Collection.
A full description of the crystallization and structure solution can be found in Supporting Information. Briefly, hRRM1–TTP–NSAH was cross-seeded with preformed hRRM1–TTP crystals (9) to form the cocrystal of hRRM1–TTP–NSAH. TTP is the only effector that yields good diffracting crystals. The crystals were cryogenized, and data were collected from the Northeastern Collaborative Access Team (NE-CAT) beamline at Advanced Photon Source (APS). A full description of the refinement and model building can be found in Supporting Information.
Cell-Free Inhibition Studies.
The IC50 was determined using the method described in ref. 29. Briefly, boronate chromatography was used to separate the product (10). Six concentrations of NSAH were studied ranging from 5 to 100 µM. The assay was repeated in triplicate. Data were fitted in GraphPad Prism 6.05 using a sigmoidal dose–response curve.
Inhibition Mechanism.
RR inhibition assays were performed as described earlier for WT hRRM1 at inhibitor concentrations of 0, 1, 5, 10, 25, and 50 µM NSAH. For all three inhibitor concentrations, the specific activity was recorded for substrate concentrations of 5.0, 3.5, 2.5, 2.0, 1.5, 1.0, 0.5, 0.25, 0.1, and 0.05 mM 14C-ADP. Global fitting was performed in Excel using the Solver add-in. All graphs were prepared in Excel. A full description of the methods may be found in Supporting Information.
Binding Studies Using Fluorescence Quenching.
As described previously (29), to assay binding NSAH was titrated into a solution of 0.5 mg of hRRM1 at 10–200 µM in 10 µM increments. The emission spectrum was recorded over 300–400 nm using a Jobin Yvon-Spex fluorescence spectrophotometer. The data were fitted by nonlinear regression using the one-site binding (hyperbola) equation Y = Bmax × X/(KD(app) + X), where Bmax is the maximum extent of quenching and KD(app) is the apparent dissociation constant, using GraphPad Prism 6.05. Measurements were recorded in duplicate.
Blood Progenitor CFU Assay.
The CFU assay was performed in the Hematopoietic Biorepository and Cellular Therapy Core Facility of the Case Comprehensive Cancer Center using a standardized protocol. More details can be found in Supporting Information.
Cancer Cell Line Growth Inhibition Assay.
The growth inhibition assay was performed in the Preclinical Drug Testing Laboratory of the Case Comprehensive Cancer Center using a standardized protocol. More details may be found in Supporting Information.
Cell Cycle and dNTP Analyses.
Cells were seeded in parallel into 15-cm (for dNTP) and 10-cm dishes (for cell cycle) at 40,000 cells per cm2. Drugs were added to dishes 2 d later, and cells were harvested at 4 and 24 h after drug addition. More details may be found in Supporting Information.
SI Materials and Methods
In Silico Screening of NSAH.
PDB ID 3HND, which is the hRRM1 dimer in complex with TTP–GDP, was first prepared in Prime using the optimized potentials for liquid simulations all-atom (OPLS) force field and the surface-generalized Born (SGB) continuum solution model for optimization and minimization (30, 31). The 3D structures of the ligands were generated from the 2D drug library using OPLS 2001 force field in Ligprep. The docking grid was defined as a 5-Å cube around the GDP substrate in the C-site. All docking was performed using Glide. The docking poses of the top ranking hits were visually analyzed in Maestro. Most of the top 100 hits were eliminated as not relevant to cancer cell growth experimentally using the cancer cell inhibition assay. Six of the top 100 hits are shown in Table S1. Furthermore, a cell inhibition IC50 of lower than 1 µM was used as an elimination cutoff for further characterization, which led to identification of NSAH and a related naphthyl acyl hydrazone.
Crystallization and Data Collection.
In our laboratory, hRRM1 crystallizes with the TTP effector to provide diffracting crystals, which led us to use the following crystallization protocol. The 20 mg/mL hRRM1 protein was incubated with 20 mM TTP and 1 mM NSAH at 4 °C for 30 min. The drop consisting of hRRM1–TTP–NSAH was cross-seeded with preformed hRRM1–TTP crystals previously reported in ref. 9 to form the cocrystal of hRRM1–TTP–NSAH. The crystals were screened by the hanging-drop method. The well solution for crystallization was composed of 100 mM Tris, pH 7.9, 200 mM Li2SO4, and 19% PEG-3350. The drop size was 2 µL in volume. Diffraction quality crystals appeared after 1 wk and were transferred to the mother liquor, with 20% glycerol as cryoprotectant, and then flash frozen in liquid nitrogen for data collection. Diffraction data were collected with crystals flash cooled at 100 K in a stream of liquid N2 using the synchrotron radiation source, NE-CAT beamline, at APS Chicago. The crystals were of space group P212121 and diffracted to 2.66-Å resolution.
Structure Solution and Refinement.
The structure of the NSAH–hRRM1 complex structure was solved by molecular replacement using a previously solved structure of hRRM1–GDP–TTP (PDB ID code 3HNC) as a search model. Molecular replacement gave a single prominent solution after rotation and translation function. The initial solution was refined by rigid-body refinement, which produced a clearly interpretable electron density map for the overall structure. The ligand density for NSAH was clearly observed at the active site of hRRM1 (Fig. S2). Manual adjustment of the backbone and sidechains of the model was conducted in Coot (42). Crystallographic refinement was carried out using the programs phenix.refine (43) and refmac 5 (44) within CCP4 program suite 6.2.0 (45). The final rounds of refinement were conducted using refmac 5. Difference Fourier maps with coefficients 2|Fo|–|Fc| and |Fo|–|Fc| were used to model NSAH interacting with amino acid residues at the catalytic site. After a few rounds of model building water molecules were added using the |Fo|–|Fc| map peaks above 3.0σ. The value of Rfree can be used as an indicator to validate the water picking and refinement and to avoid any possible overfitting of the data. Data quality and refinement statistics are presented in Table S2. Figures were prepared in PyMOL.
Inhibition Mechanism.
To test for different mechanisms of inhibition, the reaction velocities observed over a range of inhibitor and substrate concentrations were globally fit to mixed model of inhibition represented by the following equilibrium reaction scheme:
Scheme S1.
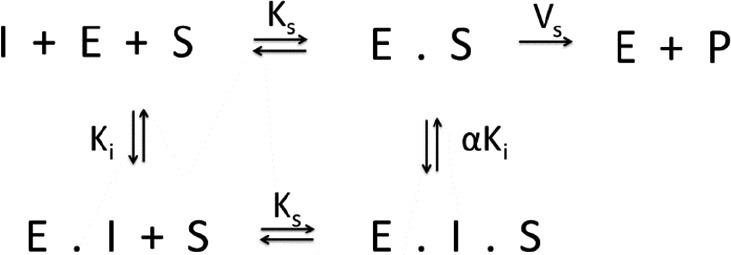
In this general mechanism, the inhibitor can bind both to the free enzyme as well as the enzyme–substrate complex with varying affinities and behaving according to the following rate equation:
In this expression, the affinity of the inhibitor for the free enzyme is defined as Ki. The magnitude of the term α allows the inhibition mechanism to be evaluated. When α approaches 1, the inhibitor is defined as noncompetitive, whereas when α approaches infinity the mechanism becomes competitive.
Blood Progenitor CFU Assay.
Briefly, mobilized peripheral blood mononuclear cells were collected from healthy donors by apheresis after Neupogen stimulation under University Hospitals IRB Protocol #09-90-195. Excess discarded cells were diluted to 1(106) cells/mL in RPMI 1640 plus 15% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin, and 50.4 units/mL GM-CSF. Drug solution was added to the cell suspension and 9× volume of complete methylcellulose (Methocult H4434 plus 50 µM hemin). The methylcellulose/cell suspension was aliquoted into triplicate 35-mm gridded tissue culture plates and incubated in a humidified 5% CO2 incubator at 37 °C for 14 d. Plates were counted visually and clusters of >50 cells were scored as surviving colonies.
Cancer Cell Line Growth Inhibition Assay.
Cell lines (human colon cancer HCT116 and human breast cancer MDA-MB-231 cells) were maintained in standard growth medium; RPMI 1640 plus 10% FBS plus 2 mM glutamine plus 100 U/mL penicillin, 100 µg/mL streptomycin, and shown to be negative for mycoplasma contamination using the MycoAlert Mycoplasma Detection kit (Lonza). Cell identity was verified by Short Tandem Repeat testing performed using the Promega StemElite kit, in the Genetic Resources Core Facility at Johns Hopkins University.
For growth inhibition assays, cells were harvested by trypsinization and seeded into 96-well tissue culture plates at 2,500 cells per mL. The following day, an equal volume of 2×-drug containing medium was added to each well. The cells were cultured for 3 additional days at 37 °C in a 5% CO2 humidified incubator. Cell growth was assessed by measuring DNA content per well using the method of Labarca and Paigen (46). Dye fluorescence was measured in a Perkin-Elmer 1420 Victor3 Multilabel plate reader using 355-nm excitation and 460-nm emission.
Cell Cycle and dNTP Analyses.
For cell cycle analysis, cells were harvested by trypsin, resuspended in complete medium, and counted. Cells were washed in PBS and fixed with 0.25% formaldehyde at 37 °C for 10 min. Cells were spun and then stored in 90% methanol/10% PBS at −20 °C until staining. Cells were washed two times in PBS and treated with 40 µg/mL RNase A for 30 min at 37 °C. Staining was accomplished by addition of 50 µg/mL propidium iodide and incubation at 4 °C for >30 min. Cell cycle distribution was determined using an Attune Acoustic Focusing Cytometer, and data analysis was performed using ModFit LT software.
For dNTP analysis, cells were harvested by trypsin, resuspended in complete medium, and counted. Cells were lysed in 0.4 M perchloric acid and neutralized with potassium hydroxide, and soluble nucleotides were separated from precipitate by centrifugation at 400 × g for 5 min. Supernatant was frozen at −20 °C until analysis. Ribonucleotides were removed from by boronate chromatography, and the resulting dNTPs were measured by high-pressure liquid chromatography on a Waters Alliance 2690 system equipped with a strong anion exchange column. Deoxynucleotides were separated with a linear gradient of 0.15–0.6 M ammonium phosphate buffer, pH 3.5, and then detected and quantitated at 254 and 281 nm. The NSAH used for fluorescence assays, enzymology, CFU assays, some of the growth inhibition assays, and the cell cycle and dNTP pool analyses was produced at the R.V. laboratory. This compound matched in spectral data with the commercially purchased NSAH.
Acknowledgments
We thank the members of the NE-CAT at APS and Advanced Light Source (ALS) with data collection. We thank Banumati Sankaran for screening crystals and collecting data at the ALS. We thank Dr. David Wald for assisting with the hematopoietic progenitor CFU assay. This work was funded by NIH Grant R01GM100887, the Council to Advance Human Health (CAHH), and Taipei Medical University/Clinical and Translational Science Center Pilot Award. This research was also supported by the Translational Research and Pharmacology Core Facility and the Hematopoietic Biorepository and Cellular Therapy Core Facility of the Case Comprehensive Cancer Center (Grant P30 CA43703). The NE-CAT beamlines are supported by National Institute of General Medical Sciences Grant P41 GM103403 from the NIH. This research used resources of the APS, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. The Berkeley Center for Structural Biology is supported in part by the NIH, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US DOE under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5TUS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620220114/-/DCSupplemental.
References
- 1.Brown NC, Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J Mol Biol. 1969;46:39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg G, Ullman B, Martin DW., Jr Mutator phenotypes in mammalian cell mutants with distinct biochemical defects and abnormal deoxyribonucleoside triphosphate pools. Proc Natl Acad Sci USA. 1981;78:2447–2451. doi: 10.1073/pnas.78.4.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 5.Yao R, et al. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci USA. 2003;100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown NC, Canellakis ZN, Lundin B, Reichard P, Thelander L. Ribonucleoside diphosphate reductase. Purification of the two subunits, proteins B1 and B2. Eur J Biochem. 1969;9:561–573. doi: 10.1111/j.1432-1033.1969.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 7.Licht S, Gerfen GJ, Stubbe J. Thiyl radicals in ribonucleotide reductases. Science. 1996;271:477–481. doi: 10.1126/science.271.5248.477. [DOI] [PubMed] [Google Scholar]
- 8.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J Biol Chem. 2006;281:27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 9.Fairman JW, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18:316–322. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine-5′-diphosphate inhibit ribonucleotide reductases. Proc Natl Acad Sci USA. 2007;104:14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aye Y, Stubbe J. Clofarabine 5′-di and -triphosphates inhibit human ribonucleotide reductase by altering the quaternary structure of its large subunit. Proc Natl Acad Sci USA. 2011;108:9815–9820. doi: 10.1073/pnas.1013274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashlan OB, Scott CP, Lear JD, Cooperman BS. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41:462–474. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- 13.Aye Y, et al. Clofarabine targets the large subunit (α) of human ribonucleotide reductase in live cells by assembly into persistent hexamers. Chem Biol. 2012;19:799–805. doi: 10.1016/j.chembiol.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie C, Plunkett W. Metabolism and actions of 2-chloro-9-(2-deoxy-2-fluoro-beta-d-arabinofuranosyl)-adenine in human lymphoblastoid cells. Cancer Res. 1995;55:2847–2852. [PubMed] [Google Scholar]
- 15.Wisitpitthaya S, et al. Cladribine and fludarabine nucleotides induce distinct hexamers defining a common mode of reversible RNR inhibition. ACS Chem Biol. 2016;11:2021–2032. doi: 10.1021/acschembio.6b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stearns B, Losee KA, Bernstein J. Hydroxyurea. A new type of potential antitumor agent. J Med Chem. 1963;6:201. doi: 10.1021/jm00338a026. [DOI] [PubMed] [Google Scholar]
- 17.Donehower RC. An overview of the clinical experience with hydroxyurea. Semin Oncol. 1992;19(3 Suppl 9):11–19. [PubMed] [Google Scholar]
- 18.Shewach DS, Lawrence TS. Radiosensitization of human solid tumor cell lines with gemcitabine. Semin Oncol. 1996;23(5) Suppl 10:65–71. [PubMed] [Google Scholar]
- 19.Loehrer PJ, Sr, et al. Gemcitabine alone versus gemcitabine plus radiotherapy in patients with locally advanced pancreatic cancer: An Eastern Cooperative Oncology Group trial. J Clin Oncol. 2011;29:4105–4112. doi: 10.1200/JCO.2011.34.8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker CH, et al. 2′-Deoxy-2′-methylenecytidine and 2′-deoxy-2′,2′-difluorocytidine 5′-diphosphates: Potent mechanism-based inhibitors of ribonucleotide reductase. J Med Chem. 1991;34:1879–1884. doi: 10.1021/jm00110a019. [DOI] [PubMed] [Google Scholar]
- 21.Gandhi V, Plunkett W. Modulatory activity of 2′,2′-difluorodeoxycytidine on the phosphorylation and cytotoxicity of arabinosyl nucleosides. Cancer Res. 1990;50:3675–3680. [PubMed] [Google Scholar]
- 22.Heinemann V, Schulz L, Issels RD, Plunkett W. Gemcitabine: A modulator of intracellular nucleotide and deoxynucleotide metabolism. Semin Oncol. 1995;22(4 Suppl 11):11–18. [PubMed] [Google Scholar]
- 23.Heinemann V, et al. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: A mechanism of self-potentiation. Cancer Res. 1992;52:533–539. [PubMed] [Google Scholar]
- 24.Honeywell RJ, Ruiz van Haperen VW, Veerman G, Smid K, Peters GJ. Inhibition of thymidylate synthase by 2′,2′-difluoro-2′-deoxycytidine (Gemcitabine) and its metabolite 2′,2′-difluoro-2′-deoxyuridine. Int J Biochem Cell Biol. 2015;60:73–81. doi: 10.1016/j.biocel.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Heinemann V. Ongoing selective internal radiation therapy-based studies in the treatment of liver-dominant metastatic colorectal cancer. Future Oncol. 2014;10(15) Suppl:37–39. doi: 10.2217/fon.14.221. [DOI] [PubMed] [Google Scholar]
- 26.Heinemann V, Plunkett W. Modulation of deoxynucleotide metabolism by the deoxycytidylate deaminase inhibitor 3,4,5,6-tetrahydrodeoxyuridine. Biochem Pharmacol. 1989;38:4115–4121. doi: 10.1016/0006-2952(89)90693-x. [DOI] [PubMed] [Google Scholar]
- 27.Pourquier P, et al. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J Biol Chem. 1997;272:26441–26447. doi: 10.1074/jbc.272.42.26441. [DOI] [PubMed] [Google Scholar]
- 28.Pourquier P, et al. Gemcitabine (2′,2′-difluoro-2′-deoxycytidine), an antimetabolite that poisons topoisomerase I. Clin Cancer Res. 2002;8:2499–2504. [PubMed] [Google Scholar]
- 29.Ahmad MF, et al. Identification of non-nucleoside human ribonucleotide reductase modulators. J Med Chem. 2015;58:9498–9509. doi: 10.1021/acs.jmedchem.5b00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halgren TA, et al. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 31.Friesner RA, et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, et al. Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation. Proc Natl Acad Sci USA. 2006;103:4022–4027. doi: 10.1073/pnas.0600443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsson KM, et al. Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase. Nat Struct Mol Biol. 2004;11:1142–1149. doi: 10.1038/nsmb838. [DOI] [PubMed] [Google Scholar]
- 34.Zimanyi CM, Chen PY, Kang G, Funk MA, Drennan CL. Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli. Elife. 2016;5:e07141. doi: 10.7554/eLife.07141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flanagan SA, Robinson BW, Krokosky CM, Shewach DS. Mismatched nucleotides as the lesions responsible for radiosensitization with gemcitabine: A new paradigm for antimetabolite radiosensitizers. Mol Cancer Ther. 2007;6:1858–1868. doi: 10.1158/1535-7163.MCT-07-0068. [DOI] [PubMed] [Google Scholar]
- 36.Zimanyi CM, et al. Tangled up in knots: Structures of inactivated forms of E. coli class Ia ribonucleotide reductase. Structure. 2012;20:1374–1383. doi: 10.1016/j.str.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker WB, Bapat AR, Shen JX, Townsend AJ, Cheng YC. Interaction of 2-halogenated dATP analogs (F, Cl, and Br) with human DNA polymerases, DNA primase, and ribonucleotide reductase. Mol Pharmacol. 1988;34:485–491. [PubMed] [Google Scholar]
- 38.Gandhi V, Huang P, Xu YZ, Heinemann V, Plunkett W. Metabolism and action of 2′,2′-difluorodeoxycytidine: Self-potentiation of cytotoxicity. Adv Exp Med Biol. 1991;309A:125–130. doi: 10.1007/978-1-4899-2638-8_28. [DOI] [PubMed] [Google Scholar]
- 39.Tan Z, et al. Small-molecule targeting of proliferating cell nuclear antigen chromatin association inhibits tumor cell growth. Mol Pharmacol. 2012;81:811–819. doi: 10.1124/mol.112.077735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, Faber C, Uchiki T, Racca J, Dealwis C. Structures of eukaryotic ribonucleotide reductase I define gemcitabine diphosphate binding and subunit assembly. Proc Natl Acad Sci USA. 2006;103:4028–4033. doi: 10.1073/pnas.0600440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmad MF, et al. Role of arginine 293 and glutamine 288 in communication between catalytic and allosteric sites in yeast ribonucleotide reductase. J Mol Biol. 2012;419:315–329. doi: 10.1016/j.jmb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labarca C, Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980;102:344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]