Significance
Primordial germ cells (PGCs) are the origin of germ cells and are critically important for continuity of multicellular organisms. Although the unique characteristics of mouse PGCs have been described in gene expression and epigenome levels, the metabolomic and proteomic profiles of PGCs and their significance for PGC properties have been unclear. Our findings in this study demonstrate not only distinct energy metabolisms in PGCs and pluripotent stem cells (PSCs), but also the essential contribution of enhanced oxidative phosphorylation and glycolysis in PGC specification from PSCs, and in reprogramming of PGCs into PSCs, respectively. The results uncover the importance of a shift in main energy metabolism for establishment and maintenance of PGC characteristics.
Keywords: primordial germ cell, metabolome, proteome, glycolysis, oxidative phosphorylation
Abstract
Primordial germ cells (PGCs), undifferentiated embryonic germ cells, are the only cells that have the ability to become gametes and to reacquire totipotency upon fertilization. It is generally understood that the development of PGCs proceeds through the expression of germ cell-specific transcription factors and characteristic epigenomic changes. However, little is known about the properties of PGCs at the metabolite and protein levels, which are directly responsible for the control of cell function. Here, we report the distinct energy metabolism of PGCs compared with that of embryonic stem cells. Specifically, we observed remarkably enhanced oxidative phosphorylation (OXPHOS) and decreased glycolysis in embryonic day 13.5 (E13.5) PGCs, a pattern that was gradually established during PGC differentiation. We also demonstrate that glycolysis and OXPHOS are important for the control of PGC reprogramming and specification of pluripotent stem cells (PSCs) into PGCs in culture. Our findings about the unique metabolic property of PGCs provide insights into our understanding of the importance of distinct facets of energy metabolism for switching PGC and PSC status.
In mouse, germ cells first develop as primordial germ cells (PGCs) from a subset of cells in late epiblasts consisting of primed pluripotent stem cells (PSCs) that differentiate from naïve PSCs, designated primitive ectoderm or early epiblast, at around embryonic day 7.25 (E7.25) in the extraembryonic mesoderm (1). Several cytokines (2) and transcription factors (3–5) have critical roles in the emergence of PGCs. Following their initial appearance, PGCs migrate and colonize the genital ridges at ∼E10.5, subsequently exhibiting sexual differentiation at ∼E11.5. After their initial development, PGCs undergo characteristic epigenetic reprogramming, including the global reduction of histone H3 lysine 9 dimethylation (H3K9me2) and DNA methylation (6–8). As a result of the dynamic changes in gene regulation and epigenetic states that occur in the course of PGC differentiation, PGCs have developmental potential distinct from that of PSCs. Notably, PGCs show dormant totipotency, although PGCs are monopotential cells for the generation of gametes. Nonetheless, PGCs and PSCs remain closely related: both cell types share the expression of several pluripotency-associated transcription factors (9–12), and PGCs are easily reprogrammed into naïve PSCs, designated embryonic germ cells (EGCs), in culture (13, 14). Therefore, the intrinsic mechanisms that control the distinct developmental potential of these two cell types are of great interest.
Recent studies have focused primarily on the transcriptome and epigenome to explain the functional difference between PSCs and PGCs. However, the differences in metabolites and proteins in these cells, which may be closely linked to their distinct developmental potential, have not been examined. Recently, exhaustive analyses of metabolites, especially related to PSCs, have been performed. These analyses have revealed that changes in the metabolic profile of PSCs have an important role in the differentiation and reprogramming of these cells. For instance, although PSCs have high glycolytic activity compared with differentiated cells and are unable to proliferate without glucose (15), ATP production is shifted to oxidative phosphorylation (OXPHOS) in the mitochondria at the time of differentiation of PSCs (16, 17). OCT4 [also known as POU domain, class 5, transcription factor 1 (POU5F1)] facilitates expression of the genes encoding the glycolysis-related enzymes hexokinase 2 (HK2) and pyruvate kinase, muscle 2 (PKM2), and overexpression of these enzymes maintains pluripotency and inhibits the differentiation of embryonic stem cells (ESCs) even if leukemia inhibitory factor (LIF) is absent in culture (18). In addition, there is a glucose concentration dependency in the reprogramming of induced PSCs (iPSCs) from mouse embryonic fibroblasts (MEFs), and this reprogramming is significantly inhibited by the addition of a glycolytic inhibitor, 2-deoxy-d-glucose (2DG) (19). These results indicate that changes in the metabolic profile do not only arise as a result of a change in cellular state, but are also responsible for the control of cellular statuses such as proliferation and maintenance of pluripotency. Whereas the metabolic profiles of PSCs have been well characterized, metabolic differences between PSCs and PGCs, and their physiological meanings, remain unclear.
In the work described here, we determined the metabolic profiles of E13.5 male PGCs by means of comprehensive metabolomic and proteomic analyses using mouse germ cells. We observed that E13.5 male PGCs exhibit several metabolic properties distinct from those of ESCs and gonadal somatic cells (somas). We were especially interested in the energy metabolism of PGCs, which exhibit increased activity of the mitochondrial aerobic respiration pathways and decreased glycolysis. The inhibition of the electron transport chain (ETC) and of glycolysis during PGC induction from PSCs suppressed different phases of the transition, and the inhibition of glycolysis also suppressed PGC reprogramming into PSCs. These results indicate that the changes in energy metabolism between PSCs and PGCs play an important role in the regulation of these processes.
Results
Metabolomic and Proteomic Analyses of PGCs.
To examine the metabolic properties of mouse PGCs, we first performed quantitative metabolomic (SI Appendix, Fig. S1 A–D, Table S1, and Dataset S1) and proteomic (SI Appendix, Fig. S1 A and E–G, and Dataset S2) analyses of sorted viable E13.5 male mouse gonadal PGCs, gonadal somatic cells (somas), and ESCs. Upon settling in embryonic gonads, PGCs complete DNA demethylation and imprint erasure by E13.5, when male PGCs enter into mitotic arrest and female PGCs enter into prophase I of meiotic division. In the case of proteomic data, we analyzed the relative amounts of proteins based on enzyme commission (EC) numbers (SI Appendix, Fig. S1 E–G).
The similarity of metabolomic and proteomic data among E13.5 male PGCs, somas, and ESCs was assessed by principal component analysis (PCA) (SI Appendix, Fig. S1 B and E) and hierarchical clustering (SI Appendix, Fig. S1 C and F). Both PCA and hierarchical clustering showed that metabolomic and proteomic data for each cell type were well separated into distinct clusters, indicating that PGCs have distinguishable metabolic and proteomic characteristics compared with somas and ESCs. To better characterize the metabolic and proteomic characteristics of E13.5 male PGCs, we extracted the metabolites and proteins with differing abundances among PGCs, somas, and ESCs by an analysis of variance (ANOVA, P < 0.05). Then, the extracted metabolites and proteins were divided into several clusters by K-mean clustering [SI Appendix, Fig. S1 D (M1–M4) and G (P1–P6)]. In metabolomic analysis, each cluster contained the metabolites that exhibited characteristic variation among cell types (SI Appendix, Fig. S2A). In the case of proteomic data, we performed functional annotation analysis of each cluster (SI Appendix, Fig. S2B). Metabolomic and proteomic analyses showed several similar tendencies for the metabolic properties of each cell type. In particular, abundance of mitochondria- and glycolysis-related enzymes in E13.5 male PGCs and ESCs, respectively, was evident.
Characteristics of Energy Metabolic Pathways in PGCs.
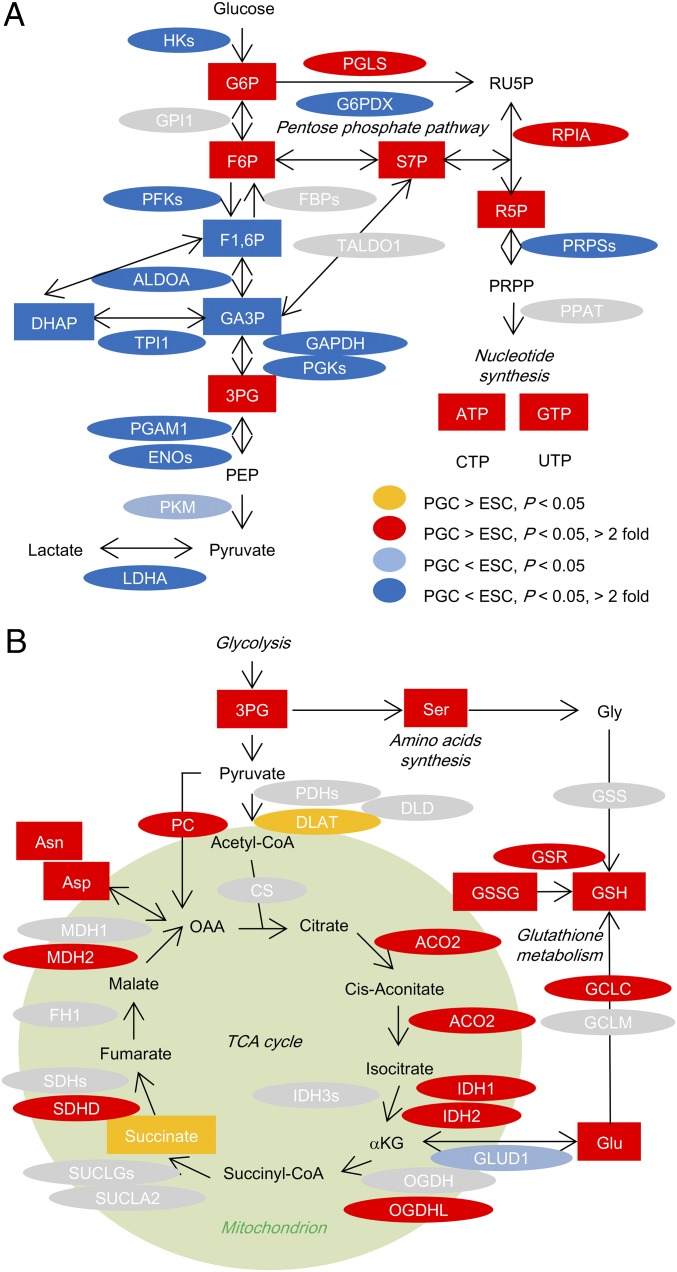
To identify the characteristics of PGCs in the main energy metabolic pathways in detail, we assessed the abundance of individual metabolites and metabolic enzymes involved in glycolysis, the tricarboxylic acid (TCA) cycle, and OXPHOS (SI Appendix, Datasets S1 and S2) and marked the compounds and enzymes that showed differing abundances between E13.5 male PGCs and ESCs (Fig. 1), or between E13.5 male PGCs and somas (SI Appendix, Fig. S3). In glycolysis, G6P and F6P (abbreviations are as shown in SI Appendix, Fig. S2A) are enriched in PGCs; as described in more detail below, levels of these metabolites correlate with activity of the pentose phosphate pathway (PPP) in PGCs. Meanwhile, the downstream metabolites F1,6P and DHAP are enriched (Fig. 1A and SI Appendix, Fig. S4A), and phosphofructokinases (PFKs) and many other glycolysis-related enzymes accumulate to high levels in ESCs compared with PGCs (Fig. 1A), indicating that ESCs highly promote glycolysis, an observation that has been reported previously (17, 19). Few glycolytic compounds and enzymes showed differing abundances between PGCs and somas (SI Appendix, Figs. S3A and S4A), suggesting that glycolytic activity does not differ greatly between these cell types.
Fig. 1.
Integrated analysis of metabolomic and proteomic data between E13.5 male PGCs and ESCs. (A and B) An integrated view of glycolysis-related (A) and TCA cycle-related (B) metabolic pathways comparing E13.5 male PGCs with ESCs. Ellipses and rectangles show enzymes and metabolites, respectively. Orange and red indicate elements that are moderately or strongly (respectively) enriched in PGCs compared with ESCs; light blue and blue indicate elements that are moderately or strongly (respectively) depleted in PGCs compared with ESCs. Gray indicates elements that were undetected or those that did not differ significantly between PGCs and ESCs. For clarity, these figures exclude several metabolites and enzymes.
Some metabolites of the TCA cycle, including citrate and succinate, had a tendency to be abundant in E13.5 male PGCs relative to ESCs (Fig. 1B and SI Appendix, Fig. S4B). PGCs notably accumulated enzymes involved in the earlier part of the TCA cycle [aconitase 2 (ACO2), isocitrate dehydrogenases (IDH1, IDH2), etc.] compared with ESCs (Fig. 1B). On the other hand, somas had lower amounts of pyruvate dehydrogenases (PDHs) than did PGCs (SI Appendix, Fig. S3B); the PDH enzymes are required for the production of acetyl-CoA from pyruvate. These results implied that E13.5 male PGCs have higher TCA cycle activity than do somas and ESCs. Oxidized nicotinamide adenine dinucleotide (NAD+) also tended to be abundant in PGCs compared with somas and ESCs (SI Appendix, Fig. S4C). NAD+ arises from the oxidation of NADH by respiratory chain complex I under aerobic respiration. In accordance with this observation, many components of the respiratory chain complexes were highly abundant in PGCs (SI Appendix, Table S2). These results indicate that E13.5 male PGCs exhibit higher mitochondrial activity than do somas and ESCs, suggesting enhanced promotion of energy production by aerobic respiration in PGCs.
Additional Distinctive Features of Metabolic Pathways in PGCs, Somas, and ESCs.
The PGC-enriched metabolite clusters (M1 and M3) contained nucleoside triphosphates (NTPs) (ATP, CTP, and GTP), and the PGC-enriched protein cluster (P4) had a term related to purine biosynthesis, indicating the promotion of nucleotide synthesis in E13.5 male PGCs (SI Appendix, Fig. S2 A and B). Actually, the levels of some metabolites and enzymes related to the PPP were elevated in PGCs compared with ESCs and somas (Fig. 1A and SI Appendix, Figs. S3A and S5 A and B). These results suggested that E13.5 male PGCs enhance the production of nucleotides through the activation of PPP or subsequent processes.
The PGC-enriched metabolite clusters (M1 and M3) also contained many amino acids and glutathiones (both oxidized and reduced forms), and the PGC-enriched protein cluster (P3) included proteins related to glutathione metabolism (SI Appendix, Fig. S2 A and B). The former part of the TCA cycle has an important role in the production of glutamate and subsequently glutathione from α-ketoglutarate (αKG). Indeed, PGCs had higher amounts of glutamate, glutathione, and glutathione-related enzymes [glutathione-disulfide reductase (GSR) and glutamate-cysteine ligases (GCLs)] than did ESCs and somas (Fig. 1B and SI Appendix, Figs. S3B and S5 C and D), indicating that amino acid biosynthesis and subsequent glutathione metabolism are enhanced in E13.5 male PGCs.
Energy Metabolic Activity in PGCs.
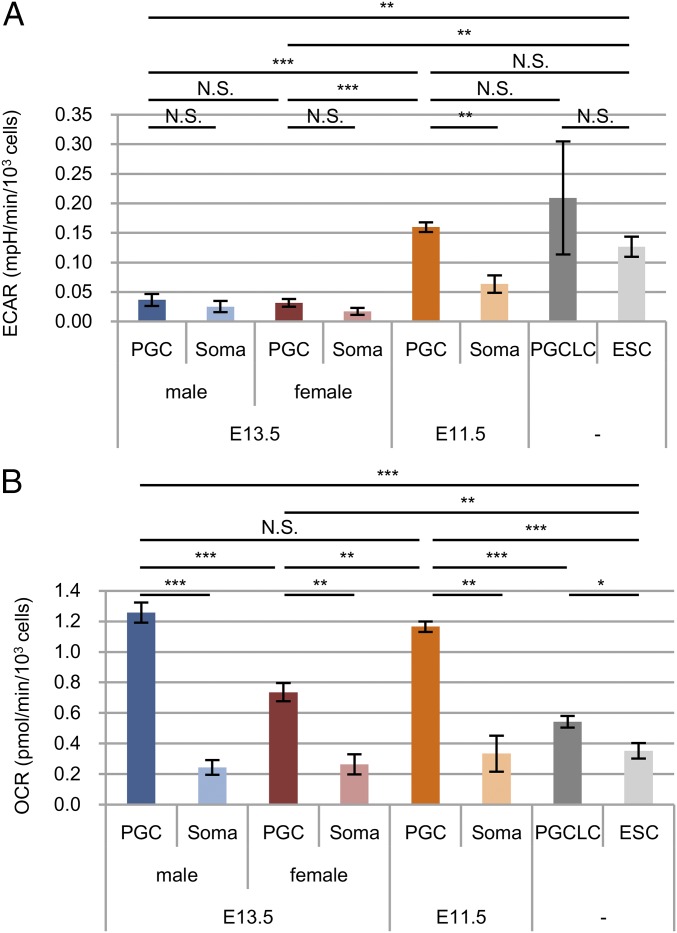
To confirm the above-mentioned features of energy metabolism in E13.5 PGCs, we examined the metabolic activities in glycolysis and mitochondrial respiration by measuring the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of each cell type (Fig. 2 and SI Appendix, Table S3). The glycolytic activity of E13.5 male PGCs was comparable to, and lower than, that of somas and ESCs, respectively (Fig. 2A), and the OXPHOS activity of E13.5 male PGCs was significantly higher than that of male somas and ESCs (Fig. 2B). These results were in accordance with the results of metabolomic and proteomic analyses (Fig. 1 and SI Appendix, Fig. S3 and Table S2). We further examined E13.5 female PGCs and found metabolic features similar to those of male PGCs (Fig. 2 A and B), although the level of OXPHOS in female PGCs was lower than that in male PGCs.
Fig. 2.
Glycolytic and OXPHOS activities of E13.5 male and female PGCs and somas, E11.5 PGCs, and somas, PGCLCs, and ESCs. (A) Glycolytic activity determined by extracellular acidification rate (ECAR), (B) basal respiration determined by subtracting the oxygen consumption rate (OCR) after the addition of antimycin and rotenone (a known inhibitor of OXPHOS complex I) from the basal OCR (three to six biological replicates). Experiments consisted of three to six biological replicates for each sample (SI Appendix, Table S3). Values are plotted as mean ± SEM. N.S., not significant, *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).
We then measured ECAR and OCR in E11.5 PGCs and somas. The results showed that glycolysis in E11.5 PGCs was enhanced compared with that in E13.5 male and female PGCs, and was comparable to that in ESCs (Fig. 2A). The OXPHOS in E11.5 PGCs was comparable to that of E13.5 male PGCs and was enhanced compared with that in ESCs (Fig. 2B). We also examined ECAR and OCR in PGC-like cells (PGCLCs) induced from ESCs via epiblast-like cells (EpiLCs) by culturing in the presence of specific cytokines. Previous work has demonstrated that this culture system faithfully reproduces the differentiation (during embryogenesis) of early PGCs (equivalent to E9.5 PGCs) from epiblast (20). Glycolysis in PGCLCs was comparable to that in ESCs and E11.5 PGCs (Fig. 2A). OXPHOS in PGCLCs was nominally higher than that in ESCs, but apparently lower than that in E11.5 PGCs (Fig. 2B). These results together indicated that glycolysis and OXPHOS in PGCLCs (corresponding to E9.5 PGCs) are maintained at levels similar to those in ESCs, but in the course of PGC differentiation, glycolysis is decreased after E11.5 and OXPHOS is enhanced after E9.5.
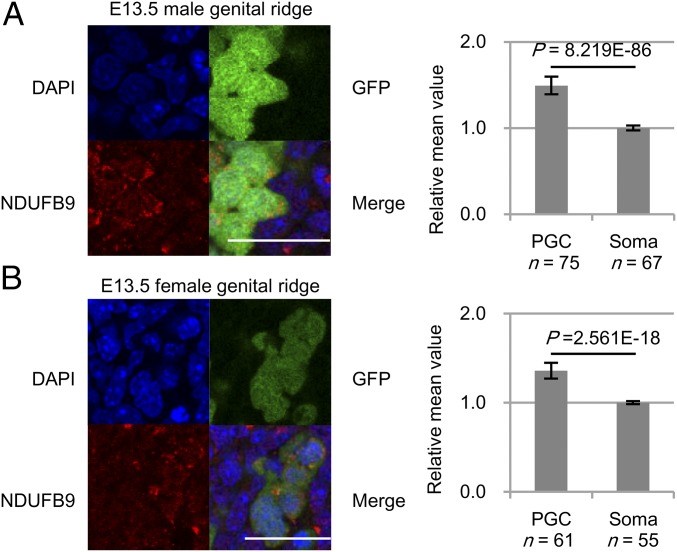
Furthermore, we verified whether the enhanced OXPHOS of PGCs could also be seen in situ by assessing the expression of OXPHOS-related enzymes. Proteomic analysis demonstrated that both NADH:ubiquinone oxidoreductase subunit B9 (NDUFB9) and ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E (ATP5K), which are subunits of OXPHOS complexes I and V, respectively, were abundant in E13.5 male PGCs compared with somas (SI Appendix, Table S2). Quantitative estimation of immunofluorescence staining of E13.5 male genital ridges showed significantly higher expression of both of these enzymes in PGCs than in somas (Fig. 3A and SI Appendix, Fig. S6A). NDUFB9 was also highly expressed in E13.5 female PGCs compared with somas (Fig. 3B). These results supported the idea that OXPHOS is elevated in E13.5 male and female PGCs in situ. We further observed that a glutathione-related enzyme, GSR, accumulated to higher levels in E13.5 male and female PGCs compared with somas (SI Appendix, Fig. S6 B and C), consistent with enhanced glutathione metabolism in E13.5 male PGCs.
Fig. 3.
The expression of OXPHOS-related enzymes in E13.5 genital ridges. (A and B) NDUFB9 immunostaining of E13.5 male (A) and female (B) genital ridges (Left). The relative intensities of NDUFB9 fluorescence in Oct4-deltaPE-GFP+ PGCs compared with those in surrounding somatic cells (Right). (A) PGC, n = 75; soma, n = 67. (B) PGC, n = 61; soma, n = 55. Experiments consisted of two biological replicates for each sample. (Scale bar: 25 μm.) Values are plotted as mean ± SEM (Student’s t test).
Glycolysis-Dependent PGC Reprogramming.
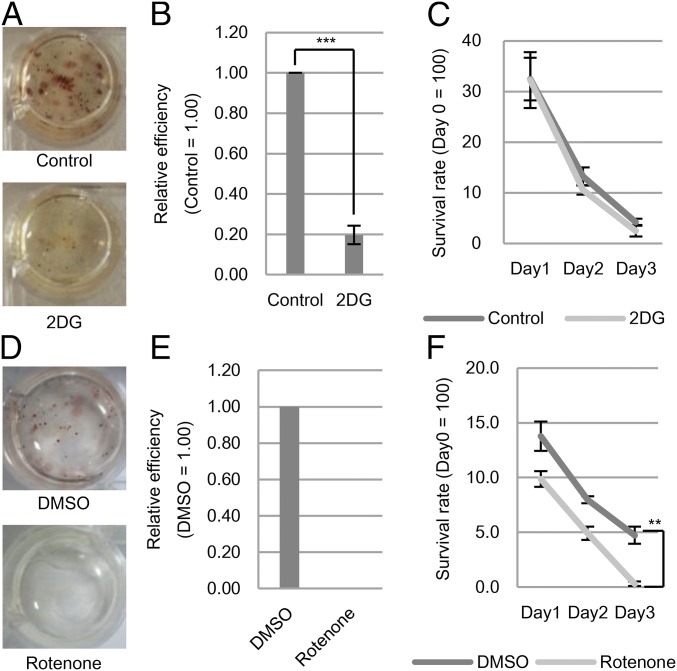
In culture, PGCs can be reprogrammed into PSCs (EGCs) (13, 14); the importance of metabolic change during this process has not been defined. To elucidate this point, we checked the reprogramming efficiency of E12.5 PGCs in the presence of a glycolytic inhibitor, 2DG. We found that the addition of 2DG at the beginning of reprogramming clearly repressed the formation of alkaline phosphatase (ALP)+ colonies, which correspond to EGCs (Fig. 4A), to about 20% of the level seen in the control culture (Fig. 4B). Treatment with 2DG did not affect the cell viability of cultured PGCs (Fig. 4C), indicating that the inhibitory effects on PGC reprogramming did not reflect nonspecific toxicity of 2DG to PGCs. Indeed, the inhibitory effect of 2DG was detectable for up to 72 h after the initiation of culture (SI Appendix, Fig. S7A), demonstrating that glycolytic activity is important for the initial phase of reprogramming of PGCs into EGCs. Moreover, reprogramming efficiency clearly exhibited glucose concentration dependency, reaching a peak at 5 mM glucose (SI Appendix, Fig. S7 B and C). On the other hand, E12.5 PGCs cultured with an ETC inhibitor, rotenone, did not form EGC colonies (Fig. 4 D and E). The survival of E12.5 PGCs was severely decreased when rotenone was added at the early stage of culture due to the elevated apoptosis, indicating that ETC activity is required for the survival of PGCs (Fig. 4F and SI Appendix, Fig. S7D). Addition of rotenone at up to 72 h after the initiation of culturing also inhibited EGC formation, presumably because of negative effects on PGC survival. In contrast, addition of rotenone after 72 h had no effect on reprogramming efficiency, but did result in a decrease in colony size (SI Appendix, Fig. S7E), an effect that may reflect inhibition of growth and/or survival of EGCs. Meanwhile the inhibitory effect of rotenone on ESC number was limited in comparison with that on PGCs (SI Appendix, Fig. S7F).
Fig. 4.
Effects of metabolic perturbations on E12.5 PGC reprogramming. (A) Representative images of alkaline phosphatase (ALP) staining of EGC colonies with or without 2DG (1 mM). (B) Relative efficiency of EGC colony formation (data from experiment portrayed in A). (C) Temporal survival rate of Oct4-deltaPE-GFP+ E12.5 PGCs with or without 2DG in culture (day 1∼day 3). (D) Representative images of ALP staining of EGC colonies with DMSO (vehicle) or rotenone (ETC inhibitor; 0.12 μM). (E) Relative efficiency of EGC colony formation (data from experiment portrayed in D). (F) Temporal survival rate of E12.5 PGCs with DMSO or rotenone (day 1–day 3) as assessed by ALP staining. Values are plotted as mean ± SEM of three biological replicates, **P < 0.01, ***P < 0.001 (Student’s t test).
To eliminate the possibility that 2DG and rotenone affect PGC reprogramming indirectly (specifically, through the feeder cells), we investigated the effect of these drugs in a feeder-free process of EGC development (21). In the feeder-free cultures, the addition of 2DG or rotenone again markedly inhibited the induction of EGCs (SI Appendix, Fig. S7G), indicating that these drugs directly affect PGCs with regard to cellular reprogramming.
Roles of OXPHOS and Glycolysis in PGC Specification.
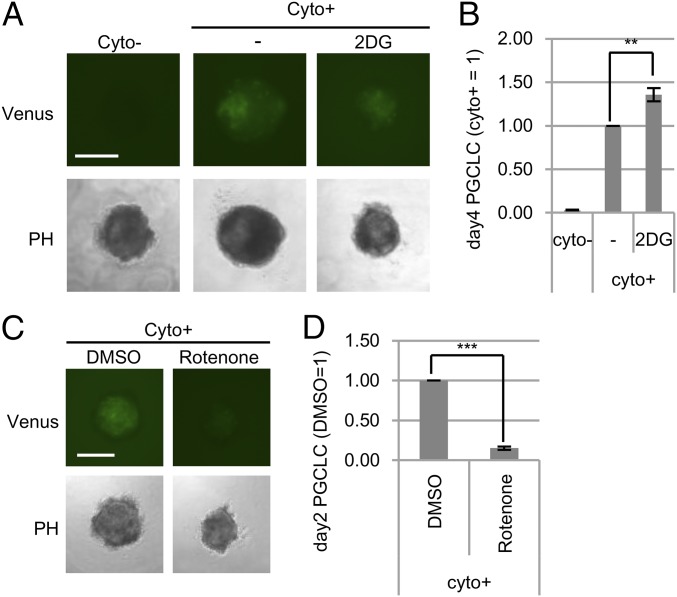
To elucidate the relationship between the generation of PGCLCs and metabolic control, we examined the induction efficiency of PGCLCs under conditions of metabolic perturbation. The addition of a glycolytic inhibitor (2DG) to the PGCLC induction resulted in decreased aggregation size (Fig. 5A), presumably due to the inhibition of growth of EpiLC-derived cells, while yielding a slight (1.4-fold) increase in the proportion of Blimp1-mVenus (BV)+ cells (corresponding to PGCLCs) (Fig. 5B and SI Appendix, Fig. S8A). In addition, BV+ cells were formed even in the absence of glucose (SI Appendix, Fig. S8 B–D). On the other hand, the addition of an ETC inhibitor, rotenone, inhibited PGCLC induction to a level about 15% of that observed under control conditions (Fig. 5 C and D and SI Appendix, Fig. S8E).
Fig. 5.
Effects of metabolic perturbations on PGCLC induction. (A) PGCLC induction with (+) or without (−) cytokines (Cyto). 2DG (1 mM) was added to the culture medium with the cytokines. BV fluorescence was applied as a marker of PGCLCs. (B) Relative proportion of PGCLCs after 4 d of induction (data from experiment portrayed in A). BV+ cells were counted using a cell sorter. (C) ETC inhibition upon PGCLC induction. DMSO (vehicle) or rotenone (ETC inhibitor; 0.03 μM) was added to the medium. (D) Relative proportion of PGCLCs with rotenone after 2 d of induction (data from experiment portrayed in C). PH, phase difference; Venus, Blimp1-mVenus. (Scale bar: 250 μm.) Values are plotted as mean ± SEM of three to four biological replicates, **P < 0.01, ***P < 0.001 (Student’s t test).
Because differentiation from EpiLCs to PGCLCs via early mesodermal-like cells always occurs in a cell aggregate, it is quite difficult to examine whether or not the observed changes reflect the effects of metabolic perturbations on PGCLCs or on the surrounding cells. To elucidate which stages of PGCLC induction were affected by metabolic perturbation, we investigated the expression of marker genes during PGCLC induction. After 2 or 4 d of glucose depletion, BV+ cells clearly expressed early PGC marker genes (Blimp1, Stella, and Nanos3), although the expression levels, especially of Stella (whose expression is normally up-regulated in the later phase of PGCLC induction), were lower than those observed in control cultures on day 4 (SI Appendix, Fig. S9 A and B). Glucose depletion did not affect the expression of an epiblast marker, Fgf5, but caused a modest increase in the expression of an early mesoderm marker, Hoxb1. The addition of 2DG caused expression changes similar to those seen upon glucose depletion (SI Appendix, Fig. S9 C and D). These results together suggested that glycolysis-related pathways are required in the later phase of PGCLC specification. On the other hand, when rotenone was added, expression of Blimp1 and Nanos3, as well as that of Hoxb1, was significantly inhibited in day-2 PGCLC aggregates (SI Appendix, Fig. S9E). These results indicated that ATP production by OXPHOS is required for the initial step of PGCLC specification.
Transcriptional Regulation of Energy Metabolic Enzymes.
To elucidate the mechanisms by which the characteristic energy metabolic profiles of PGCs are established during PGC differentiation, we examined transcriptome data for PGCs (22) to compare the expression levels of genes encoding energy metabolism-related enzymes. The accumulation of transcripts encoding glycolytic enzymes decreased between E11.5 and E13.5 (SI Appendix, Fig. S10A). However, the expression of OXPHOS-related genes did not increase, but rather decreased, during PGC differentiation from E9.5 to E13.5 (SI Appendix, Fig. S10B). These results suggested that the metabolic changes associated with glycolysis, but not those associated with OXPHOS, that occur during the PGC differentiation process are likely, at least in part, due to down-regulation of the genes encoding glycolysis-related enzymes.
Discussion
In this study, we showed the comprehensive metabolic characteristics of mouse E13.5 male PGCs and their physiological meanings. In 1977, Brinster and Harstad reported that mouse E15 germ cells have 70% lower oxidative activity for glucose, and 60% higher oxidative activity for pyruvate, than unfertilized ova (23). In chicken, transcriptome-based comparative metabolic pathway analysis revealed that chicken PGCs up-regulate several nucleotide metabolism-related genes (24). The present report shows the metabolic profiles of mammalian germ cells based on comprehensive metabolomic and proteomic analyses.
Our analyses, in combination with metabolic flux analysis, revealed that E13.5 male and female PGCs exhibit high OXPHOS activity and low glycolytic activity. We further confirmed, using immunostaining, that enzymes involved in OXPHOS accumulate to higher levels in PGCs localized to the E13.5 genital ridges compared with the levels in the surrounding somatic cells. These results indicate that the enhanced OXPHOS activity observed in isolated PGCs reflects genuine metabolic features of PGCs in situ.
In PGCs, the metabolic activities for OXPHOS and glycolysis are initially similar to those in ESCs, but OXPHOS is increased after E9.5, and glycolysis is decreased after E11.5. These observations indicate that the characteristic energy metabolism in E13.5 male and female PGCs is gradually established during differentiation (SI Appendix, Fig. S10C). These changes may be closely correlated to the cellular characteristics of PGCs. For instance, the ability of PGCs to be reprogrammed to pluripotential EGCs is gradually lost after E10.5 (25); we postulate that this change is linked to the diminished glycolytic activity in these cells.
In addition, we showed that the activities of several pathways for the synthesis of various metabolites (notably nucleic acids and amino acids) are relatively high in E13.5 male PGCs compared with those in ESCs and somas. These results also highlight the additional particularity of the metabolic properties of E13.5 PGCs, in which large amounts of ATP are synthesized via highly activated OXPHOS, followed by the syntheses of nucleic acids and amino acids (SI Appendix, Fig. S10C).
Male PGCs stop proliferating and are maintained in a mitotically quiescent state after E13.5 and until birth. In this context, the physiological implications of the metabolic changes described here remain unclear. Nonetheless, the effects of 2DG and rotenone on cultured E12.5 PGCs indicated that OXPHOS, but not glycolysis, is critical for the survival of male PGCs. Further research will be needed to clarify the possible role of these metabolic features in the differentiation of male and female germ cells during development of the embryonic gonad.
E13.5 male PGCs also accumulate high levels of proteins related to DNA repair and glutathione metabolism. These data suggest that PGCs are actively using the machinery for stress response against various environmental perturbations [e.g., reactive oxygen species (ROS) produced as byproducts of OXPHOS], which may be necessary for the maintenance of cell integrity of PGCs under the high OXPHOS condition.
Our results show a correlation of decreased glycolytic activity to down-regulation of the related enzyme-encoding genes in PGCs between E9.5 and E13.5. This observation suggested in turn that the genes encoding glycolytic enzymes are subject to transcription-based regulation. On the other hand, the expression pattern of OXPHOS-related genes was not consistent with the observed changes in OXPHOS activity between E9.5 and E11.5. Therefore, the enhanced OXPHOS during the PGC differentiation may be caused by nontranscriptional regulatory mechanisms, such as those at the level of translation or posttranslational modification.
We also show that the energy metabolic properties of ESCs and PGCs contributed to the differentiation and reprogramming of these cells. In our work, the reprogramming of PGCs to EGCs depended on the presence of glucose, as has been reported for the reprogramming of MEFs to iPSCs (19). In the iPSC reprogramming process, the metabolic shift from OXPHOS to glycolytic status occurs at the early phase of reprogramming via the activation of hypoxia-inducible factors (HIFs) (26, 27). In the present work, we showed that EGC formation was inhibited by the addition of 2DG at the early phase of reprogramming. We hypothesize that metabolic shifts in PGC reprogramming may be controlled by a mechanism similar to that used in somatic cell reprogramming. We found that glycolysis and OXPHOS in PGCLCs are maintained at levels similar to those seen in ESCs; we also found that inhibition of glycolysis and OXPHOS impaired the late and early periods (respectively) of PGCLC differentiation from EpiLCs. It will be important in future studies to elucidate how glycolysis and OXPHOS control the process of PGC specification.
Although PGCs are known to have a unique epigenetic status compared with somas and ESCs (6–8), we did not detect characteristic changes among these cell types in the profiles of metabolites involved in epigenetic regulation. Notably, the levels of S-adenosylmethionine (SAM), a donor of methyl groups, did not differ significantly among these cells (SI Appendix, Fig. S5E), and flavin adenine dinucleotide (FAD) and αKG, known cofactors of demethylases (28), were undetectable in E13.5 male PGCs in our analysis. Because epigenetic reprogramming is almost completed in E13.5 PGC, it is possible that the amounts of these metabolites change at an earlier stage of PGC differentiation, before or concomitant with epigenetic reprogramming. As to histone acetylation, acetyl-CoA was undetectable in E13.5 male PGCs, whereas NAD+, a cofactor of sirtuin-type histone deacetylases, tended to be abundant in PGCs compared with somas and ESCs. Although the relationship between histone acetylation and PGC differentiation is not well understood, further studies may determine whether the quantitative change of NAD+ levels affects the acetylation status of PGCs and the differentiation of PSCs to PGCs.
To further elucidate the importance of metabolic conversion during germ cell differentiation in the development of functional gametes, it will be critical to perform comparative metabolomic analyses of germ cells in various differentiation stages, including male spermatogonia, spermatocytes, and spermatids, and female PGCs, oogonia, and oocytes. Moreover, transomics analyses of the epigenome, transcriptome, proteome, and metabolome (29) in developing germ cells are expected to reveal novel regulatory circuits controlling germ cell properties.
Materials and Methods
Detailed methods are available in SI Appendix, SI Materials and Methods.
Oct4-deltaPE-GFP transgenic mice were maintained in a C57BL/6J genetic background. Animal protocols were reviewed and approved by the Tohoku University Animal Studies Committee.
PGCs, somas, and ESCs were purified by cell sorting. We confirmed the high survival rate (>93%) of each cell type immediately after sorting (SI Appendix, Fig. S1A). The metabolites of sorted E13.5 male PGCs, somas, and VR15 ESCs were extracted by the methanol:chloroform:water extraction method and measured by capillary electrophoresis time-of-flight mass spectrometry. Whole-cell extracts of these cells were digested to peptides in solution and analyzed by nano-liquid chromatography–tandem mass spectrometry. A Seahorse XF24 analyzer was used to measure the oxygen consumption rate and extracellular acidification rate. The significance of differences was assessed by the unpaired two-tailed Student’s t test, one-way ANOVA, or the Mann–Whitney u test. The level of significance was set at P < 0.05. Metabolomic and proteomic data are available as SI Appendix.
Supplementary Material
Acknowledgments
We thank M. Saitou for providing Blimp1-mVenus-Stella ECFP ESCs; Y. Ito-Matsuoka for technical support in the preparation of animals; K. Saito, K. Kato, H. Maki, and M. Homma for technical support in the metabolomic analysis; S. Kobayashi and Y. Hayashi for technical advice on metabolomic analysis; K. Miyasaka for technical advice on the flux analyzer; J. Gao for helpful discussion about mammalian metabolism; all the members of the Cell Resource Center for Biomedical Research for helpful discussions; the Center of Research Instruments in the Institute of Development, Aging, and Cancer; and Biomedical Research Unit of Tohoku University Hospital for use of instruments and technical support. This work was partly supported by the cooperation program of research institutes in Tohoku University (Y.H.); Grant-in-Aid for Scientific Research in the innovative area “Mechanisms Regulating Gamete Formation in Animals” (Grant 25114003) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Y.M.); and by the Japan Agency for Medical Research and Development–Core Research for Evolutional Science and Technology (Y.M., K.I., and T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620915114/-/DCSupplemental.
References
- 1.Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 2.Lawson KA, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 4.Yamaji M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 5.Weber S, et al. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod. 2010;82:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- 6.Seki Y, et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Seisenberger S, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng JH, et al. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev Cell. 2013;24:324–333. doi: 10.1016/j.devcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Kehler J, et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura D, Tokitake Y, Niwa H, Matsui Y. Requirement of Oct3/4 function for germ cell specification. Dev Biol. 2008;317:576–584. doi: 10.1016/j.ydbio.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Campolo F, et al. Essential role of Sox2 for the establishment and maintenance of the germ cell line. Stem Cells. 2013;31:1408–1421. doi: 10.1002/stem.1392. [DOI] [PubMed] [Google Scholar]
- 12.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 13.Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 14.Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 15.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandel NS, Jasper H, Ho TT, Passegué E. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol. 2016;18:823–832. doi: 10.1038/ncb3385. [DOI] [PubMed] [Google Scholar]
- 18.Kim H, et al. Core pluripotency factors directly regulate metabolism in embryonic stem cell to maintain pluripotency. Stem Cells. 2015;33:2699–2711. doi: 10.1002/stem.2073. [DOI] [PubMed] [Google Scholar]
- 19.Folmes CD, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 21.Leitch HG, et al. Rebuilding pluripotency from primordial germ cells. Stem Cell Rep. 2013;1:66–78. doi: 10.1016/j.stemcr.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinster RL, Harstad H. Energy metabolism in primordial germ cells of the mouse. Exp Cell Res. 1977;109:111–117. doi: 10.1016/0014-4827(77)90050-7. [DOI] [PubMed] [Google Scholar]
- 24.Rengaraj D, Lee BR, Jang HJ, Kim YM, Han JY. Comparative metabolic pathway analysis with special reference to nucleotide metabolism-related genes in chicken primordial germ cells. Theriogenology. 2013;79:28–39. doi: 10.1016/j.theriogenology.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Matsui Y, Tokitake Y. Primordial germ cells contain subpopulations that have greater ability to develop into pluripotential stem cells. Dev Growth Differ. 2009;51:657–667. doi: 10.1111/j.1440-169X.2009.01125.x. [DOI] [PubMed] [Google Scholar]
- 26.Prigione A, et al. HIF1α modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1-3 and PKM2. Stem Cells. 2014;32:364–376. doi: 10.1002/stem.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathieu J, et al. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014;14:592–605. doi: 10.1016/j.stem.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryall JG, Cliff T, Dalton S, Sartorelli V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell. 2015;17:651–662. doi: 10.1016/j.stem.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yugi K, Kubota H, Hatano A, Kuroda S. Trans-Omics: How to reconstruct biochemical networks across multiple ‘omic’ layers. Trends Biotechnol. 2016;34:276–290. doi: 10.1016/j.tibtech.2015.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.