Significance
Thyroid hormone (T3) controls both developmental and physiological processes. Its nuclear receptors (TR) are transcription factors. Methyl dioxygenase ten-eleven translocation protein 3 (TET3) is characterized here as a TR coregulator. It stabilizes and promotes TR chromatin association in a dioxygenase-independent manner, thus increasing the sensitivity of the cell to T3. Mutations in TR cause the resistance to thyroid hormone syndrome (RTH) symptom, the severity of which varies with the particular mutation. Only some mutated TR can be stabilized by TET3. The availability of TET3 is therefore a parameter modulating TR activity, and its differential interaction with mutated TR might explain different severity of RTH. Furthermore, TET3 is likely to function as a general coregulator for nuclear receptors, as it enhances chromatin association of additional members of this superfamily.
Keywords: thyroid hormone receptor, methylcytosine dioxygenase TET3, protein stability, chromatin recruitment, RTH syndrome
Abstract
Thyroid hormone receptors (TRs) are members of the nuclear hormone receptor superfamily that act as ligand-dependent transcription factors. Here we identified the ten-eleven translocation protein 3 (TET3) as a TR interacting protein increasing cell sensitivity to T3. The interaction between TET3 and TRs is independent of TET3 catalytic activity and specifically allows the stabilization of TRs on chromatin. We provide evidence that TET3 is required for TR stability, efficient binding of target genes, and transcriptional activation. Interestingly, the differential ability of different TRα1 mutants to interact with TET3 might explain their differential dominant activity in patients carrying TR germline mutations. So this study evidences a mode of action for TET3 as a nonclassical coregulator of TRs, modulating its stability and access to chromatin, rather than its intrinsic transcriptional activity. This regulatory function might be more general toward nuclear receptors. Indeed, TET3 interacts with different members of the superfamily and also enhances their association to chromatin.
Thyroid hormone (T3) is the main natural iodinated compound possessing a biological activity. It exerts a pleiotropic action on development and homeostasis, acting on most, if not all, cell types (1). T3 acts directly on gene transcription by binding to the thyroid hormone receptors (TRs) TRα1, TRβ1, and TRβ2. They are, respectively, encoded by the THRA and THRB genes. In humans, mutations of either THRA or THRB cause the resistance to thyroid hormone syndrome (RTH). The severity of the disease is determined by the precise location of the mutation (2), dictating the ability of the mutated TR to respond to T3 (3).
TRs, as the other members of the nuclear receptor superfamily, are ligand-regulated transcription factors consisting of three major functional domains: the amino-terminal A/B domain, the DNA-binding domain, and the ligand-binding domain. TRs can bind to DNA on a TR response element (TRE) the in absence of T3, and on most genes they repress transcription until T3 binds and leads to activation. Helix12 is the major structural element associated with this process. T3 triggers a dramatic shift of its position, leading to dissociation of corepressors and recruitment of coactivators, including coactivators that have the ability to change the chromatin microenvironment (4). T3 binding also induces a rapid proteasome-mediated degradation of TRs that is associated with T3-dependent transcriptional activity (5). TR availability and chromatin access are thus possibly important levels of modulation of T3 cellular response.
The goal of the present study was to identify epigenetic regulators that interact with, and therefore may modulate, TR transcriptional activity, using in vitro pull-down screening. This approach allowed us to identify TET3, a member of the ten-eleven translocation (TET) family proteins, as a partner for TRs. The TET proteins have been extensively studied as dioxygenase enzymes responsible for demethylation of methylated CpG dinucleotides by catalyzing the hydroxylation of 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) (6, 7). Here we demonstrate a direct interaction between TET3 and TRs, involving primarily the catalytic domain of TET3 and the helix12 of TR. This interaction stabilizes TRs in the chromatin compartment and enhances its transcriptional activity. This does not involve TET3 dioxygenase activity. Thus, we discovered a way for TET3 to regulate transcription by modulating the protein turnover and chromatin association of a transcription factor; here, TRs. Furthermore, we present evidence that TET3 may broadly enhance chromatin association of nuclear hormone receptors.
Results
TET Proteins Interact with TR.
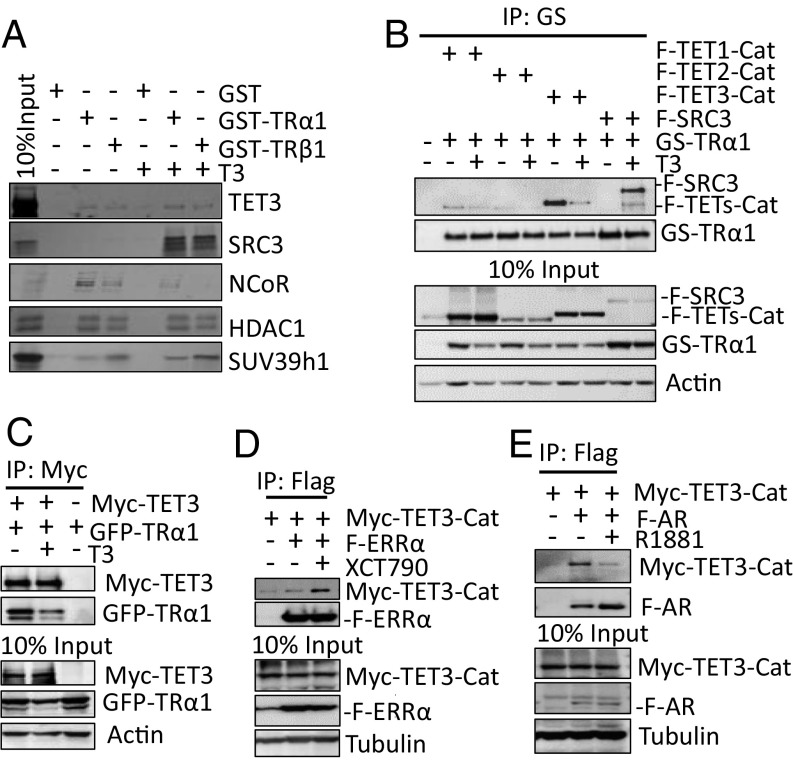
To determine epigenetic modifiers involved in modulating TR activity, the interactions between the recombinant TRα1 or TRβ1 fused to GST (GST-TRα1 or GST-TRβ1), and around 50 epigenetic modification enzymes were tested by in vitro pull down, followed by coimmunoprecipitation assay. The NCoR corepressor and SRC3 coactivator were found to interact with both GST-TRα1 and GST-TRβ1 validating the screen. Other factors such as histone lysine methyltransferase SUV39h1 and histone deacetylase HDAC1 that are known to be coregulators of other nuclear receptors were also identified (8). TET3 was an interactor that came out of the screen (Fig. 1A). Coimmunoprecipitation assays were performed for all three TETs to test whether the interaction with TRs can take place in HEK293T cells. As TETs are large proteins that are difficult to produce, only their catalytic domains were used as a first intention. The catalytic domain of TET3 interacted with TRα1 to a similar level as the interaction between SRC3 and liganded TRα1 (Fig. 1B). The interaction with full-length TET3 was also validated by coimmunoprecipitation (Fig. 1C). In contrary to T3-induced TRα1-SRC3 interaction, T3 reduced the interaction between TRα1 and TET3 in HEK293T cells, but not in the pull-down assay. This suggests TET3 can interact with both the apo- and holo-conformations of TR, but that in a cell environment and in the presence of T3, this interaction is somehow balanced and displaced by the present “classical” coactivators. Similarly, we found that TRβ1 interacts with TET3 (Fig. S1A). Furthermore, initially attempted as controls, we observed that TET3 also interacts with two other nuclear receptors: the constitutive activated estrogen-related receptor α (ERRα) (Fig. 1D) and the androgen receptor (AR) (Fig. 1E). In both cases, the interaction was modulated by ligands, respectively increased by the antagonist XCT790, and decreased by the agonist R1881. As the interaction with the catalytic domain of TET1 and TET2 was considerably weaker (Fig. 1B), the rest of the study was limited to TET3.
Fig. 1.
TET3 interacts with NR. (A) Interaction between TET3 and TR. GST pull-down assays were performed using recombinant GST-TRα1 or GST-TRβ1 proteins and lysates from HEK293T cells overexpressing TET3, SRC3, NcoR, SUV39h1, or HDAC1 in the presence or absence of T3 (5.10−7 M). (B) Interaction between TETs-Cat and TRα1. HEK293T cells extracts expressing indicated proteins were precipitated with M280 beads. Coprecipitated Flag-TETs-Cat or Flag-SRC3 were detected using an anti-Flag antibody. (C) T3 effect on TET3-TRα1 interaction. Myc-TET3 and GFP-TRα1 were transfected in HEK293T cells and immunoprecipitated using an anti-Myc antibody. Coimmunoprecipitated GFP-TRα1 was detected using anti-GFP antibody. (D and E) Effects of XCT790 (antagonist)/R1881 (agonist) on ERRα/AR-TET3Cat interactions. HEK293T cells were transfected with indicated plasmids, Flag beads were used to precipitate Flag-ERRα/Flag-AR, coimmunoprecipitated Myc-TET3-Cat was detected using anti-Myc antibody.
Fig. S1.
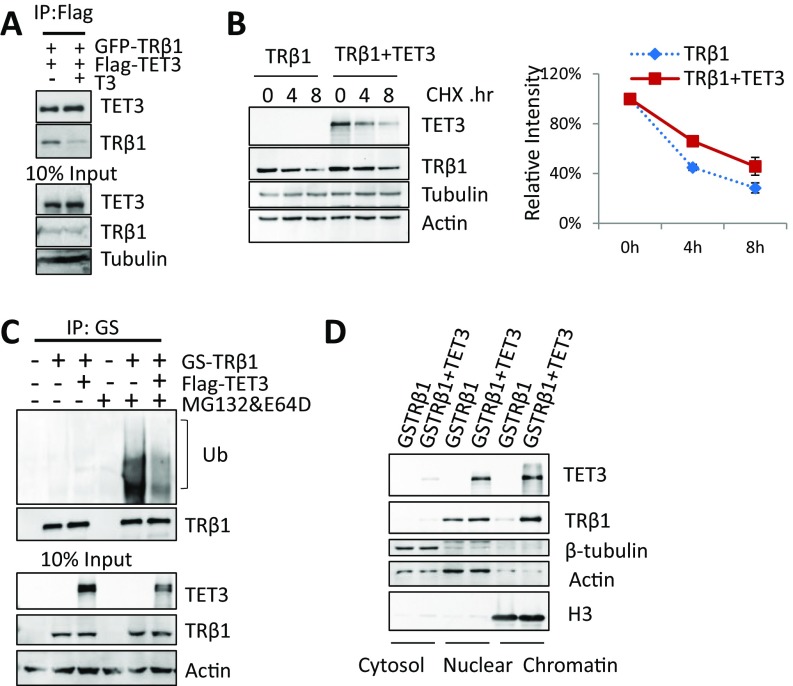
TET3 interacts with, stabilizes, and enhances the recruitment to chromatin of TRβ1. (A) Effect of T3 treatment on TRβ1–TET3 interaction. Flag-TET3 and GFP-TRβ1 were transfected in HEK293T cells and immunoprecipitated using M2 beads; coimmunoprecipitated GFP-TRβ1 was detected using anti-GFP antibody. (B) Effect of TET3 on protein turnover of TRβ1. HEK293T cells expressing GS-TRβ1 (TRβ1) with or without Flag-TET3 (TET3) were treated with CHX for 4 or 8 h. Protein levels of TET3 and TRβ1 were detected by anti-Flag and anti-GS antibodies, respectively (Left). The intensity of TRβ1 signals corrected by tubulin + actin signals were plotted on the Right as 100% was set for the intensity measured for CHX 0 h exposure. (C) Ubiquitination pattern of TRβ1 in the presence or absence of TET3. Whole-cell extracts were prepared from HEK293T cells transfected with indicated plasmids. Cells were treated with or without MG132&E64D before collection. Immunoprecipitates were obtained using M280 beads; half of the immunoprecipitated TRβ1 was subjected to Western blotting as loading control, and the other half was used to determine the ubiquitination level of TRβ1 by incubating with an anti-ubiquitin antibody. The smear indicates the poly-ubiquitination of TRβ1. (D) Effect of TET3 on the subcellular distribution of TRβ1. GS-TRβ1 was cotransfected with or without Flag-TET3 (TET3) in HEK293T cells. Cells were fractionated using NE-PER nuclear and cytoplasmic extraction reagents kit. Cytosol, nucleus, and chromatin fractions were subjected to Western blotting. Protein levels of TET3 or TRβ1 were respectively detected by anti-Flag and anti-GS antibody. β-tubulin, actin, and H3 were, respectively, the loading controls for the cytosol, nucleus, and chromatin.
TET3 and TRα1 Interact Mainly via the Catalytic Domain of TET3 and the AF2 Domain in TRα1.
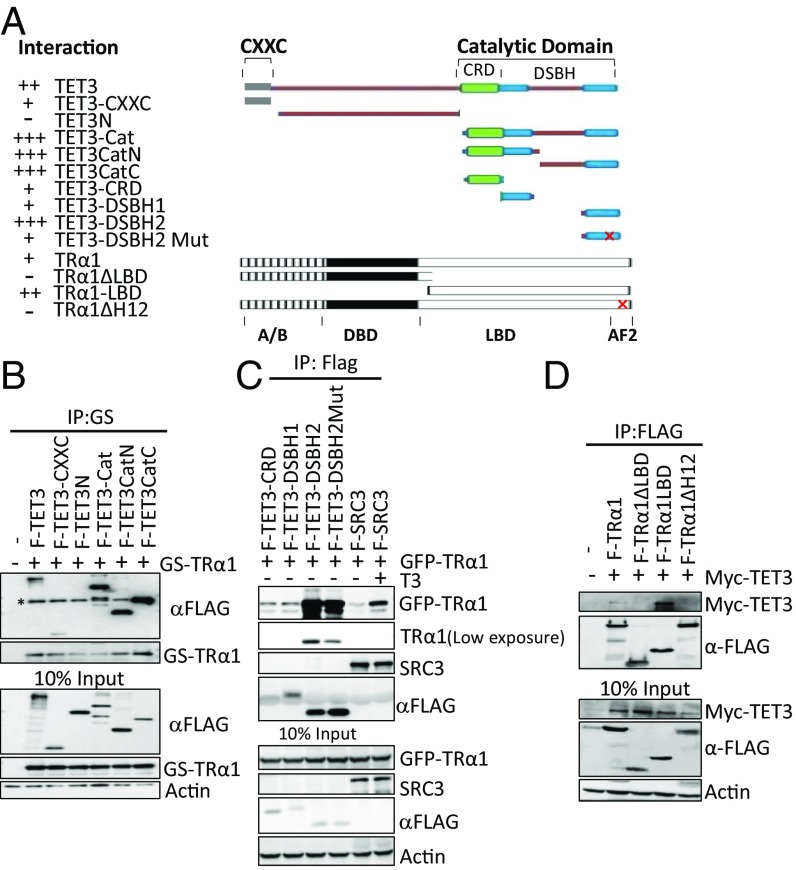
To further characterize the interaction between TET3 and TRs, a series of vectors was generated to express tagged and truncated TET3 (Flag) or TRα1 (Flag) (Fig. 2A). Coimmunoprecipitations showed that TRα1 interacted strongly with the catalytic domain of TET3 (TET3-Cat), weakly with the CXXC domain, and not with the N-terminal portion of TET3 (TET3N) (Fig. 2B). Further dissection of TET3-Cat showed that TRα1 strongly interacts with both the N-terminal half (TET3CatN) and C-terminal half (TET3CatC) of the catalytic domain (Fig. 2B). When structured domains within TET3Cat were analyzed, DSHB2 conferred the most robust interaction (Fig. 2C). Mutation of the only putative LXXLL, a sequence that is frequently present in the interaction domain of NR cofactors, in this domain reduced but did not abolish the interaction (Fig. 2C). Conversely, the presence of the C-terminal helix12, commonly called AF2, of the ligand binding domain of TRα1 was found to be necessary for interaction with TET3 (Fig. 2D).
Fig. 2.
The catalytic domain of TET3 and the AF2 domain of TRα1 confer the interaction between these two proteins. (A) Schematic representation and summary of interactions of full-length and truncation mutants of TET3 and TRα1. AF2, activation function domain 2; Cat, catalytic domain; CRD, cysteine-rich domain; CXXC, CXXC domain; DSBH, double-stranded beta-helix domain; DBD, DNA binding domain; LBD, ligand binding domain. “+,” more “+,” and “−” represent, respectively, interaction, stronger interaction, and no interaction. (B and C) Characterization of the interacting regions in TET3 with TRα1. Whole-cell extracts were prepared from HEK293T cells expressing indicated proteins. (B) GS-TRα1 was precipitated with M280 beads; coprecipitated F-TET3 mutants were detected using anti-Flag antibody. Asterisk indicates bands for TRα1 that are recognized thanks to the cross-reaction between anti-Flag antibody and its GS tag. (C) Flag beads were used to precipitate TET3 truncation mutants; coprecipitated TRα1 was detected with anti-GFP antibody. (D) Characterization of the interacting regions in TRα1 with TET3. Flag beads were used to precipitate TRα1 truncation mutants; coprecipitated TET3 was detected with anti-Myc antibody.
TET3 Modulates T3 Response and Regulates TRα1 Protein Levels.
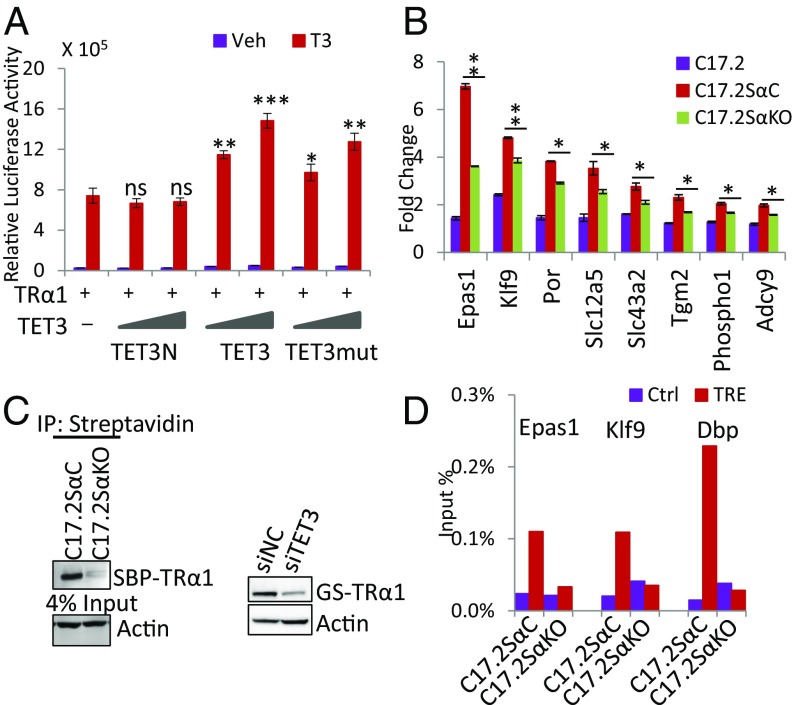
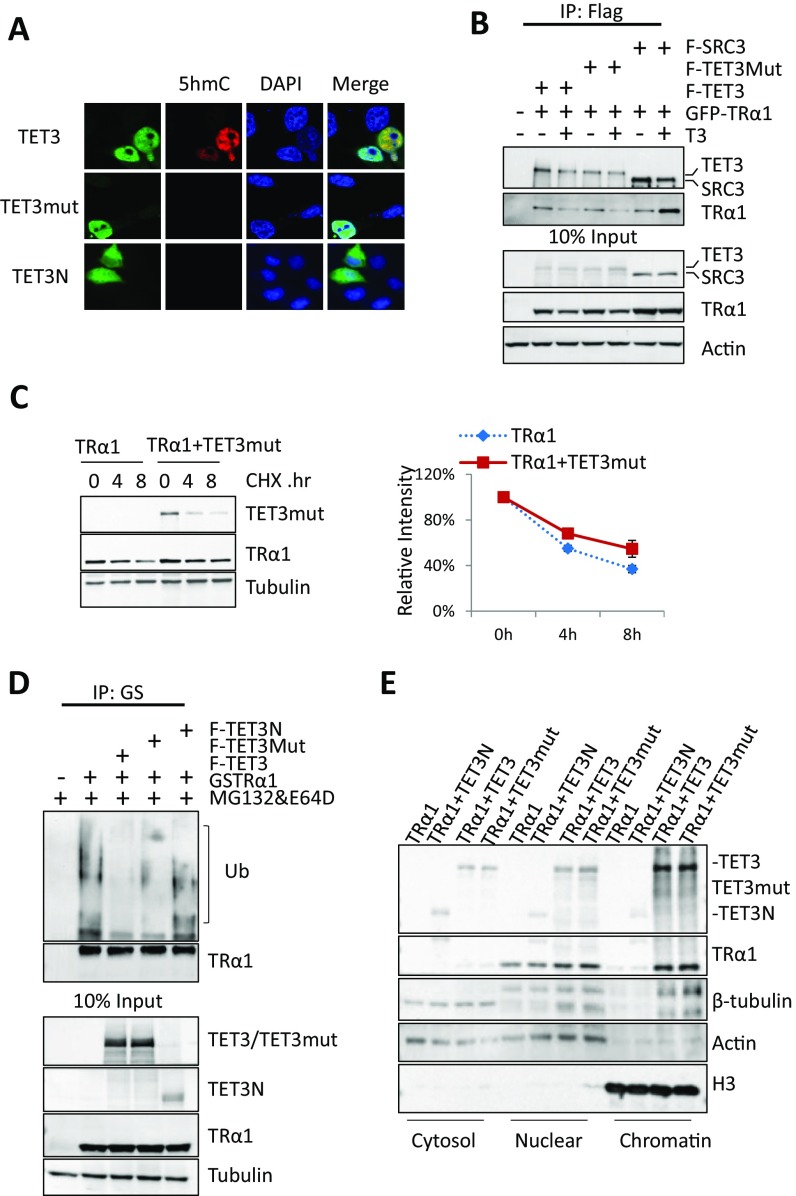
After identifying the interaction between TET3 and TRα1, we evaluated whether TET3 affected TRα1 activity. We first examined the effect of TET3 expression on TRα1 transcription capacity in a transient expression assay performed in HEK293T cells. Full-length TET3 enhanced TRα1 transcriptional activity in a dose-dependent manner, whereas a TET3N mutant that cannot interact with TR failed to do so (Fig. 3A). The catalytic activity was not required for this effect, as demonstrated by using the TET3 mutant (TET3mut) (Fig. 3A) that lacks the dioxygenase activity (Fig. S2A), but retains the ability to interact with TRα1 (Fig. S2B).
Fig. 3.
TET3 regulates TRα1 activity and TRα1 protein level. (A) TET3 regulation of TRα1 activity. HEK293T cells were transfected with TRα1, a luciferase reporter plasmid, and 2 doses of TET3 constructs: TET3N that lost interaction with TR; TET3 and enzymatic dead mutant (TET3mut). Relative luciferase activities were measured 24 h after T3 (10−8 M) treatment; the triangles represent increasing amount of TET3 constructs. The asterisks indicate the significance of the differences between each condition and TRα1 alone in the presence of T3. (B) TR target gene expression is regulated by TET3 levels in C17.2Sα cells. Expressions of TRα1 target genes in indicated cells treated or not with T3 (10−8 M) were examined by relative qRT-PCR. Relative induction triggered by T3 in each cell line was presented. The asterisks indicate the significance of the differences between two indicated conditions. (C) Effect of TET3 inhibition on TRα1 protein level in C17.2 cells. TRα1 was detected using an anti-TRα1 antibody in C17.2GSα, C17.2SαC, and C17.2SαKO cells. Streptavidin beads were used to precipitate SBP-TRα1 in C17.2SαC and C17.2SαKO. (D) TRα1 binding to target genes after TET3 KO in C17.2Sα. ChAP of TRα1 on TRE or control (Ctrl) regions of indicated genes. Results are presented as percentage of input.
Fig. S2.
Enzymatic dead mutant of TET3 retains the modulation effect on TRα1. (A) TET3mut lost its enzyme activity measured by immunofluorescence. 5hmC levels were analyzed in HeLa cells transfected with Flag-TET3 (TET3), Flag-TET3mut (TET3mut), or Flag-TET3N (TET3N). Expression of TET3 constructs were revealed with the Flag antibody and presence of 5hmC with the 5hmC antibody. (B) Both TET3 and TET3mut interact with TRα1, as identified by coimmunoprecipitation. Whole-cell extracts were prepared from HEK293T cells cotransfected with GFP-TRα1 (TRα1) and Flag-TET3 (F-TET3) or Flag-TET3mut (F-TET3mut) or Flag-SRC3 (F-SRC3) treated or not by T3 (5.10–8 M) before collection. Immunoprecipitates were obtained using M2 magnetic beads. Coimmunoprecipitated GFP-TRα1 was detected by Western blotting, using anti-GFP antibody. (C) Effect of TET3mut on TRα1 protein turnover. HEK293T cells expressing GS-TRα1 (TRα1) with or without Flag-TET3mut (TET3mut) were treated with CHX for 4 or 8 h. Whole-cell lysates were subjected to Western blotting (Left). Protein levels of TRα1 were detected by anti-GS antibody. The intensity of TR signals corrected by tubulin signals were plotted on the Right as 100% was set for the intensity measured for CHX 0h exposure. (D) Ubiquitination pattern of TRα1 in the presence or absence of TET3mut. Whole-cell extracts were prepared from HEK293T cells transfected with indicated plasmids. Cells were treated or not with MG132&E64D before collection. Immunoprecipitates were obtained using M280 beads; half of the immunoprecipitated TRα1 was subjected to Western blotting as loading control, whereas the other half was used to determine the ubiquitination level of TRα1 by incubating with an anti-ubiquitin antibody. The smear indicates the polyubiquitination of TRα1. (E) Effect of TET3, TET3mut, or TET3N on TRα1 subcellular distribution. Cell fractionation experiment was performed for HEK293T cells cotransfected with GS-TRα1 (TRα1) and different Flag-tagged mutants of TET3. After cell fractionation cytosol, nucleus and chromatin fractions were subjected to Western blotting. TET3 or TRα1 were, respectively, detected by anti-Flag and anti-GS antibody. β-tubulin, actin, and H3 were, respectively, the loading controls for the cytosol, nucleus, and chromatin.
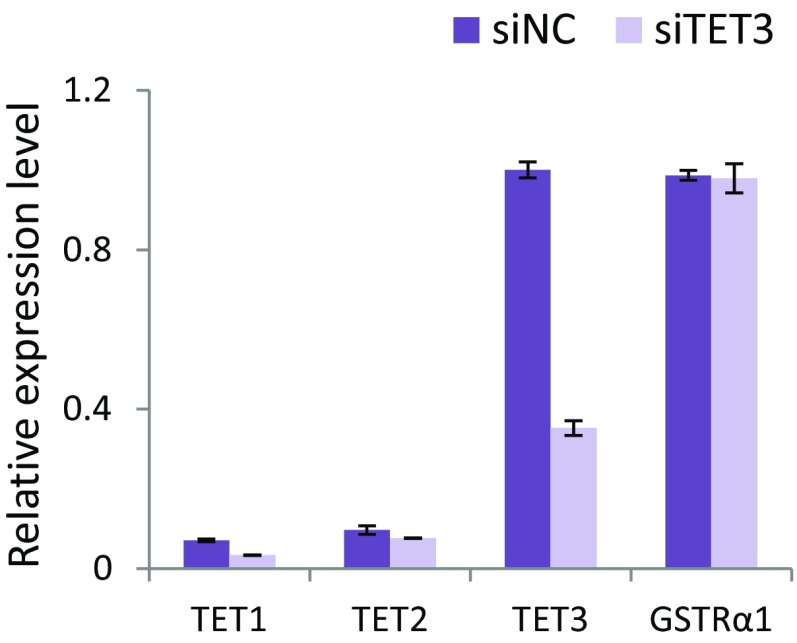
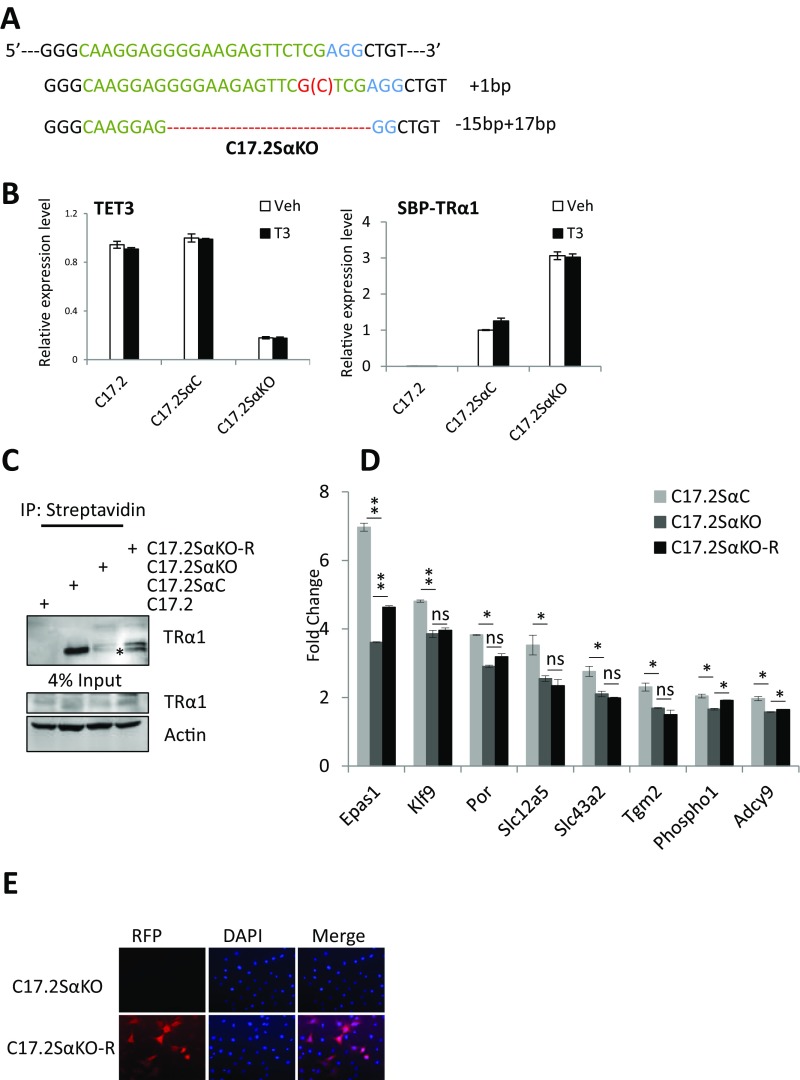
Then we moved to cellular systems to look at the regulation of endogenous target genes. We used a neural stem cell line, namely, C17.2GSα, in which a murine GS-tagged TRα1 is stably expressed and TRα1 target genes have been fully identified (9). RT-PCR analyses revealed that these cells express endogenous TET3 at higher level than TET1 and TET2 (Fig. S3), but we failed to detect TET3 proteins by Western blotting, using commercial TET3 antibodies. To investigate the potential role of TET3 in TR transcriptional regulation, we used the CRISPR/Cas9 technology to knockout both copies of the TET3 gene, in equivalent cells, called C17.2Sα, expressing a streptavidin-binding protein (SBP)-tagged TRα1 protein. A cell clone was identified (C17.2SαKO) with frameshift mutations on both alleles (Fig. S4A). The absence of TET3 expression in this clone was confirmed by qRT-PCR (Fig. S4B). A cell clone without TET3 mutation and with a comparable level of TRα1 expression (Fig. S4B) served as a control cell line (C17.2SαC) in the following experiments. TET3 KO led to a decreased induction by T3 of all TR target genes (Fig. 3B). The sensitivity to the KO ranged from high (Epas1, Slc43a2, Tgm2) to low (Klf9, Phospho1, Adcy9). Importantly, TET3 KO severely compromised the level of SBP-TRα1 protein (Fig. 3C, Left), even though more SBP-TRα1 transcript was detected in C17.2SαKO than in C17.2SαC (Fig. S4B). To rule out potential off-target effects of CRISPR/Cas9, we knocked down TET3 in C17.2GSα by siRNA and observed that siRNA-based TET3 knockdown (Fig. S3) also resulted in substantial reduction of GS-TRα1 proteins (Fig. 3C, Right). Consistent with reduced TRα1 proteins after TET3 knockout, chromatin affinity precipitation assay (ChAP) revealed that TRα1 recruitment to several of its TREs is severely impaired in C17.2SαKO (Fig. 3D). The destabilization of TRα1 in C17.2SαKO is most likely the direct consequence of TET3 KO, as TRα1 protein level (Fig. S4C) and T3 induction of target genes (Epas1, Phospho1, and Adcy9) (Fig. S4D) were partially rescued after reintroduction of TET3 by lentiviral infection. The rescue is only partial, as less than 30% of the cells could be transduced by the lentivirus vector (Fig. S4E). These results indicate TET3 plays a critical role in regulating TR transcriptional activity. Furthermore, these results reveal a function for TET3 in modulating TRα1 protein level.
Fig. S3.
SiTET3-mediated knockdown of TET3 expression levels of TET1-2–3 and TRα1 after siTET3-mediated knockdown. RNA were extracted from C17.2GSα cells transfected with siRNA against TET3 (siTET3) or negative control siRNA (siNC). Expression levels of indicated genes were examined by relative qRT-PCR. The expression levels were normalized to the level of 36B4 mRNA, with the amount of TET3 in siNC transfected cells set to 1.
Fig. S4.
TET3 regulates TRα1 activity by modulating TRα1 protein level in C17.2SαKO cells. (A) Scheme of CRISPR strategy and mutation identification. The top line represents the sequence of 21 bp CRISPR guiding RNA used for genomic editing (underlined) to delete TET3 followed by an XGG PAM motif necessary for CRISPR action. Sequences below are the DNA sequencing results of the two alleles of TET3 in TET3 knockout C17.2Sα clone (C17.2SαKO). Two different frame shifts were successfully introduced on the two alleles. (B) Expression level of endogenous TET3 and SBP-TRα1 in control (C17.2SαC) and TET3KO (C17.2SαKO) cells measured by qRT-PCR. RNA were extracted from indicated cells treated or not with T3 (10–8 M), expression levels of SBP-TRα1 and TET3 were examined by relative qRT-PCR. Expression levels were normalized to the level of 36B4 mRNA, with the amount in the C17.2SαC (Veh) set to 1. (C) Effect of reintroduction of TET3 on TRα1 protein level in C17.2Sα cells. Streptavidin beads were used to precipitate SBP tagged TRα1 from indicated cell lines (lentivirus-infected C17.2SαKO cells, C17.2SαKO-R). Western blot for TRα1 was detected by TRα1 antibody. Asterisk indicates band for TRα1. (D) Effect of reintroduction of TET3 on TRα1 target genes expressions in C17.2Sα cells. RNAs were extracted from indicated cells treated or not with T3 (10–8 M), expression levels of indicated TRα1 target genes were examined by relative qRT-PCR. Expression levels were normalized to the level of 36B4 mRNA. Relative induction triggered by T3 in each cell line was presented. The asterisks indicate the significance of the differences between indicated two conditions. (E) Estimation of the efficiency of lentivirus infection in C17.2SαKO by immunofluorescence. C17.2SαKO cells were infected with lentivirus encoding human TET3 and RFP. RFP-positive cells theoretically express human TET3.
TET3 Stabilizes TRs by Inhibiting Their Ubiquitination.
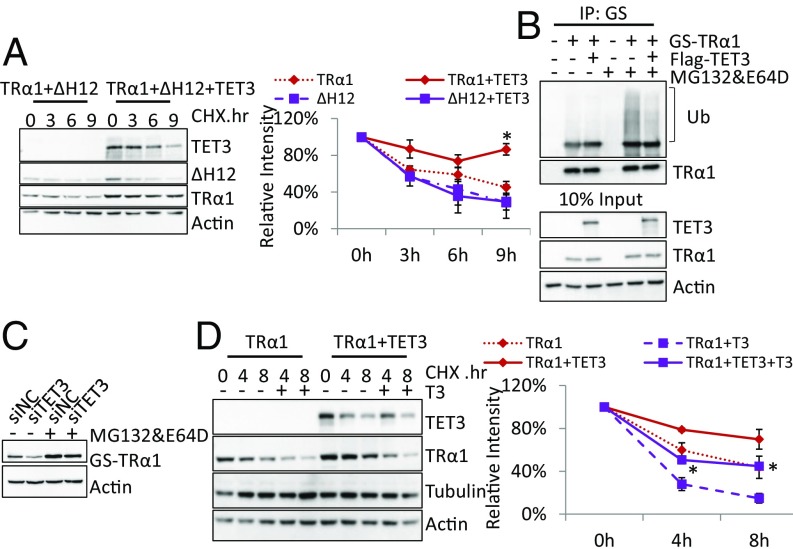
To further evaluate the ability of TET3 to regulate TRα1 protein level, we examined the effect of TET3 on TRα1 protein stability in transfected HEK293T cells by adding cycloheximide (CHX), an inhibitor of protein translation. As expected, the protein level of TRα1 and TRα1ΔH12, a mutant with deletion of helix12, which corresponds to the AF2 domain, quickly decreased over time on addition of CHX. Coexpression of TET3 reduced the degradation of TRα1, but not TRα1ΔH12, with which TET3 cannot interact (Fig. 4A). Similarly, coexpression of TET3 enhanced the stability of TRβ1 (Fig. S1B). TET3mut retains its capacity to stabilize TRα1 protein (Fig. S2C). These results show that TET3 regulates TR protein stability and that this stabilization requires the direct interaction between the two proteins, but not the enzymatic activity of TET3.
Fig. 4.
TET3 stabilizes TRα1 by inhibiting its ubiquitination. (A) Effect of TET3 on protein turnover of wild-type TRα1 (TRα1) and Helix12 deletion mutant (ΔH12). HEK293T cells were cotransfected with Flag-TRα1 (TRα1) and GS-TRα1ΔH12 (ΔH12), with or without Flag-TET3 (TET3). Lysates were prepared from cells treated with CHX for indicated periods; protein levels of TRα1 or ΔH12 were detected by anti-Flag and anti-GS antibodies, respectively (Left). Band intensities of TR corrected by actin signals were plotted (Right). The abundance at each point was calculated relative to the abundance at T0. The asterisk indicates the significance of the differences between TRα1 and TRα1+TET3. (B) TET3 effect on the ubiquitination pattern of TRα1. Whole-cell extracts from HEK293T cells transfected with indicated plasmids and treated or not with MG132&E64D were immunoprecipitated using M280 beads. Half of the immunoprecipitated TRα1 served as loading control, the other half was used to determine the ubiquitination level of TRα1 with an anti-ubiquitin antibody. The smear indicates the poly-ubiquitination of TRα1. (C) Effect of MG132&E64D treatment on TRα1 protein level in C17.2GSα after TET3 knocking down. C17.2GSα were pretreated or not with MG132&E64D before collection. TRα1 was detected in the cell lysates using an anti-GS antibody. Actin serves as a loading control. (D) Effect of TET3 expression on TRα1 protein turnover. HEK293T cells cotransfected with GFP-TRα1 (TRα1) in the presence or not of Flag-TET3 (TET3) were treated with T3 (5.10−8 M) and/or CHX for indicated periods of time. Lysates were subjected to Western blot (Left), protein level of TET3 and TRα1 were respectively detected by anti-Flag and anti-GFP antibody. The intensity of TRα1 signals corrected by tubulin + actin signals were plotted on the right panel as 100% was set for the intensity measured for CHX 0h exposure. The asterisks indicate the significance of the differences between TRα1+T3 and TRα1+TET3+T3.
As TRα1 is degraded via the ubiquitin-mediated proteasome, we then examined whether the overexpression of TET3 could modify the ubiquitination pattern of TRα1. Ubiquitin-dependent degradation can be prevented by a mixture of MG132 and E64D inhibiting, respectively, the proteasome per se and the lysosome-mediated degradation of ubiquitinated proteins that might occur when proteasome is blocked. As expected, this resulted in an accumulation of polyubiquitinated TRα1 in transfected HEK293T cells (Fig. 4B). Importantly, coexpression of TET3 limited the amount of poly-ubiquitinated TRα1 (Fig. 4B), and this effect was independent of TET3 enzymatic activity (Fig. S2D). Furthermore a similar blockade of degradation could prevent the decrease of TRα1 protein level trigged by TET3 knockdown in C17.2GSα (Fig. 4C). Similarly, TET3 also inhibited TRβ1 ubiquitination (Fig. S1C). Altogether, these results suggest TET3 protects TRs from degradation by limiting its polyubiquitination; this effect is independent of TET3 enzymatic activity.
Because T3 attenuates TET3/TRα1 interaction, as demonstrated by coimmunoprecipitation, we tested whether TET3 stabilizes TRα1 in the presence of T3. In agreement with published results (5), we observed that T3 accelerates the degradation of TRα1 in transfected HEK293T cells (Fig. 4D). In this system, however, TET3 extended the half-life of TRα1 in both the absence and the presence of T3 (Fig. 4D), implying TET3 stabilizes TR even in the presence of T3.
TET3 Stabilizes TRs in the Chromatin Fraction.
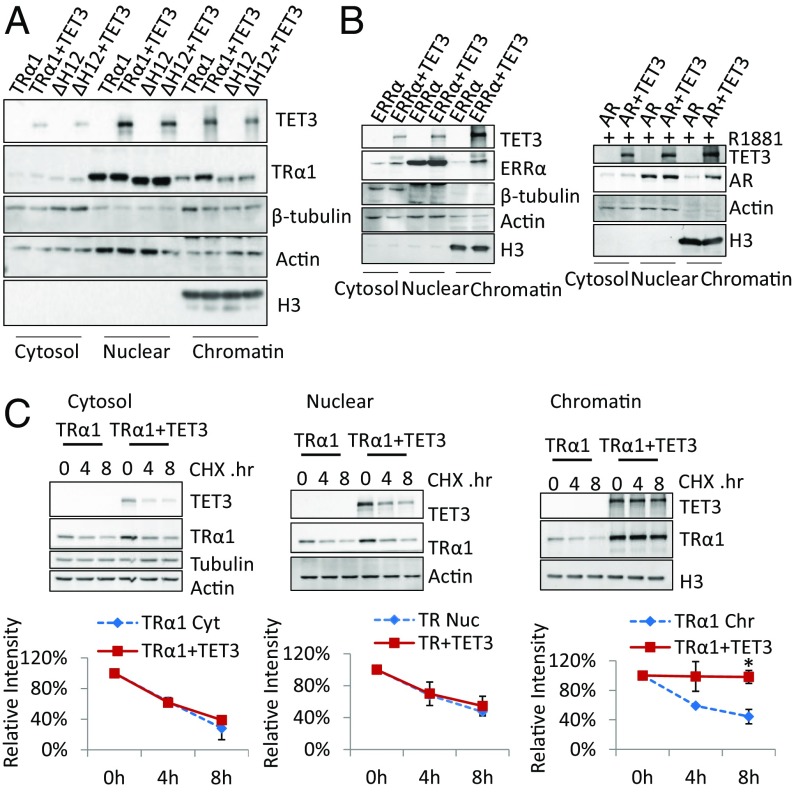
We next evaluated the possibility that TET3 may influence TRα1 subcellular localization and/or chromatin association. Immunofluorescent staining showed that TET3 and TRα1 are both nuclear proteins, and their coexpression has no obvious effect on nuclear localization (Fig. S5). Biochemical fractionation of transfected HEK293T cells confirmed that TRα1 is mainly recovered in the nucleus, but only a subfraction is chromatin associated (Fig. 5A). Coexpression of TET3 substantially increased the chromatin fraction of TRα1, but not cytosol and nuclear TRα1 (Fig. 5A). This effect requires direct interaction, as increased chromatin fraction was not observed for TRα1ΔH12 (Fig. 5A). In addition, TET3N, a truncated form of TET3 that does not interact with TRα1, could not promote TRα1 enrichment in the chromatin (Fig. S2E). In contrast, TET3mut enhanced TRα1 chromatin enrichment to the same extent as the wild-type TET3 (Fig. S2E). Similarly, both ERRα and AR that interact with TET3 showed an increased presence on chromatin with TET3 coexpression (Fig. 5B). Strikingly, even though TRα1 and TET3 were strongly coexpressed in both nucleus and chromatin, the stabilization effect measured after CHX treatment was only observed in the chromatin fraction (Fig. 5C). These results argue for a stabilization of TRα1 on the chromatin via its interaction with TET3. Similarly, TET3 also substantially enhanced TRβ1 chromatin association (Fig. S1D). Thus, TET3 has a marked effect in enhancing TR chromatin association and protecting chromatin-associated TRs from ubiquitination-mediated degradation.
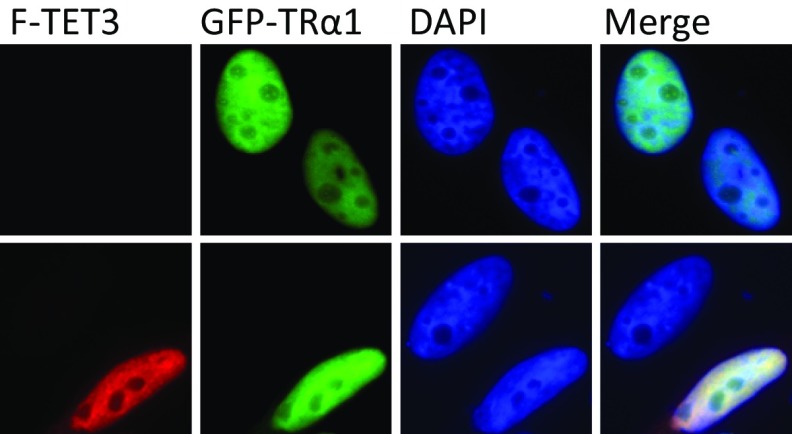
Fig. S5.
Estimation of the influence of TET3 expression on TRα1 subcellular localization by immunofluorescence HeLa cells were transfected with GFP-TRα1 with or without Flag-TET3 (F-TET3). Expression of TET3 was revealed with the Flag antibody.
Fig. 5.
TET3 increases and stabilizes TRα1 chromatin association. (A) Effect of TET3 expression on the subcellular distribution of TRα1 or TRα1ΔH12. GS-TRα1 (TRα1) or GS-TRα1ΔH12 (ΔH12) were transfected with or without Flag-TET3 (TET3) in HEK293T cells. Anti-Flag and anti-GS antibodies were respectively used to detect TET3 and TRα1 in the different compartments after cell fractionation. β-tubulin, actin, and H3 were respectively the loading controls for the cytosol, nucleus, and chromatin. (B) Effect of TET3 expression on the subcellular distribution of ERRα or AR. Same fractionation experiment as in A. (C) Effect of TET3 expression on TRα1 protein in the different cell fractions. Same fractionation experiment as in A was performed for HEK293T cells with CHX treatment for indicated periods of time. Protein levels of TET3 and TRα1 were respectively detected by anti-Flag and anti-GS antibody. The intensity of TRα1 signals corrected by tubulin + actin/actin/H3 signals were plotted on the lower panels as 100% was set for the intensity measured for CHX 0h exposure. The asterisk indicates the significance of the differences between the two conditions.
The Potential Role of TET3 in Modulating the Dominant-Negative Effect of TR Mutants.
One situation in which TET3/TR interaction may have significant consequences is in patients with RTHα/β. The missense or frameshift mutations found in these patients, often located in the AF2 domain of TR, confer dominant-negative properties toward the wild-type receptor. This can be evidenced in transient expression assays, where the coexpression of mutant and WT TRα1 results in impaired transactivation capacity, mimicking the situation found in cells of heterozygous patients (10).The dominant-negative effect varies with the type of mutation. The mechanisms responsible for wide spectrum of dominant-negative action are not entirely clear, and probably involve mutant protein stability and balance between corepressor and coactivator interactions. Because TET3 interacts with and stabilizes TR in a helix12-dependent manner, we tested the possibility that the interaction of the mutant receptors with TET3 could determine the stoichiometry between mutant and WT receptors, and thus influence the dominant-negative activity of the mutant receptors, and consequently the disease severity.
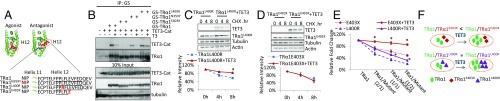
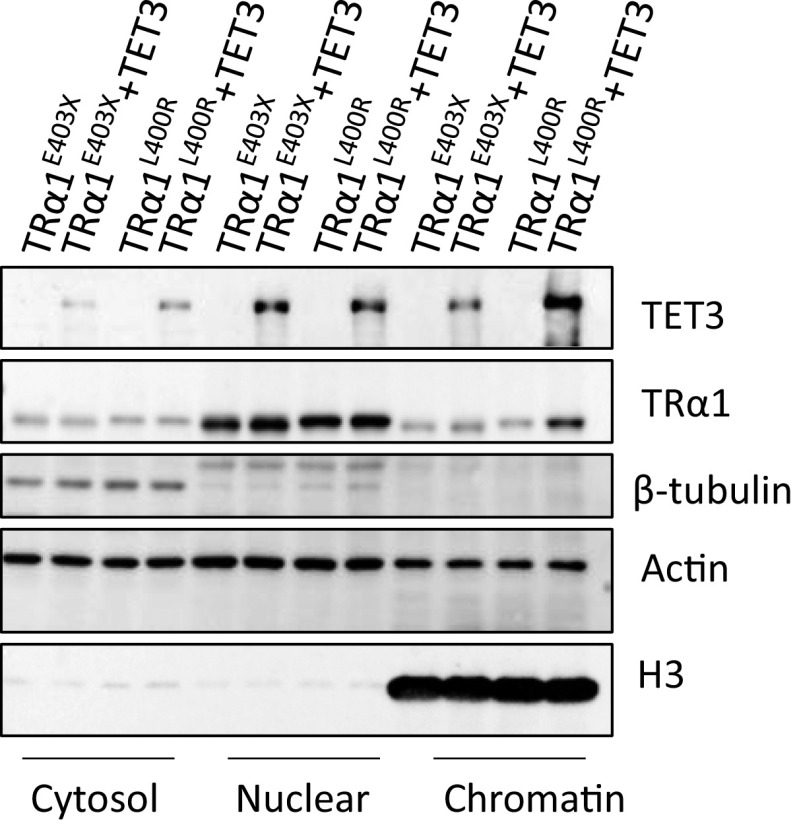
We used here a panel of natural and artificial mutations altering helix12 (Fig. 6A) and assess both the influence of the mutation on TET3 interaction and dominant-negative property. TRα1E403X (11) and TRα1N359Y (12) have been found in two patients, and TRα1L400R is lethal in a mouse knock-in model (13). As expected TRαN359Y and TRα1L400R, but not TRα1E403X, interacted with TET3-Cat (Fig. 6B) because helix12 is required for TET3/TRα1 interaction. Aa a consequence, TET3 stabilized TRα1L400R (Fig. 6C), but not TRα1E403X (Fig. 6D), and increased the presence of TRα1L400R, but not TRα1E403X, on chromatin (Fig. S6). Transfections were performed to test whether the ability to interact with, and thus be stabilized by, TET3 of TRα1L400R, but not TRα1E403X, affect their dominant-negative activity. As expected, increasing the mutant/WT receptor ratio decreased TR activity for both TRα1L400R and TRα1E403X (Fig. 6E). However, coexpression of a fixed amount of TET3 strongly attenuates the dominant-negative potential of TRα1E403X, but not TRα1L400R, as illustrated by the changes in the slopes (Fig. 6E). A simple explanation would be that TET3 stabilizes TRα1 and TRα1L400R, but not TRα1E403X, and thus influences the stoichiometry and the capacity of the cells to respond to T3, as illustrated on the scheme (Fig. 6F).
Fig. 6.
Role of TET3 mediated stabilization of TRα1 on the dominant-negative effect of TRα1 mutants. (A) Schematic of different TRα1 mutants. TRα1N359Y has a mutation before helix11 and an intact helix12; TRα1L400R has a point mutation in helix12, and TRα1E403X has a truncated helix12. (B) Identification of the interaction between TET3 and TRα1 mutants. Flag-TET3Cat (TET3-Cat) and GS-tagged mutants of TRα1 (TRα1) were transfected in HEK293T cells treated or not with T3 (5.10−8M), TRα1 mutants were immunoprecipitated with M280 beads, coprecipitated TET3-Cat was detected with anti-Flag antibody. (C and D) Identification of differential stabilization of TRα1L400R (C) and TRα1E403X (D) by TET3. HEK293T cells cotransfected with TRα1L400R or TRα1E403X and Flag-TET3 (TET3) were treated with CHX for indicated periods of time. Protein levels of TET3 or TRα1 mutants were detected by anti-Flag and anti-GS antibodies respectively (upper panels). The intensity of TR mutant signals corrected by tubulin + actin signals were plotted on the lower panels. 100% was set for the intensity measured for CHX 0h exposure. The asterisk indicates the significance of the differences between the two conditions. (E) TET3 modulation of the dominant-negative effects of TRα1L400R or TRα1E403X. HEK293T cells were transfected with luciferase reporter, TRα1, varying ratios of TRα1/ΤRα1 mutants (2/1; 1/1; 1/2) and TET3. Luciferase activities were measured 24 h after T3 (10−8M) treatment. The relative fold induction upon T3 treatment (taking the fold change of transfecting TRα1 alone as 100%) was plotted on the graph. Each transfection condition was performed as triplicates, error bars of the three independent experiments were indicated in the graph. The asterisks indicate the significance of the differences between TRα1 mutant and TRα1 mutant+TET3. (F) Working model for TET3 modulation of the dominant-negative effects of TRα1 mutants. The ratio of TRα1/ TRα1E403X and TRα1/ TRα1L400R changes differently after addition of TET3, thus the cell responsiveness to T3 treatment is differently affected by TET3 between the two mutants.
Fig. S6.
TET3 facilitates chromatin recruitment of TRα1L400R, but not TRα1E403X, Effect of TET3 on the subcellular distribution of TRα1E403X or TRα1L400R. GS-TRα1E403X (TRα1E403X) or GS-TRα1L400R (TRα1L400R) were cotransfected with or without Flag-TET3 (TET3) in HEK293T cells, cells were fractionated using NE-PER nuclear and cytoplasmic extraction reagents kit. Cytosol, nucleus, and chromatin fractions were subjected to Western blotting. TET3 and TRα1 were detected by anti-Flag and anti-GS antibodies, respectively. β-tubulin, actin, and H3 were, respectively, the loading controls for the cytosol, nucleus, and chromatin.
Discussion
In this study, we demonstrate that TET3 proteins can interact with four nuclear receptors: TRα1, TRβ1, AR, and ERRα. Focusing on TET3/TRs interaction, we found that the presence of TET3 has three consequences: it increases the half-life of TRs by reducing ubiquitination and degradation, it stabilizes TRs presence on chromatin, and it increases TRα1 capacity to mediate transcriptional activation on ligand binding. These three effects do not rely on the catalytic activity of TET3, thus revealing a DNA demethylation-independent function for TET3, as well as a mode of regulation for TRs. In addition, we observed that TET1 and TET2 also interact with TR, even though interactions with them are weaker than with TET3. However, given the sequence similarities, TET1 and TET2 might also modulate TR function in a similar manner. Proper experiments are needed to ascertain this hypothesis. The interaction with additional nuclear receptors such as AR and ERRα also suggests TET3, and potentially TET1 and TET2, may have a broad regulatory role in the function of nuclear receptors.
Although the initial in vitro pulldown assay suggests a weak protein–protein interaction between TR and TET3 (Fig. 1A), subsequent coimmunoprecipitation (co-IP) experiments reveal an interaction that is comparable to or even stronger than that with the classical coactivator SRC3 (Figs. 1B and 2C). The observed robust interaction may be explained by the presence of multiple TR interaction regions in TET3 regions (CXXC, CatN, and CatC) that can interact with TRα1 independently in co-IP assay (Fig. 2). It is noteworthy that TET3 stabilizes and enhances TRα1 chromatin association in a TET3-TRα1 interaction-dependent manner (Figs. 4 and 5). Thus, the striking reduction of TRα1 protein, but not transcript levels, on TET3 knockdown or knockout (Fig. 3C) nicely manifests the physiological relevance and function significance of this interaction. In support of this notion, TET3 KO in C17.2 markedly impairs the binding of TRα1 to three previously described TRE (9) in the Epas1, Klf9, and Dbp promoters (Fig. 3D). We noted that the interaction between TET3 and TR is ligand-independent in vitro (Fig. 1A) and reduced in T3-treated cells (Fig. 1 B and C and Fig. S1A). The reduced interaction observed in T3-treated cells is likely a consequence of competition from other proteins (coactivators) that are able to interact with TR in a T3-dependent manner and displace TET3. Nevertheless, even weaker interaction is likely functionally relevant, as TET3 stabilizes TR even in presence of T3. This T3-dependent competition may also allow a switch of TR interacting partners from TET3 to coactivators, activation of the target genes, and recycling of the receptor via its degradation.
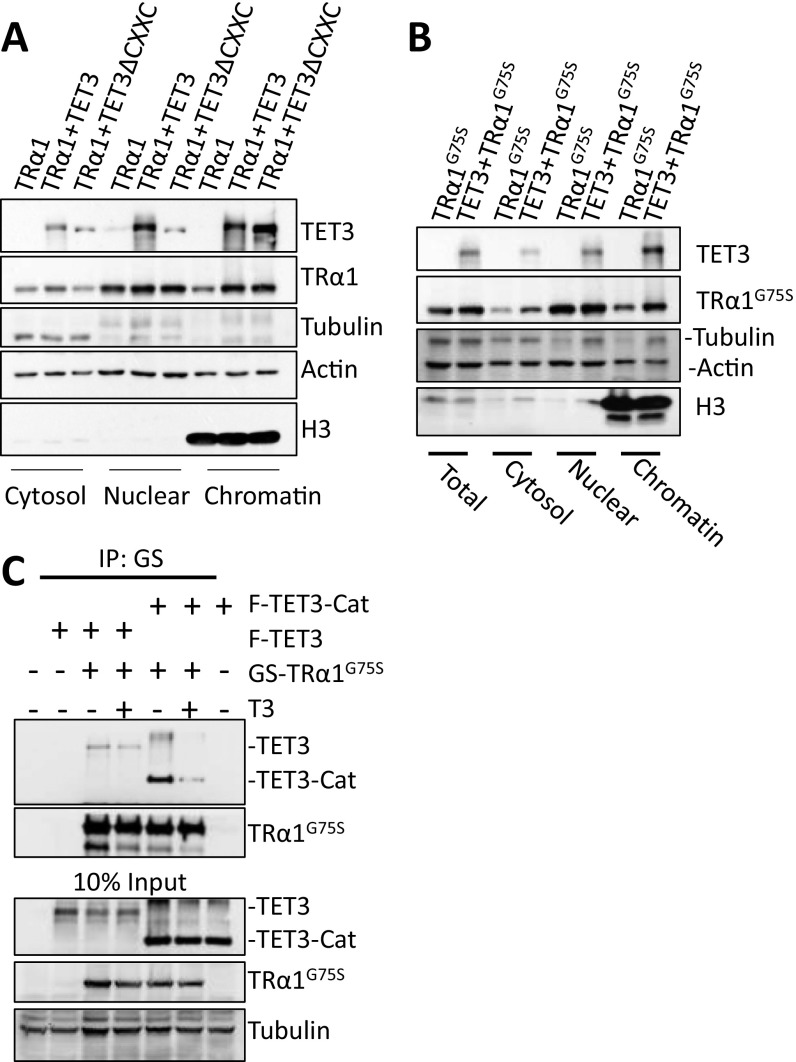
A surprising finding in our study is that the stabilization of TR by TET3 is limited to the chromatin compartment (Fig. 5), even though TET3 and TR interact (co-IP) and colocalize in the soluble fraction of the nucleus. At this stage, the underlying mechanism is not known, but presumably involves enhanced recruitment of TR to chromatin by TET3 and/or protection of chromatin-associated TR from ubiquitination and subsequent degradation. Our ChAP assay clearly demonstrated that TET3 is required for efficient binding of TRα1 to TRE in TR target genes (Fig. 3D), although it remains to be determined whether TET3 enhances TRα1-specific enrichment at TRE and/or other genomic sites. In an effort to decipher the underlying mechanism, we demonstrated that the CXXC domain, which mediates TET3 direct binding of genomic DNA (14), is dispensable for stabilization of TRα1 recruitment in chromatin (Fig. S7A). In addition, we demonstrated that TET3 stabilized and promoted chromatin association of a TRα1 mutant (TRα1G75S) defective in DNA binding (15) as a result of a mutation in DNA binding domain (Fig. S7B). This TR mutant, as expected, maintained an interaction with TET3 (Fig. S7C). Thus, our data indicate that stabilization of chromatin-associated TRα1 by TET3 depends neither on TET3’s nor on TRα1’s DNA-binding activity, but on the interaction between TET3 and TR. Future work is needed to elucidate the detailed mechanism by which TET3 selectively stabilizes chromatin-associated TR.
Fig. S7.
Modulation effect of TET3 on TRα1 is independent of DNA binding abilities of both TRα1 and TET3. (A) Effect of TET3/TET3ΔCXXC on the subcellular distribution of TRα1. HEK293T cells were cotransfected with indicated plasmids. After cell fractionation cytosol, nucleus and chromatin fractions were subjected to Western blotting. TET3/TET3ΔCXXC or TRα1 were, respectively, detected by anti-Flag and anti-GS antibody. β-tubulin, actin, and H3 were, respectively, the loading controls for the cytosol, nucleus, and chromatin. (B) Effect of TET3 on the subcellular distribution of TRα1G75S. Same cell fractionations were performed as in (A) for cells transfected with indicated plasmids. TET3 and TRα1G75S were respectively detected by anti-Flag and anti-GS antibody. (C) Interaction between TET3 and TRα1G75S. Whole-cell extracts from HEK293T cells cotransfected with indicated plasmids, treated or not with T3 (5.10–8 M) before collection, were used for Immunoprecipitation using M280 beads that retain GS tag. Coprecipitated TET3-Cat or TET3 were detected using an anti-Flag antibody.
In the present study, we reveal a role of TET3 on transcriptional regulation by nuclear receptors that does not rely on its catalytic activity. As anticipated from their hydroxymethylase activity, TETs can modulate transcription by adjusting levels of DNA methylation at promoters. Accordingly, both TET1 and 5hmC often localize to transcriptional start sites (16, 17). With regard to nuclear receptors, it was reported previously that peroxisome proliferator-activated receptor-γ has the ability to direct local demethylation around its binding sites via recruitment of TET1 through peroxisome proliferator-activated receptor-γ-induced PARylation (18). In addition, TET3 up-regulation was shown to be responsible for glucocorticoid receptor-induced DNA hypomethylation in neural stem cells (19). TET proteins have also been reported to regulate transcription via interacting proteins such as mSin3A (16), MBD3/NuRD complex (20), polycomb repressive complex PRC2 (21), and the O-linked N-acetylglucosamine transferase (22). However, to our knowledge, TET protein has not been described to specifically stabilize a chromatin-associated protein, and in so doing enhance its transcriptional function. As the stability and chromatin association of wild-type and TR mutants can be differentially affected, depending on the presence or absence of their TET3 interaction (Fig. 6), we also provide proof of principle that TET3 may potentially modulate the clinical outcome in patients resistant to T3, as a result of TR mutation.
In sum, in this study we uncover a TET3 catalytic activity-independent mechanism for enhancing TR function. The mechanism involved (i.e., stabilization of TR on chromatin) is also very different from the one classically described for nuclear receptor coactivators. By interacting with and stabilizing TR binding to chromatin, TET3 protects it from ubiquitination and proteasome degradation and favors the activation of gene expression in the presence of T3. The presence of TET3 would thus increase the cellular sensitivity to T3 stimulation. This role of TET3 may not be limited to TR. TET3 may regulate the hormone sensitivity of the cell to a host of different nuclear receptors, as the AF2 domain involved in the interaction is well conserved in this family of transcription factors and TET3 has been observed to interact with and promote the stabilization of AR and ERRα in chromatin.
Methods
Plasmids, Antibodies, and Drugs.
Plasmids encoding TET1, TET2, TET3 (23), and TRα1/TRβ1 (9) were previously described. TET3/TRα1 mutants were generated by PCR and are described as antibodies and drugs used in SI Methods.
Immunoprecipitation.
Immunoprecipitations were carried out as described (23). Magnetic M2 (Sigma), magnetic M280 (Dynabeads M-280, Invitrogen; used to retain the GS tag), or Streptavidin beads (Agilent Technologies) were used when indicated.
RNA Interference and CRISPR/Cas9 in C17.2 Cell Lines.
Knocking down and knocking out TET3 in C17.2 lines stably expressing TRα1 were respectively obtained by siRNA and CRISPR/Cas9 technology. Detailed information is provided in SI Methods.
RNA Extraction, qPCR Measurements, and ChAP.
Protocols were described before (9). The sequences of the primers used are provided in Table S1.
Table S1.
Primers for qRT-PCR or ChAP
| Genes | Forward | Reverse |
| Primers for qRT-PCR | ||
| Epas1 | TGGACATCCCCCTGGACAGCAA | GGTCATGTTCTCCGAATCCAGGGCA |
| Klf9 | ACATCGGGGAGAATGGGTGGGA | TTGTCCAACGAGCGCCAGACAC |
| Tgm2 | GTCCCTGAAGAACCCACTTT | TTGTGGAGGCCAATATCAGT |
| Slc43a2 | CTCCACACAGTTTGGTAGCC | CCGGTAGCAGATGAGGTAAA |
| Adcy9 | CCCTGGCTGTCTATTCACAC | GCTGAGCTGGCAGAAGTTAC |
| Slc12a5 | GAAGAATAAAGGCCCCAGTC | CCCGAGATTTATTCACGATG |
| Por | GGAGATCACGTGGCTGTGTA | GGGGAACGGATGCTTCTTAT |
| Phospho1 | TGACAGCTGGAGACCAACAG | CATCCTGCCGTCCCTAGATA |
| 36B4 | ACCTCCTTCTTCCAGGCTT | CCCACCTTGTCTCCAGTCTTT |
| SBPTRα1 | CTAGCGGCCATCAAACAAGT | CTGTTCTCCTCTGGGTCTGA |
| TET1 | GCTGGATTGAAGGAACAGGA | GTCTCCATGAGCTCCCTGAC |
| TET2 | GTCAACAGGACATGATCCAGGAG | CCTGTTCCATCAGGCTTGCT |
| TET3 | ACTCATGGAGGATCGGTATG | GCTTCTCCTCCAGTGTGTGT |
| Primers for ChAP | ||
| Epas1 TRE | GTCACCCCGGGGCTCAAGGA | CACCAGCCGAATGTGACAGGCA |
| Epas1 ctrl | ACAGTCTATGGGCCTGCAGCGA | TTAGGGAGCCAGCTGCCAGTCA |
| Klf9 TRE | TGCACGAGTTTGGGGCGGATTC | TGGGCCTGGCATCGCCCTTTTA |
| Klf9 ctrl | CACGGGAAAGGCTGGGTTGTGA | TTACTGTCTCTACCTCTGGGCCTGC |
| Dbp TRE | TCGGCTGACCCGGGCTTATTTG | TTTGTTCCCACTGCTGTGTCCGAG |
| Dbp Ctrl | ACGGGAGGGATGTGCAACCAGA | TAGGACTCACGTTGGCAGGGCA |
Cell Fractionation.
Nuclear-cytoplasmic fractionation was performed using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Cell Culture and Transient Transfection Assays.
C17.2 cells, human HEK293T, and HeLa cells were cultured in recommended medium. TransIT-LT1 (Mirus) was used for transfection according to the manufacturer’s instructions. Luciferase assay was carried out as described (24).
SI Methods
Plasmids, Antibodies, and Drugs.
Monoclonal anti-FLAG M2-Peroxidase (HRP) (A8592, Sigma), monoclonal anti–β-Actin (A5316, Sigma), anti-GFP (ab290, Abcam), anti–β-tubulin (ab6046, Abcam), anti-TRα1/β1 (SC-739, SantaCruz), anti-Histone3 (ab1791, Abcam), anti-Myc (ab9106, Abcam) and anti-Ubiquitin (Z0458, Dako and VU101, Life Sensor) were used for Western blot or coimmunoprecipitation. Chemicals used in this study include: 3,3′,5-Triiodo-l-thyronine sodium salt (T3, sigma), CHX (Calbiochem), MG132 (Calbiochem), E64D (Enzo Life Sciences). When indicated, 5.10−8 M or 10−8 M T3, 80 μg/mL CHX, 5.10−6 M MG132, and 2.10−5 M E64D were used. The TET3 (H1077Y/D1079A) mutant, TET3-DSBH2 (V1692A/F1693A) mutant, pCEMM-GS-TRα1L400R and pCEMM-GS-TRα1E403X were created by PCR-directed mutagenesis.
TET3 KD by siRNA, TET3 KO by CRISPR/Cas9, and TET3 Rescue by Lentiviral Vector in C17.2 Cell Line.
Small interference RNAs (siRNA) against TET3 were purchased from Dharmacon (siGenome TET3 siRNA smart pool and nontargeting pool) and were transfected using lipo3000 (Invitrogen) according to the manufacturer’s instructions. RNA interferences were performed in C17.2GSα cells that express in a stable manner a murine TRα1 with a double GS N-terminal tag (protein G fragment followed by streptavidin binding peptide).
The pCEMM-STRα1 vector encodes a SBP (streptavidin binding peptide) tagged murine TRα1 and downstream IRES-GFP cassette that ensures coexpression of GFP from the same CMV transcription promoter. pX459-TET3 guide RNA vector contains a single guide RNA against TET3, a cas9 protein coding sequence, and a puromycin resistance gene. The guide RNA against TET3 was designed as described before, and no off-target was identified (25, 26). The two constructs were transfected into C17.2 cells. Puromycin-resistant cells (1 μg/mL) were FACS sorted to select cells expressing both GFP and SBP-TRα1. Sorted cells were cloned and amplified; genotypes were determined by sequencing. One clone with frameshift mutations for TET3 on both alleles was chosen (C17.2SαKO) for further analyses. A clone with comparable TR level (C17.2Sα), issued from a transfection with a PX459 empty vector and pCEMM-STRα1, was used as a control (C17.2SαC).
Lentiviral particles for hTET3 over expression (amsbio; LVP876) were used to rescue TET3 expression in C17.2SαKO cells. A blasticidin-RFP fusion dual marker was expressed under the same promoter. Blasticidin-resistant (1.5 μg/mL) infected cells (C17.2Sα-R) were used for further experiments.
Immunofluorescence Staining.
For immunofluorescence staining of 5hmC, HeLa cells were washed with PBS before fixation in 4% fresh paraformaldehyde in PBS for 15 min. Then cells were treated 30 min at 37 °C 2N HCl for 20 min at room temperature with 100 mM Tris⋅HCl (pH 8.0) for neutralization and blocked with 5% BSA in PBS for 1 h at 37 °C. 5hmC primary antibody was incubated overnight at 4 °C, and secondary antibody at room temperature for 1 h. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). For immunofluorescence staining in C17.2 cells, C17.2Sα or C17.2SαKO cells were washed with PBS before fixation in 4% fresh paraformaldehyde in PBS for 15 min. Fixed cells were treated with 1% triton-100 for 10 min, followed by DAPI staining for 15 min at room temperature. Images were acquired using Zeiss Inverted fluorescence microscope Axioobserver 7.
Data Analysis.
Data are presented as mean of 2 or 3 independent experiments. Comparisons between 2 groups were performed using an unpaired Student’s t test. Statistical relevance was determined using the one-way variable ANOVA method for QRT-PCR, luciferase assay, and band intensity quantifications. The error bars represent ± SD. Differences of P < 0.05 were considered significant. *0.005 < P < 0.05; **0.0005 < P < 0.005.
Acknowledgments
We thank Q. Zhang for construction of full-length TET expression vectors; S. Richard, P. Godement, B. Py, and W. Bourguet for their technical help and advice; and C. Couturier and S. Dussurgey from Structure Fédérative de Recherche Biosciences (UMS3444/CNRS, US8/Inserm, Ecole Normale Supérieure de Lyon, Université Claude Bernard Lyon) for FACS. The work is supported by the Ministry of Science and Technology of China (Grant 2015CB910402 to J.W.), the National Natural Science Foundation of China (Grants 81530078 and 31571325 to J.W.), the Science and Technology Commission of Shanghai Municipality (Grants 14XD1401700 and 11DZ2260300 to J.W.), the French Agence Nationale de la Recherche (Thyromut2; Grant ANR-15-CE14-0011-01 to F.F.), and region Rhône-Auvergne (Mobilité Internationale Rhône-Alpes to K.C.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702192114/-/DCSupplemental.
References
- 1.Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 2.Moran C, et al. Contrasting phenotypes in resistance to thyroid hormone α correlate with divergent properties of thyroid hormone receptor α1 mutant proteins. Thyroid. May 4, 2017 doi: 10.1089/thy.2017.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortiga-Carvalho TM, Sidhaye AR, Wondisford FE. Thyroid hormone receptors and resistance to thyroid hormone disorders. Nat Rev Endocrinol. 2014;10:582–591. doi: 10.1038/nrendo.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dace A, et al. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. Proc Natl Acad Sci USA. 2000;97:8985–8990. doi: 10.1073/pnas.160257997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato S, Yokoyama A, Fujiki R. Nuclear receptor coregulators merge transcriptional coregulation with epigenetic regulation. Trends Biochem Sci. 2011;36:272–281. doi: 10.1016/j.tibs.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Chatonnet F, Guyot R, Benoît G, Flamant F. Genome-wide analysis of thyroid hormone receptors shared and specific functions in neural cells. Proc Natl Acad Sci USA. 2013;110:E766–E775. doi: 10.1073/pnas.1210626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran C, Chatterjee K. Resistance to thyroid hormone due to defective thyroid receptor alpha. Best Pract Res Clin Endocrinol Metab. 2015;29:647–657. doi: 10.1016/j.beem.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochukova E, et al. A mutation in the thyroid hormone receptor alpha gene. N Engl J Med. 2012;366:243–249. doi: 10.1056/NEJMoa1110296. [DOI] [PubMed] [Google Scholar]
- 12.Espiard S, et al. A novel mutation in THRA gene associated with an atypical phenotype of resistance to thyroid hormone. J Clin Endocrinol Metab. 2015;100:2841–2848. doi: 10.1210/jc.2015-1120. [DOI] [PubMed] [Google Scholar]
- 13.Quignodon L, Vincent S, Winter H, Samarut J, Flamant F. A point mutation in the activation function 2 domain of thyroid hormone receptor α1 expressed after CRE-mediated recombination partially recapitulates hypothyroidism. Mol Endocrinol. 2007;21:2350–2360. doi: 10.1210/me.2007-0176. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibusawa N, Hollenberg AN, Wondisford FE. Thyroid hormone receptor DNA binding is required for both positive and negative gene regulation. J Biol Chem. 2003;278:732–8. doi: 10.1074/jbc.M207264200. [DOI] [PubMed] [Google Scholar]
- 16.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011;10:2428–2436. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiki K, et al. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat Commun. 2013;4:2262. doi: 10.1038/ncomms3262. [DOI] [PubMed] [Google Scholar]
- 19.Bose R, et al. Tet3 mediates stable glucocorticoid-induced alterations in DNA methylation and Dnmt3a/Dkk1 expression in neural progenitors. Cell Death Dis. 2015;6:e1793. doi: 10.1038/cddis.2015.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q, et al. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT) J Biol Chem. 2014;289:5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauthier K, et al. Thyroid hormone receptor beta (TRbeta) and liver X receptor (LXR) regulate carbohydrate-response element-binding protein (ChREBP) expression in a tissue-selective manner. J Biol Chem. 2010;285:28156–28163. doi: 10.1074/jbc.M110.146241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang Y. Regulation of TET protein stability by calpains. Cell Reports. 2014;6:278–284. doi: 10.1016/j.celrep.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]