Significance
Mutations are the raw material for evolution. However, complex evolutionary dynamics make it challenging to identify which mutations drive adaptation. During adaptation in asexual populations, multiple mutations move synchronously through the population as mutational cohorts. Here we quantify the fitness effect of 116 mutations from 11 laboratory-evolved yeast populations. We show that only a fraction of genome evolution is strongly adaptive. We map driver and hitchhiker mutations to 31 mutational cohorts, and we identify 1 cohort in which mutations combine to provide a fitness benefit greater than the sum of their individual effects. Our analysis uncovers the roles of genetic hitchhiking and epistasis in determining which mutations ultimately succeed or fail in the context of a rapidly evolving microbial population.
Keywords: experimental evolution, cohorts, fitness, epistasis
Abstract
Beneficial mutations are the driving force of adaptive evolution. In asexual populations, the identification of beneficial alleles is confounded by the presence of genetically linked hitchhiker mutations. Parallel evolution experiments enable the recognition of common targets of selection; yet these targets are inherently enriched for genes of large target size and mutations of large effect. A comprehensive study of individual mutations is necessary to create a realistic picture of the evolutionarily significant spectrum of beneficial mutations. Here we use a bulk-segregant approach to identify the beneficial mutations across 11 lineages of experimentally evolved yeast populations. We report that nearly 80% of detected mutations have no discernible effects on fitness and less than 1% are deleterious. We determine the distribution of driver and hitchhiker mutations in 31 mutational cohorts, groups of mutations that arise synchronously from low frequency and track tightly with one another. Surprisingly, we find that one-third of cohorts lack identifiable driver mutations. In addition, we identify intracohort synergistic epistasis between alleles of hsl7 and kel1, which arose together in a low-frequency lineage.
Adaptation is a fundamental biological process. The identification and characterization of the genetic mechanisms underlying adaptive evolution remains a central challenge in biology. To identify beneficial mutations, recent studies have characterized thousands of first-step mutations and systematic deletion and amplification mutations in the yeast genome (1, 2). These unbiased screens provide a wealth of information regarding the spectrum of beneficial mutations, their fitness effects, and the biological processes under selection. However, this information alone cannot predict which mutations will ultimately succeed in an evolutionary context as genetic interactions and population dynamics also impart substantial influence on the adaptive outcomes.
Early theoretical models assume that beneficial mutations are rare, such that once a beneficial mutation escapes drift, it will fix (3–5). For most microbial populations, however, multiple beneficial mutations will arise and spread simultaneously, leading to complex dynamics of clonal interference and genetic hitchhiking (6–9), and in many cases, multiple mutations track tightly with one another through time as mutational cohorts (10–13). The fate of each mutation is therefore dependent not only on its own fitness effect, but on the fitness effects of and interactions between all mutations in the population. Many beneficial mutations will be lost due to drift and clonal interference, whereas many neutral (and even deleterious) mutations will fix by hitchhiking. The influence of clonal interference and genetic hitchhiking on the success of mutations makes it difficult to identify beneficial mutations from sequenced clones or population samples. The extent of genetic hitchhiking and its evolutionary significance has been investigated theoretically, but few empirical studies have contributed to our understanding of the phenomenon (14–17). Identifying beneficial mutations and quantifying their fitness effects ultimately requires assaying each individual mutation independent of coevolved mutations.
Here we present a comprehensive, large-scale survey that quantifies the fitness effects of 116 mutations from 11 evolved lineages for which high-resolution knowledge of the dynamics of genome sequence evolution is known. We describe a strategy for constructing bulk-segregant pools that enables high-throughput quantification of fitness effects of individual evolved alleles. We find that large-effect mutations in common targets of selection drive adaptation, whereas deleterious alleles rarely reach appreciable frequencies. We report that nearly 80% of genome sequence evolution is either neutral or has a fitness effect less than 0.5%, well below the median effect of 3.3% for demonstrable driver mutations. Furthermore, one-third of cohorts lack identifiable driver mutations, and the dynamics of these “driverless” cohorts can be explained by genetic hitchhiking alone. Through the extensive characterization of evolved mutations, we begin to explore the mechanisms responsible for the observed cohort dynamics and we identify one cohort that suggests that rare mutations and epistatic interactions represent evolutionarily relevant genetic mechanisms of adaptation.
Results
Fitness of 11 Representative Lineages from Nine Evolved Populations.
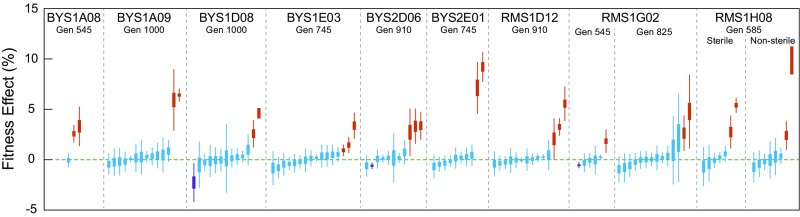
Previously we evolved nearly 600 haploid populations for 1,000 generations in a rich glucose medium. The populations were derived from two ancestral strains that were isogenic other than a single SNP in GPA1 that modifies the fitness effect of mutations in the mating pathway (8, 18). The populations were also divided by population size, which was controlled through dilution frequency and bottleneck size. We selected clones from populations for which we previously followed the frequencies of all mutations at high temporal resolution, selecting only those evolved at the smaller population size (Ne ∼105) to maintain uniformity across our experiments in this study. Because the number and identity of mutations in each population is known, we sampled across the range of biological pathways under selection (i.e., mating pathway, Ras pathway, cell wall assembly/biogenesis) and captured clones that exhibit a range of genome divergence, from 3 mutations in BYS1A08 to 16 mutations in BYS1E03. In two instances (RMS1G02 and RMS1H08), clones were isolated from two coexisting subpopulations to investigate how particular mutations and their fitness effects impact the ultimate fate of competing lineages. We identify clones by their population and the timepoint from which they were isolated (e.g., BYS1A08-545). The evolutionary dynamics corresponding to each of the 11 selected clones are shown in their entirety in SI Appendix, Fig. S1. We measured the fitness of each clone against its fluorescently labeled ancestor. Fitness values ranged from 2.7% in RMS1G02-545 to 8.6% in RMS1D12-910 (SI Appendix, Fig. S1 and Dataset S1).
A Strategy for Constructing Bulk-Segregant Pools.
We developed a bulk-segregant approach to rapidly and efficiently generate large pools of haploid segregants that contain random combinations of evolved mutations (SI Appendix, Fig. S2). We backcrossed evolved MATa clones to a MATα version of the ancestor, gene converted the mating-type locus, and sporulated the resulting MATa/a diploid by complementing the MATα2 gene on a plasmid. Our method has three key advantages over the commonly used yeast “magic-marker” approach, which selects on auxotrophic markers driven by mating type-specific promoters (19). First, our strategy ensures that the segregant pool is strictly MATa through the removal of the MATα gene during pool construction. Second, the approach produces segregants that are nearly isogenic to the evolved strain, thereby avoiding undesirable genetic interactions with the magic-marker machinery. Finally, the method can be applied to any strain background without requiring the incorporation of the magic-marker machinery and coordination of auxotrophic reporters.
Fitness Effect of 116 Mutations Across 11 Evolved Lineages.
For each of our 11 evolved clones we generated a pool of ∼105 segregants. We propagated each pool for 90 generations and determined the fitness effect of each mutation by sequencing to a depth of ∼100 reads per site every 20 generations (SI Appendix, Fig. S2). The background-averaged fitness effect of a mutation is equal to the slope of the natural log ratio of allele frequencies over time. In addition, we determined the segregation of fitness within each of the 11 crosses by isolating 192 segregants from each pool and quantifying fitness using a flow cytometry-based competitive fitness assay for a total of 2,112 fitness assays on individual segregants (Dataset S2).
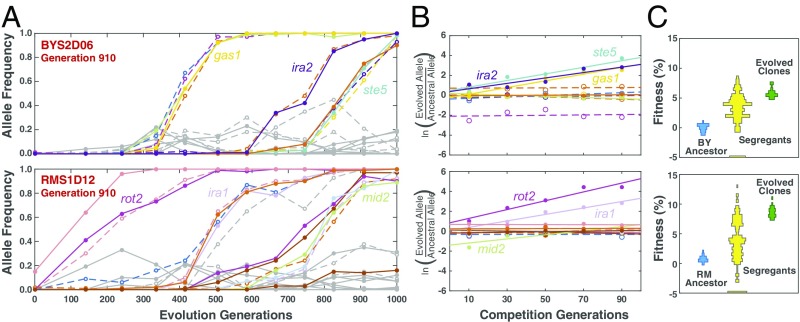
Two representative lineages are shown in Fig. 1, and all 11 lineages are shown in SI Appendix, Fig. S1. We analyzed single clones from populations BYS2D06 and RMS1D12, each isolated from generation 910. The evolutionary history of population BYS2D06 shows that 11 mutations swept through the population as three independent cohorts spaced several hundred generations apart (Fig. 1). Bulk-segregant analysis revealed the presence of three beneficial mutations (gas1, 3.4%; ira2, 2.7%; and ste5, 3.3%) in the clone BYS2D06-910, each driving a single cohort (Fig. 1). The fitness distribution of the individual segregants shows distinct modes that fall within the range of fitness bounded by the ancestral and evolved parental strains. Population RMS1D12 exhibits more complicated dynamics (Fig. 1). The clone RMS1D12-910 contains 14 mutations grouped into six cohorts (SI Appendix, Fig. S3). Bulk-segregant analysis identified three beneficial mutations (ira1, 3.3%, rot2, 5.6%, and mid2, 2.1%), each driving a different cohort. Again, the fitness values of the individual segregants fall between the fitness of the two parental strains, though the distribution appears more bimodal and less continuous than the distribution of fitness among the BYS2D06-910 segregants. These two major modes likely represent those segregants with and without the 5.6% fitness-effect rot2 mutation.
Fig. 1.
Genetic dissection of mutations from two evolved lineages. (A) Genome evolution of each population was previously tracked through time-course, whole-genome sequencing (10). An evolved clone was isolated from each population at defined time points. Each trajectory represents a unique mutation within the isolated clone (colored by chromosome), whereas gray trajectories indicate mutations detected in competing lineages within a population. (B) The background-averaged fitness effect of each evolved mutation is measured through a bulk-segregant fitness assay where a segregant pool is propagated in the selective environment and allele frequencies are tracked by whole-genome time-course sequencing. Fitness is calculated as the linear regression of the natural log ratio of evolved to ancestral allele frequency over time. The color scheme remains consistent between the evolutionary trajectories and bulk-segregant fitness assay. The dynamics of each mutation during the evolution experiment and the bulk-segregant fitness assay are in SI Appendix, Fig. S1 and Dataset S3. (C) Individual clones isolated from a bulk-segregant pool are assayed for fitness against an ancestral reference in a flow cytometry-based competition to determine how fitness segregates in the cross (yellow). The fitness distribution of segregants derived from an ancestral cross (Top, BY, Bottom, RM) provides a baseline fitness in the absence of beneficial alleles. The fitness distribution of the individual segregants is compared with the fitness of the evolved clone from which they arose (green). The fitness for all 192 segregants from each of the 11 lineages is available in Dataset S2.
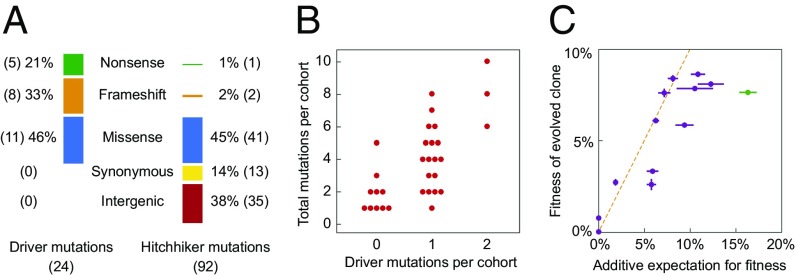
In total, we measured the fitness of 116 mutations originating from evolved clones across 11 lineages. For four of the mutations we were unable to quantify fitness because all of the segregants in the pool contain the same allele due to gene conversion events during strain construction (Discussion). Across the 11 lineages, we find that 24 of the 116 mutations are beneficial. As only 20% of mutations confer a detectable fitness advantage, a large fraction of genome evolution appears to be nonadaptive. The fitness effects of the driver mutations range from ∼1 to ∼10%, with most drivers conferring upwards of a 3% advantage. As expected, all evolved clones possess at least one beneficial mutation. In addition, evolved clones contain up to 13 hitchhiker mutations (Fig. 2 and Dataset S3). Three mutations are reported as deleterious, though only one, a read-through mutation in the stop codon of gcn2 in population BYS1D08, has greater than a 1% effect on fitness. We find no evidence of synonymous or intergenic mutations increasing fitness. When parsed by mutation type, driver mutations are enriched for nonsense and frameshift mutations compared with hitchhiker mutations (P < 0.001; Fisher–Freeman–Halton exact test); by contrast, synonymous and intergenic mutations are exclusively hitchhikers (Fig. 3A). This finding is consistent with adaptation driven by loss-of-function or alteration-of-function mutations in coding sequences (20).
Fig. 2.
Comprehensive quantification of fitness effects. Background-averaged fitness effects for 116 evolved mutations are quantified by the bulk-segregant fitness assay. Mutations are separated by lineage of origin and ordered by mean fitness effect. Fitness effect is represented as the SE of regression (thick lines) and 95% confidence interval (thin lines). Mutations are considered neutral (blue) if the confidence interval encompasses zero. Beneficial (orange) and deleterious (purple) mutations possess confidence intervals that fall completely above or below zero, respectively.
Fig. 3.
Mutational signatures, cohort composition, and additivity of fitness effects. (A) Mutations were divided into categories based upon their protein coding effect. The mutational signature of driver mutations is distinct from that of hitchhiker mutations (P < 0.001; Fisher–Freeman–Halton exact test). (B) Hierarchical clustering identified 31 cohorts among the 11 evolved lineages. Cohorts vary in size from 1 to 10 mutations and contain between zero and two drivers (SI Appendix, Fig. S3). We observe a positive relationship between the number of drivers within a cohort and cohort size (ρ = 0.70; Pearson correlation). (C) Fitness of all 11 evolved clones correlates with the sum of the fitness effects of their underlying evolved mutations, as quantified through the bulk-segregant fitness assay. Vertical error bars reflect the SE between replicate competitions of a common clone, and horizontal error bars reflect the propagation of error corresponding to the summation of individual background-averaged fitness effects. Deviation from the dashed line indicates nonadditive genetic interactions. The BYS2E01-745 clone (green) deviates furthest from the expectation.
Mapping Fitness Effects to the Dynamics of Adaptation.
Mutations often move through populations as cohorts, synchronously escaping drift and tracking tightly with one another through time (10). Cohorts are a recent observation (10–13) and the evolutionary dynamics that drive their formation are yet to be explained. In general, mutations within a cohort are genetically and functionally unrelated, suggesting that cohorts are a random collection of mutations that accumulate while at low frequencies on a common background. A deeper understanding of these dynamics requires resolving the number of driver and hitchhiker mutations for a large sampling of cohorts. We used hierarchical clustering to objectively partition the 116 mutations into distinct cohorts based on correlated changes in allele frequency over time. Across the 11 selected lineages, we identify 31 distinct cohorts (SI Appendix, Fig. S3). Each cohort contains up to 11 mutations and each evolved lineage possesses up to six cohorts. Eighteen cohorts have a single driver mutation. Three cohorts each possess two unique driver mutations. These two-driver cohorts have twice as many hitchhiker mutations compared with single-driver cohorts (6.0 ± 2.0 and 3.1 ± 1.9, respectively, P = 0.02, Wilcoxon rank sum, one tailed), contributing to the overall positive correlation between number of beneficial mutations per cohort and cohort size (Fig. 3B, ρ = 0.70, Pearson correlation). Surprisingly, no beneficial mutations were detected within 10 of the 31 cohorts. These “driverless” cohorts typically contain only one or two mutations, and their mutational signature closely resembles that of the genetic hitchhikers from “driven” cohorts. We propose that driverless cohorts represent a form of genetic hitchhiking, where collections of neutral mutations move through an adapting population due to one or more rounds of genetic associations with adaptive cohorts (SI Appendix, Fig. S4).
Synergistic Epistasis Between hsl7 and kel1, Two Genes Whose Protein Products Localize to Sites of Polarized Growth.
In the absence of epistasis, the fitness of an evolved clone is equivalent to the sum of the individual fitness effects of all constituent mutations. Indeed, we find a positive correlation (ρ = 0.82, Pearson correlation) between the additive expectation of all mutations affecting fitness and the measured fitness of each evolved clone (Fig. 3C). One clone in particular, BYS2E01-745, deviates strongly from this additive expectation, implying an epistatic interaction. In addition, the BYS2E01-745 cross exhibits a highly asymmetric fitness distribution with a large proportion of low fitness segregants (SI Appendix, Fig. S1).
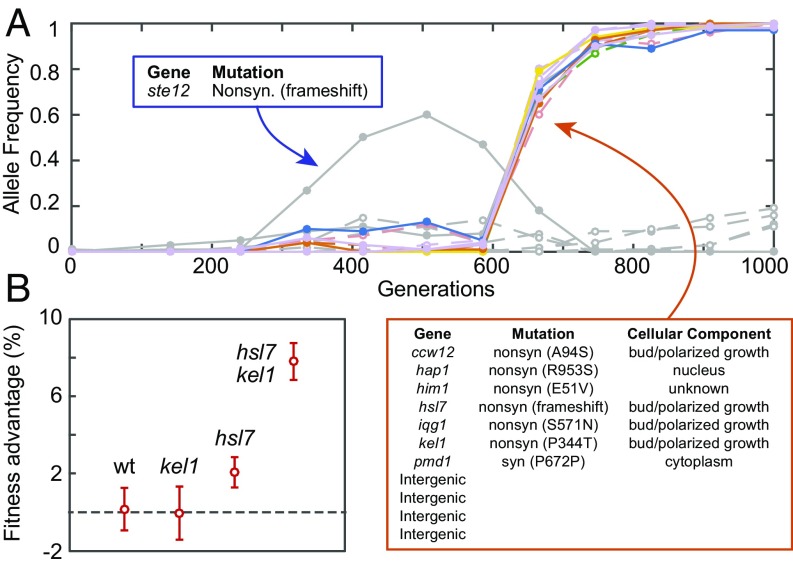
The dynamics of the BYS2E01-745 lineage are simple: an abrupt sweep of a single cohort of 11 mutations (Fig. 4A). However, the genetic basis of adaptation is unclear because BYS2E01-745 is the only lineage in this study without a mutation in a putative target of selection (10). Interestingly, the BYS2E01-745 lineage is enriched for mutations in genes whose products localize to the cellular bud and site of polarized growth (P = 0.0012 and P = 0.0016, respectively; GO Term Finder) (21). Further, four genes (hsl7, kel1, iqg1, and ccw12) whose protein products localize to sites of polarized growth each contain either a missense or frameshift mutation (Fig. 4A). Taken together, these data suggest that mutations in the BYS2E01-745 lineage may interact epistatically.
Fig. 4.
Adaptation mechanisms include rare mutations and epistatic interactions. (A) Evolutionary dynamics of population BYS2E01 as tracked through whole-genome time-course sequencing (10). A beneficial ste12 mutation (gray) was outcompeted by an 11-member cohort (colored) that is enriched for mutations in genes whose protein products localize to the cellular bud and site of polarized growth. (B) Bulk-segregant individuals from the BYS2E01-745 cross were genotyped and assayed for fitness, producing a genotype-to-phenotype map. Two evolved alleles, hsl7 and kel1, are associated with fitness gain. Shown are the average fitness and SD of segregants when parsed by HSL7 and KEL1 alleles. The kel1 mutation confers a benefit only in the hsl7 background, and the fitness advantage of the hsl7 kel1 double mutant is greater than the sum of the hsl7 and kel1 single mutants.
To test for epistatic interactions in the BYS2E01-745 lineage, we genotyped all 192 segregants that were previously assayed for fitness against an ancestral reference (Dataset S4). Of the 11 mutations in the BYS2E01-745 lineage, only two loci significantly affect fitness: HSL7 and KEL1 [P < 0.0001; N-way ANOVA, type III sum of squares (SS)], corroborating the results of the bulk-segregant fitness assay (SI Appendix, Fig. S1). Furthermore, the pairwise interaction between the hsl7 and kel1 alleles significantly impact fitness (P < 0.0001; N-way ANOVA, type III SS). The hsl7 mutation confers a modest benefit, 2.1 ± 0.5% (95% CI), in the KEL1 wild-type background, whereas it confers a significantly larger advantage, 7.8 ± 0.4%, when paired with the kel1 allele (Fig. 4B). The kel1 mutation is nearly neutral, −0.1 ± 0.3%, in the HSL7 wild-type background, but provides a substantial benefit in the hsl7 background (5.7 ± 0.6%). As initially suspected from the bulk-segregant fitness assay (SI Appendix, Fig. S1), the IQG1 allele is absent from all individual segregants, the result of a gene conversion event that occurred during the construction of the parental diploid strain (SI Appendix, Fig. S5). Through allelic replacement of IQG1 in individual segregants, we found that the evolved iqg1 allele has no effect on fitness. These results demonstrate that the rise of the BYS2E01-745 lineage is driven by intracohort epistasis between the hsl7 and kel1 mutations that combine to produce a high fitness genotype. The pairwise epistatic interaction between hsl7 and kel1 impacts fitness in a synergistic manner, representing one of few such examples to emerge from a long-term evolution experiment (22–26).
Discussion
Recurrence-based models represent a widely accepted statistical approach for inferring which genes and biological processes are under selection by identifying targets that are mutated more often than expected by chance (27–31). Such probability-based methods, however, will inherently neglect rare driver mutations and are unsuitable for quantifying fitness as occurrence is not necessarily indicative of fitness effect. Many variables, e.g., mutational target size, can impact how often mutations in a particular target are detected. Indeed, we find no apparent relationship between the fitness effect that a driver confers and its prevalence (SI Appendix, Fig. S6). For instance, both ira1 and ira2 provide similar effects on fitness (∼2.7% each); however, ira1 mutations were observed 23 times across replicate populations, yet ira2 only once (10). This stands in contrast to a recent study of paralogous genes where a correlation was observed between the frequency at which a mutation is observed and the fitness effect it confers (2). Of our 24 driver mutations, the recurrence model (10) did not detect 6 because they were mutated only twice; these include 4 modest-effect mutations (between 1 and 2%) as well as the 2 epistatic interactors, hsl7 and kel1, in population BYS2E01 (SI Appendix, Fig. S8).
The power behind identification of driver mutations through the bulk-segregant approach lies in the ability to screen lineages with numerous divergent loci for adaptive alleles. In only 1,000 generations, our populations fixed up to 19 mutations and in many cases, multiple cohorts of mutations existed simultaneously in the population leading to complex evolutionary dynamics. In addition, factors such as mutational target size, epistasis, genetic hitchhiking, and clonal interference strongly influence the identity of mutations that arise over the course of evolution. Therefore, directly measuring the fitness effects of all mutations, as we do here, is necessary to unambiguously identify and quantify the fitness effects of driver mutations that could otherwise be missed by recurrence-based methods.
There is increasing evidence that large-effect beneficial mutations drive adaptation in microbial evolution experiments (1, 2) as well as in natural microbial populations (32) and clinical infections (33). Here we identify 24 driver mutations ranging from roughly 1 to 10% effect, consistent with populations evolving under the strong-selection strong-mutation paradigm in which small fitness effects (<1%) are unlikely to contribute to fitness. Because our populations are asexual, it is possible for deleterious mutations to fix by hitchhiking on the background of strong driver mutations. We were surprised, however, to find that deleterious mutations are nearly absent from our dataset. Only 3 of 116 mutations were identified as deleterious, 2 of which are weak mutations (−0.6%) that could represent false positives. The lack of deleterious mutations is in contrast to an earlier study that found frequent hitchhiking of deleterious mutations in similar populations (12). We note that our use of 95% confidence intervals rather than SE is more conservative and may account for most of the discrepancy. We do have compelling evidence for at least one significant deleterious mutation: a stop codon readthrough of gcn2 in population BYS1D08-1000. This mutation converts a stop codon (TAG) into a lysine residue (AAG), resulting in a predicted 26-amino acid extension of the C terminus of the protein. The background-averaged fitness effect of this mutation is −2.3%, which is likely an underestimate of its cost because the mutant gcn2 allele was not detected in generation 90 of the bulk-segregant fitness assay (of the 107 reads that mapped to this position). It is unclear from our data whether this gcn2 mutation is deleterious in all backgrounds; therefore, it is possible that this mutation was beneficial on the background in which it arose.
Among the 116 mutations across the 11 evolved lineages in this study, we measured the fitness effect of multiple evolved alleles of the same gene, such as ira1, and multiple mutations in the same pathway, such as the mating pathway. The fitness effect of alternate adaptive alleles is remarkably consistent for the ira1/2, gas1, and yur1 mutations, all of which differ by less than 1% in background-averaged fitness effect (SI Appendix, Fig. S6). However, there are exceptions. A single neutral ste12 mutation (frameshift at position 472) from the RMS1G02-825 lineage falls outside the narrow range (2.6–3.3%) of the four adaptive mutations in the mating pathway. Additionally, the neutral kel1-Q107K mutation in the RMS1G02-825 lineage stands in contrast to the net-beneficial kel1-P344T mutation from BYS2E01-745 (SI Appendix, Fig. S6). The Q107K substitution is predicted to be tolerated, whereas the P344T substitution is predicted to impact protein function (PROVEAN) (34).
Whereas common targets of selection may represent a substantial fraction of realized adaptive mutations, it is evident in this study and elsewhere (22) that some lineages may owe their success to the acquisition of a rare beneficial mutation or an assembly of epistatically interacting mutations. Here, we identify synergistic epistasis between two mutations, kel1 and hsl7, that arose within a single cohort. This observation raises additional questions regarding the prevalence of such epistatic interactions; the context in which these interactions arise; and the extent to which epistasis, in addition to neutral genetic hitchhiking, gives rise to cohort dynamics.
In addition to quantifying fitness effects and identifying epistatic interactions, our data reveal a more detailed view of the dynamics of adaptation. Mapping driver and hitchhiker mutations to each of our 31 mutational cohorts shows that most cohorts consist of one to two driver mutations and zero to eight hitchhiker mutations. Curiously, we observe that approximately one-third of cohorts lack an identifiable driver mutation. The genetic composition of these driverless cohorts is unique for two reasons. First, driverless cohorts have an average of 1.9 mutations, which is significantly fewer than the average number of mutations in adaptive cohorts (4.6; P < 0.01, Wilcoxon rank sum, Fig. 3). Second, driverless cohorts are enriched for low impact (intergenic and synonymous) mutations and are devoid of high impact (frameshift and nonsense) mutations. Driverless cohorts, therefore, closely resemble hitchhiker mutations (P = 0.90, Fisher–Freeman–Halton exact test; SI Appendix, Fig. S7) yet differ significantly from mutations in adaptive cohorts (P = 0.09) and driver mutations (P < 10−6).
We propose that driverless cohorts move through an adapting population due to one or more rounds of genetic associations with adaptive cohorts. These mutations assemble on the background of an adaptive cohort while at low frequency. As the adaptive cohort increases in frequency, the driverless cohort is “pulled” up until the adaptive cohort fixes, at which point its fate is dependent upon the next beneficial mutation. In three populations (BYS1D08, BYS1E03, and RMS1D12), we observe that the next beneficial mutation “pushes” the driverless cohort to fixation (SI Appendix, Fig. S4). If the next beneficial mutation were to arise in the opposing lineage, the driverless cohort would be driven to extinction. Although not observed in this study, such dynamics do appear to occur in several populations studied previously (see populations BYS1F05 and BYS2C03 in ref. 10). An alternative hypothesis is that the presumptive driverless cohorts could be driven by undetected mutations. We extensively analyzed the evolved genomes for point mutations and copy number variants, but found no evidence of the latter. Furthermore, we find four-spore viability in crosses between evolved clones and their ancestor, suggesting that large-scale rearrangements are not common. Therefore, in our opinion, the most parsimonious explanation is that driverless cohorts are not adaptive and that one or more rounds of genetic associations govern their dynamics.
Fitness dependence, which can arise through cross-feeding or spatial structuring, often complicates fitness estimates (35–39). One of our evolved clones (BYS1E03-745) exhibits negative frequency-dependent fitness when competed against an ancestral reference: the clone plateaued at frequency of 0.75 regardless of starting proportion (SI Appendix, Fig. S9 and Dataset S1). We suspect the erg11 mutation is responsible for the plateau because defects in ergosterol biosynthesis (or drug-based inhibition of the pathway) have been shown to result in negative frequency dependence in yeast (35). We observed abnormal well morphology in erg11-containing cultures mirroring the morphology of the “adherent” A-type cells described previously (35). Because our bulk-segregant pool contained random combinations of evolved mutations, both with and without the erg11 mutation, we could successfully quantify the fitness effect of all mutations in this clone with the exception of the erg11 allele itself, which maintained a frequency between 0.8 and 0.9 for the entirety of the 90-generation bulk-segregant fitness assay. Curiously, the erg11 allele did not impart frequency dependence according to the adaptive dynamics of the evolving BYS1E03 population (10). Instead, erg11 rapidly fixed in the evolution experiment. The erg11 mutation may have been pushed through the population by the ira1-driven cohort. In other words, the ira1 allele may have masked the frequency dependence of the erg11 allele.
This study demonstrates the power of experimental evolution to identify epistatic interactions. Much of our understanding of epistasis in budding yeast comes from the systematic analysis of double mutants (40). Although informative for constructing large-scale genetic interaction networks, these studies have thus far been restricted to gene amplifications and deletions, which fail to capture a significant portion of the mutational spectrum. The observed kel1–hsl7 interaction has not been identified by these systematic approaches, and several lines of evidence suggest that the interacting hsl7 allele results from a rare mutational event. First, no other hsl7 mutations were detected in any of the other 40 time-course sequenced populations (10) despite the BYS2E01-hsl7 allele conferring a 2.1% advantage in the ancestral background. This hsl7 allele encodes a truncated version of the protein that lacks only the C-terminal domain, which is required for phosphorylation of Hsl7p by Hsl1p in a cell cycle-dependent manner to relocate Hsl7p from the spindle-pole body to the bud neck (41). Deletion of HSL7 is deleterious under a wide range of conditions, including the rich glucose media used here (42, 43); thus our data suggest that the evolved hsl7 allele bestows a novel function or alters an existing function. Extensive characterization of such rare beneficial mutations requires long-term high-replicate evolution experiments followed by comprehensive analysis linking genotype to phenotype. Likely due to their large target size, loss-of-function mutations dominate adaptive evolution experiments, though rare beneficial mutations and epistatic interactions may provide the raw material for molecular innovation in natural populations.
Methods
Strain Construction.
We generated a MATα strain with the following genotype: ade2-1, ura3::CAN1, his3-11, leu2-3,112, trp1-1, can1, bar1Δ::ADE2, and hmlαΔ::LEU2. This strain (yGIL737) differs from the ancestral strains from the original evolution experiment (8) in that it is MATα, it has a loss-of-function mutation in can1, it has a wild-type copy of CAN1 at the ura3 locus, and it does not contain the NatMX cassette linked to GPA1, which is variable between the two ancestors (DBY15095/yGIL429 and DBY15092/yGIL432).
We crossed our MATa evolved clones to yGIL737 and, when necessary, we complemented sterile mutations in the evolved clones using the appropriate plasmid from the MoBY ORF plasmid collection (44). The resulting diploid strains were converted to MATa/a by a 3-h galactose induction using a plasmid harboring a Gal-driven HO. MATa/a convertants were identified by the formation of shmoos following a 3-h αF-induction and were verified by PCR of the mating-type locus using primers MATα-internal-F (5′ GCACGGAATA TGGGACTACT TCG 3′), MATa-internal-F (5′ ACTCCACTTC AAGTAAGAGT TTG 3′), and MAT-external-R (5′ AGTCACATC AAGATCGTTT ATGG 3′). Each MATa/a diploid was transformed with a URA3 plasmid, pGIL071, which harbors the MATα2 locus needed for sporulation. Note that using the full-length MATα locus produces rare MATα spores, presumably due to gene conversion at the MAT locus. Therefore, the MATα1 gene, which is required for MATα mating but not sporulation, was not included on our plasmid.
Generating Segregant Pools and Isolating Individual Segregants.
To create segregant pools, a single colony of each MATa/a diploid containing the plasmid-borne MATα2 was transferred into 10 mL of YPD and grown overnight to saturation. Overnight cultures were spun down and resuspended in 20 mL SPO++ (1.5% potassium acetate, 0.25% yeast extract, 0.25% dextrose, supplemented with 1× CSM −Arg; Sunrise Science) and sporulated on a room temperature roller drum for 3–4 d until ∼75% sporulation efficiency was reached. Sporulated cultures were spun down and resuspended in 200 µL H2O for a final volume of 500–750 µL. Ascus walls were digested by adding 5 µL of zymolyase (150 mg/mL) and incubating for 1 h at 30 °C. Next, 50 µL glass beads and 50 µL 10% Triton were added and the asci were disrupted by vortexing for 2 min. This step was followed by an additional 40 min at 30 °C and again by vortexing for 2 min. Disrupted asci were brought up to 5 mL with H2O and were sonicated using a microtip sonicator for 4 s at full power. A liquid hold was performed by inoculating 500 µL of the spore prep into 5 mL YPD and incubating for 24 h at 30 °C.
Segregants were isolated by plating onto 150-mm Petri dishes containing solid BSP medium (CSM −Arg, yeast nitrogen base, 2% dextrose, with 1 g/L 5-FOA, 60 mg/L canavanine, and 100 mg/L ClonNat). To make segregant pools, 2 mL of the spore preparation were plated onto duplicate BSP plates (resulting in ∼105 segregants across the two plates). After ∼3 d of growth at 30 °C, 5 mL of YPD was added to the first plate and a sterile glass spreader was used to remove yeast from the surface of the agar plate. This volume (2–3 mL) was transferred to the second plate and the process was repeated; the liquid was removed with a pipette and transferred to a 5-mL tube. To remove residual yeast cells, a second plate wash was done with 2.5 mL. This liquid from the second wash (1–2 mL) was added to the same 5-mL tube for a total of ∼4 mL. To isolate individual segregants, 1 mL of the spore prep was plated onto duplicate BSP plates. After ∼3 d of growth at 30 °C, 192 colonies were picked and inoculated into 130 µL YPD in two 96-well plates. Plates were grown for 1–2 d at 30 °C. All segregants were phenotyped for growth on 5-FOA and canavanine, for the absence of growth on SC −Ura, and for mating type. Segregants were stored at −80 °C in 15% glycerol.
Bulk-Segregant Fitness Assay.
For each bulk-segregant pool, seven replicate populations were set up in a single 96-well plate. The bulk-segregant pools were propagated for 100 generations in conditions identical to the original evolution experiment (8). In brief, 132 µL cultures were maintained in YPD at 30 °C and transferred to fresh media every 24 h at a dilution of 1:1,024. The assay lasted for a total of 10 d, corresponding to 100 generations. Following each transfer, the remaining culture was pooled across replicates (for a total volume of ∼900 µL). Cells were pelleted and washed with 1 mL of H2O. Frozen pellets were stored at −20 °C. Genomic DNA was prepared from the frozen pellets using a modified glass bead lysis method (10).
Library Preparation and Whole-Genome Sequencing.
Multiplexed genomic DNA libraries were prepared using a modified variation of the Nextera protocol (45) with the following modification: to conserve Nextera Index primers, the Index PCR was performed for eight cycles, followed by amplification of the libraries via a five-cycle reconditioning PCR using primers P1 (5′ AATGATACGG CGACCACCGA 3′) and P2 (5′ CAAGCAGAAG ACGGCATACGA 3′). Libraries were quantified by NanoDrop and pooled at equal concentration. The multiplexed pool was excised from an agarose gel (QIAquick Gel Extraction Kit, Qiagen) to size select for fragments between 400 and 700 bp, and the collected fragments were analyzed by BioAnalyzer on High-Sensitivity DNA Chip (BioAnalyzer 2100, Agilent). The samples were run on an Illumina HiSeq 2500 sequencer with 250-bp single-end reads by the Sequencing Core Facility within the Lewis-Sigler Institute for Integrative Genomics at Princeton University. Sequence data were analyzed as described in SI Appendix, SI Methods.
Flow Cytometry-Based Competitive Fitness Assays.
Fitness of the evolved clones and bulk-segregant individuals was determined in flow cytometry-based competitions as described previously (8, 10). Briefly, segregants were mixed 1:1 with a ymCitrine-labeled ancestral reference and passaged in YPD broth at 1:1,024 dilution on a 24-h cycle to mimic evolution conditions. Every 10 generations, an aliquot was transferred to PBS and assayed by flow cytometry (BD FACSCanto II). Cytometric data were analyzed by FlowJo. Fitness was calculated as the linear regression of the log ratio of experimental-to-reference frequencies over the 40-generation assay. Extremely low-fitness clones, which presumably acquired a deleterious mutation, were not included in downstream analysis of the aggregate data.
Supplementary Material
Acknowledgments
We thank Michael Desai, Andrew Murray, and members of the G.I.L. laboratory for their comments on the manuscript. This work was supported by the Charles E. Kaufman Foundation of The Pittsburgh Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The short-read sequencing data reported in this paper have been deposited in the NCBI BioProject database (accession no. PRJNA388205).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702314114/-/DCSupplemental.
References
- 1.Payen C, et al. High-throughput identification of adaptive mutations in experimentally evolved yeast populations. PLoS Genet. 2016;12:e1006339. doi: 10.1371/journal.pgen.1006339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkataram S, et al. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell. 2016;166:1585–1596. doi: 10.1016/j.cell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atwood KC, Schneider LK, Ryan FJ. Periodic selection in Escherichia coli. Proc Natl Acad Sci USA. 1951;37:146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paquin C, Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- 5.Bryson V, Szybalski W. Microbial selection. Science. 1952;116:45–51. [PubMed] [Google Scholar]
- 6.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102–103:127–144. [PubMed] [Google Scholar]
- 7.Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang GI, Botstein D, Desai MM. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics. 2011;188:647–661. doi: 10.1534/genetics.111.128942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perfeito L, Fernandes L, Mota C, Gordo I. Adaptive mutations in bacteria: High rate and small effects. Science. 2007;317:813–815. doi: 10.1126/science.1142284. [DOI] [PubMed] [Google Scholar]
- 10.Lang GI, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maddamsetti R, Lenski RE, Barrick JE. Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics. 2015;200:619–631. doi: 10.1534/genetics.115.176677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald MJ, Rice DP, Desai MM. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature. 2016;531:233–236. doi: 10.1038/nature17143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moura de Sousa JA, Alpedrinha J, Campos PR, Gordo I. Competition and fixation of cohorts of adaptive mutations under Fisher geometrical model. PeerJ. 2016;4:e2256. doi: 10.7717/peerj.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dykhuizen DE. Experimental studies of natural selection in bacteria. Annu Rev Ecol Syst. 1990;21:373–398. [Google Scholar]
- 15.Hedrick PW. Genetic hitchhiking: A new factor in evolution? Bioscience. 1982;32:845–853. [Google Scholar]
- 16.Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 18.Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci USA. 2009;106:5755–5760. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 20.Lang GI, Desai MM. The spectrum of adaptive mutations in experimental evolution. Genomics. 2014;104:412–416. doi: 10.1016/j.ygeno.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyle EI, et al. GO:TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332:1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 24.Quan S, et al. Adaptive evolution of the lactose utilization network in experimentally evolved populations of Escherichia coli. PLoS Genet. 2012;8:e1002444. doi: 10.1371/journal.pgen.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quandt EM, Deatherage DE, Ellington AD, Georgiou G, Barrick JE. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc Natl Acad Sci USA. 2014;111:2217–2222. doi: 10.1073/pnas.1314561111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quandt EM, et al. Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife. 2015;4:4. doi: 10.7554/eLife.09696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou HH, Berthet J, Marx CJ. Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet. 2009;5:e1000652. doi: 10.1371/journal.pgen.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong J, Gresham D. Molecular specificity, convergence and constraint shape adaptive evolution in nutrient-poor environments. PLoS Genet. 2014;10:e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kvitek DJ, Sherlock G. Whole genome, whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLoS Genet. 2013;9:e1003972. doi: 10.1371/journal.pgen.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 32.Bailey SF, Bataillon T. Can the experimental evolution programme help us elucidate the genetic basis of adaptation in nature? Mol Ecol. 2016;25:203–218. doi: 10.1111/mec.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brockhurst MA. Experimental evolution can unravel the complex causes of natural selection in clinical infections. Microbiology. 2015;161:1175–1179. doi: 10.1099/mic.0.000107. [DOI] [PubMed] [Google Scholar]
- 34.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frenkel EM, et al. Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations. Proc Natl Acad Sci USA. 2015;112:11306–11311. doi: 10.1073/pnas.1506184112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friesen ML, Saxer G, Travisano M, Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution. 2004;58:245–260. [PubMed] [Google Scholar]
- 37.Levin BR. Coexistence of two asexual strains on a single resource. Science. 1972;175:1272–1274. doi: 10.1126/science.175.4027.1272. [DOI] [PubMed] [Google Scholar]
- 38.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 39.Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: Genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baryshnikova A, Costanzo M, Myers CL, Andrews B, Boone C. Genetic interaction networks: Toward an understanding of heritability. Annu Rev Genomics Hum Genet. 2013;14:111–133. doi: 10.1146/annurev-genom-082509-141730. [DOI] [PubMed] [Google Scholar]
- 41.Cid VJ, Shulewitz MJ, McDonald KL, Thorner J. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol Biol Cell. 2001;12:1645–1669. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown JA, et al. Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol Syst Biol. 2006;2:2006.0001. doi: 10.1038/msb4100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian W, Ma D, Xiao C, Wang Z, Zhang J. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Reports. 2012;2:1399–1410. doi: 10.1016/j.celrep.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho CH, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol. 2009;27:369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baym M, et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One. 2015;10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.