Significance
Although biological diversity is being lost at an alarming rate, a mimetic butterfly from the Amazon reveals a selective mechanism that can lead to rich adaptive diversity. In these toxic and vividly colored butterflies, several forms are maintained within populations, accurately mimicking distinct local butterflies, and controlled by differentiated alleles of a supergene. Such diversity is not expected because selection by predators promotes the fixation of a single morph. We show that the mating behavior favors pairings of dissimilar forms and boosts allelic diversity at the mimicry supergene. Therefore, sexual selection maintains a phenotypic diversity that predators channel and refine into a diversity of warning phenotypes, which correspond to the signals used by other defended species of butterflies in the local community.
Keywords: disassortative mating, selection conflicts, Müllerian mimicry, aposematism, warning signals
Abstract
Explaining the maintenance of adaptive diversity within populations is a long-standing goal in evolutionary biology, with important implications for conservation, medicine, and agriculture. Adaptation often leads to the fixation of beneficial alleles, and therefore it erodes local diversity so that understanding the coexistence of multiple adaptive phenotypes requires deciphering the ecological mechanisms that determine their respective benefits. Here, we show how antagonistic frequency-dependent selection (FDS), generated by natural and sexual selection acting on the same trait, maintains mimicry polymorphism in the toxic butterfly Heliconius numata. Positive FDS imposed by predators on mimetic signals favors the fixation of the most abundant and best-protected wing-pattern morph, thereby limiting polymorphism. However, by using mate-choice experiments, we reveal disassortative mate preferences of the different wing-pattern morphs. The resulting negative FDS on wing-pattern alleles is consistent with the excess of heterozygote genotypes at the supergene locus controlling wing-pattern variation in natural populations of H. numata. The combined effect of positive and negative FDS on visual signals is sufficient to maintain a diversity of morphs displaying accurate mimicry with other local prey, although some of the forms only provide moderate protection against predators. Our findings help understand how alternative adaptive phenotypes can be maintained within populations and emphasize the need to investigate interactions between selective pressures in other cases of puzzling adaptive polymorphism.
Local adaptation generally leads to the fixation of locally beneficial alleles and is often associated with directional or stabilizing selection that maintains populations at distinct phenotypic optima in different localities (1, 2). Similarly, sexual selection can lead to the fixation and reinforcement of divergent characters between populations (3). Natural and sexual selection, when operating in the same direction on the same trait, may accelerate trait fixation within populations and divergence between populations (3–5). When traits are subject to antagonistic selection regimes, however, it is hard to predict the trajectory and equilibria of population differentiation and adaptation. Here, we investigate how the interplay of opposing natural and sexual selection may account for spectacular adaptive polymorphism in butterfly wing patterns.
Prey warning signals and defensive mimicry are well-known examples of traits where the best protected phenotypes are well described and are determined by the most abundant local phenotype (6, 7). In such cases, defended prey display warning signals that are recognized by predators through avoidance learning, such that the protection associated with a warning signal increases with its frequency of encounter by predators (6). Predator education therefore generates positive frequency-dependent selection (FDS) favoring common warning signals, and whose slope depends on total prey numbers (6, 8, 9). This selection regime restricts the conditions allowing the emergence of polymorphisms and promotes the evolutionary convergence of signals among defended species exposed to the same predator communities [i.e., Müllerian mimicry (6)].
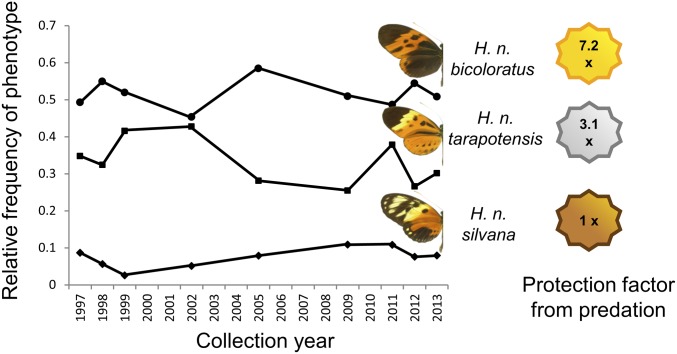
However, polymorphism is sometimes found within populations of warningly colored species (10–13). One spectacular example is the mimetic polymorphism found in the Amazonian butterfly Heliconius numata. This unpalatable species, whose wing-pattern variation is controlled by a single supergene locus [called P (14–16)], always displays multiple discrete mimetic color patterns within populations (6, 17, 18). This coexistence of distinct phenotypes within a locality involves the maintenance of one well-protected form and a diversity of rarer forms suffering up to sevenfold more predation (6) (Fig. 1). The ubiquity of polymorphism throughout the range of this species suggests that powerful balancing mechanisms are countering the strong positive FDS acting on local warning signals.
Fig. 1.
Stability of mimicry polymorphism and associated protection within the natural population of H. numata from Tarapoto (Peru). The most common morph (bicoloratus) is well protected from predator attacks, followed by tarapotensis and then the rarest, silvana, which suffers high attack rates (6).
Warning color patterns may also be used as mate recognition cues (3, 19–23). Mate preference generally causes positive assortative mating between individuals with the same phenotype and may reinforce local monomorphism in the warning signal. However, sexual preferences may maintain color polymorphism through a preference for rare phenotypes (24) or from disassortative mating [i.e., between individuals of different phenotypes (25–27)], generating negative FDS (3). Indeed, under disassortative mating, individuals with a rarer phenotype enjoy increased reproductive success because a larger proportion of the population will prefer them. Negative FDS is therefore a consequence of disassortative mating behavior and has been well described for bisexual reproduction, leading to a 1:1 sex ratio (28), and for the self-incompatibility system of plants (29), resulting in the maintenance of a high level of allelic polymorphism at the self-incompatibility locus (29, 30). Here, for the case of mating preferences associated with wing pattern in H. numata, we examine whether sexual selection may generate negative FDS on mimetic morphs opposing the positive FDS due to mimicry and explain the maintenance of stable polymorphism in this species.
Results
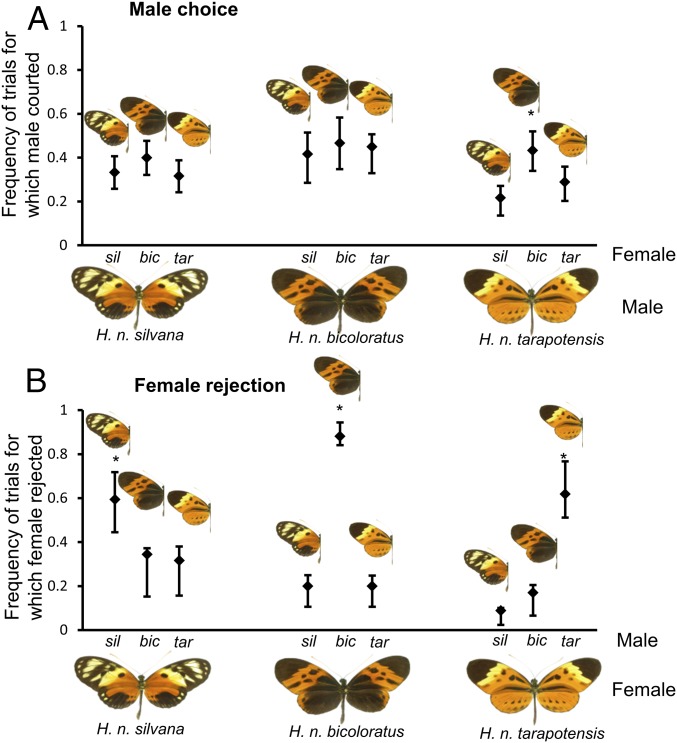
To assess the role of sexual selection in stabilizing the wing color pattern frequencies of H. numata, we first tested for morph-specific mate preferences that might prevent the fixation of one phenotype. First, male and female preferences were assessed independently by performing trials recording (i) the frequency of male courtship and (ii) female aversion toward different wing color patterns. Overall, males showed a slightly higher propensity to court females of morph bicoloratus, the most common morph in the local populations studied in northeastern Peru [χ2(2) = 16.94, P < 0.001; post hoc ǀZǀ ≥ 2.89 for bicoloratus discrimination, P ≤ 0.011; Figs. 1 and 2A]. When analyzing the response of each male morph separately, this preference was only significant for tarapotensis males [χ2(2) = 22.31, P < 0.001] that courted bicoloratus females slightly more frequently (post hoc ǀZǀ ≥ 2.93 for bicoloratus discrimination, P ≤ 0.009). In contrast to males, females of all morphs showed a clear aversion toward males displaying their own phenotype [response of each female morph analyzed separately: χ2(2) ≥ 44.09, P < 0.001; post hoc ǀZǀ ≥ 5.50 for own phenotype discrimination, P ≤ 0.001; Fig. 2B]. Overall, 88% of females reject their own morph more often than dissimilar ones. The genetics of mate preference are unknown in this species, but male preference loci causing assortative mating have been mapped in other Heliconius species and shown to be linked to color patterning loci, such as K and optix (10, 31, 32). Moreover, in the Heliconius cydno-melpomene clade, the Yb color locus, a homolog to the wing-patterning supergene of H. numata, was associated with female mate preference (33). It is therefore conceivable that disassortative mating in polymorphic populations of H. numata could be determined by a locus linked to, or contained within, the supergene P.
Fig. 2.
Sex-specific mate preference in H. numata. (A) Male courtship events toward distinct live female phenotypes (n = 30 per male morph). (B) Female rejection events toward artificial male courtship behavior (n = 30 per female morph). In both A and B, dots and bars indicate the means and 95% binomial CIs of the proportion of trials for which a preference (courting for males, rejection for females) was displayed. Significant differences in mate preference are indicated with an asterisk.
Next, to estimate actual mating patterns resulting from strongly disassortative preferences in females and slightly directional preferences in males, we performed mating experiments in which the full spectrum of courtship and preferences of both sexes could be expressed (Materials and Methods). Using a 2 × 2 (tetrad) design in which males and females of two morphs were left together until one mating occurred, we showed that mating was over twice as frequent (72%) between distinct morphs than between same morphs [for each combination of morphs: χ2(1) ≥ 7.501, P ≤ 0.006; Table 1]. Female preferences therefore override male preferences and lead to strong disassortative mating.
Table 1.
Number of mating events in tetrad mating trials
| Female morph | Male morph | |
| tarapotensis | silvana | |
| tarapotensis | 10 | 13 |
| silvana | 30+* | 7−* |
| bicoloratus | silvana | |
| bicoloratus | 10 | 19 |
| silvana | 26+* | 5−* |
| bicoloratus | tarapotensis | |
| bicoloratus | 6−* | 28+* |
| tarapotensis | 15 | 11 |
Cells for which the number of mating events significantly (P < 0.05) differs (is either higher or lower as shown by the sign) from the value expected assuming random mating.
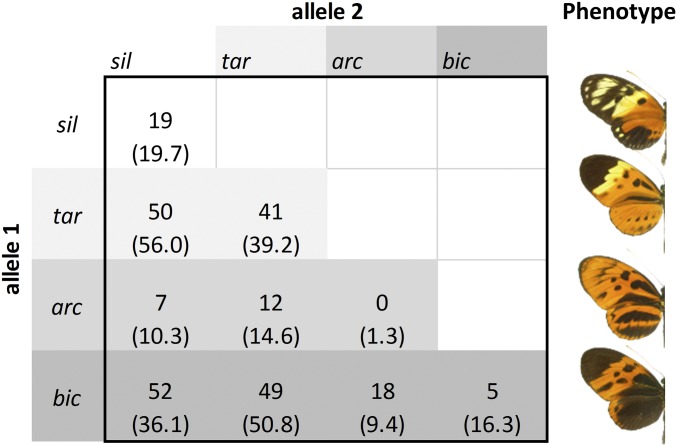
To test for the effects of disassortative mating in nature, we assessed deviations from Hardy–Weinberg genotype frequencies in natural populations of H. numata. The wing pattern supergene P displayed an excess of heterozygote genotypes (He = 0.699, Ho = 0.720, P = 0.040; Fig. 3). In contrast, 20 unlinked microsatellite markers distributed across the genome showed allele frequencies fitting Hardy–Weinberg expectations or even showing some heterozygote deficits (34). This discrepancy between heterozygote excess at P relative to the rest of the genome is expected if disassortative mating acts specifically on P genotypes and generates negative FDS. The overall heterozygote excess is largely driven by a strong deficit of homozygotes for the dominant allele Pbic (P < 0.001) underlying the most abundant and best protected phenotype (bicoloratus). Bicoloratus individuals constitute 49% of this population, of which only 4% are Pbic homozygotes (Fig. 3). Under the sole effect of positive FDS generated by predators, this allele is expected to be quickly driven to fixation (6). However, because this allele is dominant over all other mimetic alleles, Pbic homozygotes can only be formed through the mating of two individuals displaying the same bicoloratus phenotype, which rarely happens when mating occurs preferentially between dissimilar forms. Disassortative mating is therefore expected to be especially efficient in hampering the formation of homozygotes for the dominant allele, which may account for their scarcity and explain why the Pbic allele does not reach fixation, despite its strong frequency-dependent advantage with respect to mimicry.
Fig. 3.
Genotypic composition at the P mimicry supergene of H. numata from the Tarapoto population. Within each cell are the numbers of each genotype observed and expected under Hardy–Weinberg equilibrium (in parentheses). Alleles are presented following their hierarchical dominance (Psil < Ptar < Parc < Pbic), such that all of the genotypes within a row produce the same phenotype, which is presented at its end.
In contrast, homozygotes for the more recessive alleles (Ptar or Psil) may be formed through the mating of individuals with dissimilar phenotypes [e.g., genotypes Psil/Ptar (phenotype tarapotensis) mated with Ptar/Pbic (phenotype bicoloratus) will produce a progeny with 25% of Ptar/Ptar], such that deficits of homozygotes for more recessive alleles are expected to be smaller than for the dominant allele. Disassortative mating with different effects on the frequency of each genotype as a result of the dominance relations among alleles is also found in the self-incompatibility locus in Arabidopsis halleri, in which obligate disassortative mating forbids the formation of homozygotes for the dominant alleles but allows the production of homozygotes for the recessive alleles (35).
Overall, we have shown that the warning signals of H. numata are subject to two antagonistic selection regimes: sexual selection causing negative FDS and promoting polymorphism and mimicry selection causing positive FDS (6) and promoting the fixation of the most abundant mimetic wing pattern.
Discussion
Many traits are subject to different selective forces, sometimes with opposing effects. Communication signals are often shaped by their dual roles as antipredator signals (local adaptation) as well as signaling to conspecifics [social or sexual interactions (3)]. In H. numata, such dual roles generate antagonistic frequency-dependent regimes, with one favoring a diversity of mating signals independent of whether they are mimetic or not and the other favoring mimicry of the best local warning phenotypes (6). This antagonistic relationship leads to the evolution and maintenance of several coexisting accurate mimetic forms. Paradoxically, some of these mimetic forms obtain little survival benefit from mimicry because their corresponding comimics are locally rare. Nevertheless, forms mimicking warning signals with low abundance are still better off than nonmimics (6, 19). We conclude that the action of negative FDS on wing-pattern forms filtered by local mimicry explains how populations maintain a stable polymorphism for mimetic phenotypes.
Disassortative mating may seem maladaptive in the context of positive FDS for mimicry because it fosters the production of offspring with polymorphic warning signals. Therefore, the mechanism leading to the evolution of such mating behavior in a mimetic species remains an open question. Disassortative mating is often found in systems where homozygotes are at a selective disadvantage (36, 37); indeed, in certain populations of H. numata, some survival benefits are reported for larvae with heterozygous genotypes at P (17). Mimicry polymorphism in H. numata is controlled by a supergene characterized by distinct chromosomal inversions maintaining the different haplotypes underlying the various wing patterns (14). As documented in an increasing number of cases, inversions may be associated with deleterious effects, either through the disruption of gene regulation by the breakpoints or due to deleterious mutations present within the segment at the time of the inversion, causing reduced fitness in homozygotes (27, 38–40). By minimizing the frequency of homozygotes for the inversion in offspring, disassortative mating protects from the deleterious effects associated with the inversions. Both processes may contribute to the observed deficit of homozygotes in natural populations and reinforce the persistence of polymorphism.
In several adaptive radiations, selection on differentiated color phenotypes is thought to precipitate speciation through its roles in mate choice and premating reproductive isolation (23, 41, 42). In the mimicry radiation of Heliconius, preference for a similar mate (positive assortative mating) may evolve in response to the poor mimicry of heterozygous genotypes at mimicry loci (42), and it operates as a key driver of speciation along the entire continuum of differentiation, from the early stages of ecological divergence (10) to the maintenance and strengthening of species differences (42). Our results contrast with this general trend and may drive drastically different adaptive dynamics in H. numata. First, disassortative mate preferences associated with adaptive color polymorphism enhance the homogenization of genomic backgrounds across color phenotypes and hamper ecological speciation promoted by selection on alternative mimicry associations. Second, disassortative mating provides a mating advantage to rare forms, partially offsetting their poor protection from predation. This mating advantage may explain how local H. numata populations maintain relatively high frequencies of the recessive Psil allele coding for the form silvana (30% of alleles and 7% of phenotypes in Tarapoto) even though its comimetic species are vanishingly rare in this region of Peru. Because rare recessive homozygotes enjoy both a mating advantage and good mimicry protection to their offspring through mating with abundant local forms, disassortative mating could therefore promote effective migration of recessive alleles and foster genetic exchange between populations characterized by different mimicry phenotypes. Disassortative mating in mimetic species could thus act against parapatric differentiation across transition zones and limit speciation by local adaptation.
Our results provide important insights into how the interplay of antagonistic selection regimes acting on a trait enables the persistence of adaptive diversity. Similar mechanisms might provide an explanation for long-standing puzzles, such as the maintenance of certain polymorphisms in African Danaeus and Acraea butterflies (43), or more recent discoveries of other disassortative mating systems (24, 25). This study highlights the importance of understanding both the genetic architecture and frequency-dependent benefits to predict population responses to variations in local environmental conditions.
Materials and Methods
This study was performed on the H. numata population of Tarapoto in northern Peru [6°27′30′′S and 76°21′00′′W in the locality of Ahuashyacu (6)]. The study population was mainly composed of three discrete morphs (silvana, tarapotensis, and bicoloratus) (6, 14) determined by three distinct alleles (Psil, Ptar, and Pbi, respectively) at the supergene P (16). The alleles are associated with three different chromosomal arrangements and arrayed along a strict hierarchy of dominance (Psil < Ptar < Pbi) (14). Predation intensity on these morphs tightly follows the abundance of the distinct warning signal in the habitat, with the most common (bicoloratus) being well protected from predators, followed by tarapotensis, and with the rarest, silvana, suffering high attack rates as a result of positive FDS acting on their respective mimicry rings (6). A fourth morph, arcuella, is also occasionally seen in this population, but the allele controlling it (Parc) is rare (Fig. 3), so we did not include it in the study.
All behavioral experiments were conducted with unrelated sexually mature virgin males (10–18 d old) and receptive females (3–5 d old) in outdoor insectaries in Tarapoto (42). To control for relatedness and to maximize the genetic diversity of the butterflies tested, each sex was drawn from the captive raised progeny of two distinct H. numata colonies. Each colony was maintained at a minimum of 15 fertilized wild-caught females from the population studied. Tested individuals were acclimated to the insectary for at least 2 d before the experiment and had ad libitum access to sugared water, fresh flowers for pollen (Gurania sp., Psiguria sp., and Lantana sp.), and bee pollen. All experiments were conducted on sunny days between 09:00 and 16:00.
Male Preference Using Live Females.
In each trial, we placed six virgin males consisting of two bicoloratus, two tarapotensis, and two silvana phenotypes in a 4.0-m long × 3.0-m wide × 2.5-m high outdoor insectary. Each individual could be identified by a distinct dot of paint on the hind wing. Because of the strong genetic basis of preference in Heliconius butterflies (31, 32), and because all males received an individual tag, we assumed that these marks would not interact with our preference records. A single virgin female was introduced into the cage, and the presence/absence of courtship by each individual male (sustained hovering by the male over the female for a minimum of 10 s) was recorded over a period of 8 min. The female phenotype was then substituted until all three phenotypes had been presented to the same group of males. Females of the different morphs were introduced in a random order. When a mating occurred, pairs were quickly and gently separated, which did not disrupt the subsequent behavior. Fifteen different groups of males were formed (i.e., 30 males per phenotype), and each group was tested once a day on six consecutive days. Females were randomly drawn from a pool of 10 to 20 individuals per phenotype. Testing each male six consecutive times enables one to quantify the frequency at which a given male will court each of the three female phenotypes upon an encounter (represented by a trial). This within-subject measure of sexual preference, available for 30 males per phenotype (i.e., 90 observations), was used to assess whether female phenotype had an influence on the odds of male courtship. All analyses were performed using generalized linear mixed models followed by a Tukey’s honestly significant difference post hoc test [package “lme4” (44) in R version 3.1.3 (45)], with the number of trials in which a female was courted vs. not courted as the response variable (binomial response with logit link). The significance of the predictors was tested using likelihood ratio tests. Female phenotype was defined as a fixed effect and male identity as a random effect to control for within-subject repeated measure.
Female Rejection with Artificial Males.
We placed three virgin females simultaneously, one for each of the three phenotypes, in a 2.6-m long × 1.8-m wide × 2.2-m high outdoor insectary. Artificial male butterflies, made daily from the wings of real fresh males attached with clear tape to a 50-cm-long wire stick, were agitated 5 cm above a landed female to simulate male hovering. Once the female lowered her antennae, indicating her receptivity, the presence/absence of rejection behavior, characterized by the female quickly flapping her wings while rapidly lifting her abdomen, was assessed during 20 s. After 10 min of resting, the artificial male phenotype was changed, until all three male phenotypes had been tested on each female. The artificial males displaying different morphs were introduced in a random order. Thirty groups of females were formed (i.e., 30 females per phenotype), and each group was tested once a day on six consecutive days. Statistical analysis was similar to the male preference test, with within-female frequency of rejection toward the three distinct male phenotypes used for conducting generalized linear mixed models, followed by a post hoc test.
Overall Mate Choice Outcome with Live Butterflies.
Realized mating behaviors were assessed using tetrad experiments in 2.6-m long × 1.8-m wide × 2.2-m high outdoor insectaries (46). These mate choice experiments consisted of placing two virgin pairs (male-female) of different phenotypes together and ended with the first mating event. Sixty replicates, using new virgin individuals naive to the setup, were performed for each of the three combinations of morphs. For each combination, a χ2 test of independence was performed to assess whether there are differences between assortative and disassortative morph pairing. When significant, a test of standardized residuals, corrected for multiple comparisons using the Bonferroni method, was performed to identify which combination of morph pairing was significantly more or less frequent than expected under the null hypothesis of equal pairing probabilities with P = 0.05 (ǀ standardized residual ǀ ≥ 2.734 criteria).
Population Genotyping at the Supergene P.
To assess the genotypic composition at the supergene P for the Tarapoto area population, 254 wild-caught individuals captured between 2011 and 2012 were genotyped. Supergene genotypes were assessed by genotyping the H. numata ortholog of HM00025 (cortex) (GenBank accession no. FP236845.2) included in the supergene P and one of the major genes controlling wing color and pattern variation in Lepidoptera (47). Genotypes were derived from indel and single-nucleotide polymorphisms fully associated with wing color pattern phenotype (47). First, total genomic DNA was extracted with the DNeasy Blood and Tissue Kit from Qiagen according to the manufacturer’s protocol. Then, cleaved amplified polymorphic sequences markers allowed us to separate the different alleles. PCR assays were carried out in a 25-μL final volume composed of 2 μL of gDNA, 1× DreamTaq buffer, 200 μM dNTP, 2.5 μM forward primers (5′ CGCAACGTTATCGCCTAGATAGGTTCG 3′), 2.5 μM reverse primers 1 (5′ TAGTGTTAAAGCGAAAGCAC 3′), and 2.5 μM reverse primers 2 (5′ AANGCGAAASMACTGAYAACACGWG 3′) (all three primers from Eurofins), and 0.25 units of DreamTaq DNA polymerase (Thermo Scientific). The PCR amplification temperature profile consisted of an initial denaturation at 94 °C for 2 min followed by 20 cycles at 94 °C for 30 s; a step down from 60 °C to 50 °C for 30 s and 72 °C for 1 min and 30 s followed by another 20 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min and 30 s; and a final elongation at 72 °C for 15 min. Five microliters of the amplification product was run on 1.5% agarose gel electrophoresis. The presence of insertion/deletion between the different allelic sequences in the HM00025s ortholog enabled us to discriminate between the distinct mimetic alleles by PCR product size: Pbic (∼1,200 bp); Psil (∼600 bp); and other alleles, including Ptar and Parc (∼800 bp). In the second step, the cleavage by PstI restriction enzyme (New England Biolabs) according to the manufacturer’s protocol, carried with 10 μL of amplification product, allowed us to discriminate between Ptar, when cleaved, and Parc, when resistant to PstI digestion. Genepop (48) was used to test for exact population genotypic deviation from Hardy–Weinberg equlibrium and to test for either an excess or deficiency of heterozygotes. Because an excess of heterozygotes was detected, to assess which allele was more often found in a heterozygote state, we tested for an excess of heterozygotes for three modified versions of the genotypic dataset (one for each of the alleles Psil, Ptar, and Pbic) for which alleles were restrained, with the allele of interest coded as 001 (present) and all others as 002 (absent).
Acknowledgments
We thank M. McClure and M. Tuatama for assistance with butterfly raising, E. Lievens for help with data analysis, C. Haag for comments on manuscript drafts, and J. R. G. Turner and an anonymous reviewer for their reviews. We also thank the Peruvian government (Grants 236-2012-AG-DGFFS-DGEFFS, 201-2013-MINAGRI-DGFFS/DGEFFS, and 002-2015-SERFOR-DGGSPFFS) and the Proyecto Especial Huallaga Central y Bajo Mayo (PEHCBM)-Area de Conservacion Regional Cordillera Escalera (Grants 086-2012, 020-2014 and 023-2016/GRSM/PEHCBM/DMA/ACR-CE) for providing the necessary research permits. This research was supported by fellowships from the Natural Sciences and Engineering Research Council of Canada and a Marie Sklodowska-Curie Fellowship (FITINV, N 655857) (to M.C.), a French National Agency for Research (ANR) Grant (DOMEVOL, ANR-13-JSV7-0003-01) and an “Emergence” grant from the Paris City Council (to V.L.), and a European Research Council Grant (MimEvol, StG-243179) and an ANR Grant (HYBEVOL, ANR-12-JSV7-0005) (to M.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Endler JA. Natural Selection in the Wild. Princeton Univ Press; Princeton: 1986. [Google Scholar]
- 2.Williams GC. Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought. Princeton Univ Press; Princeton: 1996. [Google Scholar]
- 3.Wellenreuther M, Svensson EI, Hansson B. Sexual selection and genetic colour polymorphisms in animals. Mol Ecol. 2014;23:5398–5414. doi: 10.1111/mec.12935. [DOI] [PubMed] [Google Scholar]
- 4.Servedio MR, Kopp M. Sexual selection and magic traits in speciation with gene flow. Curr Zool. 2012;58:510–516. [Google Scholar]
- 5.Servedio MR, Van Doorn GS, Kopp M, Frame AM, Nosil P. Magic traits in speciation: ‘Magic’ but not rare? Trends Ecol Evol. 2011;26:389–397. doi: 10.1016/j.tree.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Chouteau M, Arias M, Joron M. Warning signals are under positive frequency-dependent selection in nature. Proc Natl Acad Sci USA. 2016;113:2164–2169. doi: 10.1073/pnas.1519216113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller F. Über die vortheile der mimicry bei schmetterlingen. Zool Anz. 1878;1:54–55. German. [Google Scholar]
- 8.Mallet J, Joron M. Evolution of diversity in warning color and mimicry: Polymorphisms, shifting balance, and speciation. Annu Rev Ecol Syst. 1999;30:201–233. [Google Scholar]
- 9.Turner JRG. Why male butterflies are non-mimetic: Natural selection, sexual selection, group selection, modification and sieving. Biol J Linn Soc. 1978;10:385–432. [Google Scholar]
- 10.Chamberlain NL, Hill RI, Kapan DD, Gilbert LE, Kronforst MR. Polymorphic butterfly reveals the missing link in ecological speciation. Science. 2009;326:847–850. doi: 10.1126/science.1179141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chouteau M, Angers B. Wright’s shifting balance theory and the diversification of aposematic signals. PLoS One. 2012;7:e34028. doi: 10.1371/journal.pone.0034028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comeault AA, Noonan BP. Spatial variation in the fitness of divergent aposematic phenotypes of the poison frog, Dendrobates tinctorius. J Evol Biol. 2011;24:1374–1379. doi: 10.1111/j.1420-9101.2011.02258.x. [DOI] [PubMed] [Google Scholar]
- 13.Kapan DD. Three-butterfly system provides a field test of müllerian mimicry. Nature. 2001;409:338–340. doi: 10.1038/35053066. [DOI] [PubMed] [Google Scholar]
- 14.Joron M, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477:203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber B, et al. Conservatism and novelty in the genetic architecture of adaptation in Heliconius butterflies. Heredity (Edinb) 2015;114:515–524. doi: 10.1038/hdy.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joron M, et al. A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 2006;4:e303. doi: 10.1371/journal.pbio.0040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown KS, Benson WW. Adaptive polymorphism associated with multiple Müllerian mimicry in Heliconius numata (Lepid. Nymph) Biotropica. 1974;6:205–228. [Google Scholar]
- 18.Joron M, Wynne IR, Lamas G, Mallet J. Variable selection and the coexistence of multiple mimetic forms of the butterfly Heliconius numata. Evol Ecol. 1999;13:721–754. [Google Scholar]
- 19.Arias M, et al. Crossing fitness valleys: Empirical estimation of a fitness landscape associated with polymorphic mimicry. Proc Biol Sci. 2016;283:20160391. doi: 10.1098/rspb.2016.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkbeiner SD, Briscoe AD, Reed RD. Warning signals are seductive: Relative contributions of color and pattern to predator avoidance and mate attraction in Heliconius butterflies. Evolution. 2014;68:3410–3420. doi: 10.1111/evo.12524. [DOI] [PubMed] [Google Scholar]
- 21.Maan ME, Cummings ME. Sexual dimorphism and directional sexual selection on aposematic signals in a poison frog. Proc Natl Acad Sci USA. 2009;106:19072–19077. doi: 10.1073/pnas.0903327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merrill RM, Chia A, Nadeau NJ. Divergent warning patterns contribute to assortative mating between incipient Heliconius species. Ecol Evol. 2014;4:911–917. doi: 10.1002/ece3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Twomey E, Vestergaard JS, Summers K. Reproductive isolation related to mimetic divergence in the poison frog Ranitomeya imitator. Nat Commun. 2014;5:4749. doi: 10.1038/ncomms5749. [DOI] [PubMed] [Google Scholar]
- 24.Hughes KA, Houde AE, Price AC, Rodd FH. Mating advantage for rare males in wild guppy populations. Nature. 2013;503:108–110. doi: 10.1038/nature12717. [DOI] [PubMed] [Google Scholar]
- 25.Hedrick PW, Smith DW, Stahler DR. Negative-assortative mating for color in wolves. Evolution. 2016;70:757–766. doi: 10.1111/evo.12906. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi T, Hori M. Evidence of disassortative mating in a Tanganyikan cichlid fish and its role in the maintenance of intrapopulation dimorphism. Biol Lett. 2008;4:497–499. doi: 10.1098/rsbl.2008.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuttle EM, et al. Divergence and functional degradation of a sex chromosome-like supergene. Curr Biol. 2016;26:344–350. doi: 10.1016/j.cub.2015.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press; Oxford: 1930. [Google Scholar]
- 29.Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castric V, Vekemans X. Plant self-incompatibility in natural populations: A critical assessment of recent theoretical and empirical advances. Mol Ecol. 2004;13:2873–2889. doi: 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- 31.Kronforst MR, et al. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc Natl Acad Sci USA. 2006;103:6575–6580. doi: 10.1073/pnas.0509685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrill RM, Van Schooten B, Scott JA, Jiggins CD. Pervasive genetic associations between traits causing reproductive isolation in Heliconius butterflies. Proc Biol Sci. 2011;278:511–518. doi: 10.1098/rspb.2010.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrill RM, et al. Disruptive ecological selection on a mating cue. Proc Biol Sci. 2012;279:4907–4913. doi: 10.1098/rspb.2012.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chouteau M, Whibley A, Angers B, Joron M. Development of microsatellite loci from a reference genome for the Neotropical butterfly Heliconius numata and its close relatives. Entomol Sci. 2015;18:283–287. [Google Scholar]
- 35.Billiard S, Castric V, Vekemans X. A general model to explore complex dominance patterns in plant sporophytic self-incompatibility systems. Genetics. 2007;175:1351–1369. doi: 10.1534/genetics.105.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mays HL, Jr, Hill GE. Choosing mates: Good genes versus genes that are a good fit. Trends Ecol Evol. 2004;19:554–559. doi: 10.1016/j.tree.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Tregenza T, Wedell N. Genetic compatibility, mate choice and patterns of parentage: Invited review. Mol Ecol. 2000;9:1013–1027. doi: 10.1046/j.1365-294x.2000.00964.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, et al. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493:664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- 39.Küpper C, et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 2016;48:79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner CE, Harmon LJ, Seehausen O. Ecological opportunity and sexual selection together predict adaptive radiation. Nature. 2012;487:366–369. doi: 10.1038/nature11144. [DOI] [PubMed] [Google Scholar]
- 42.Merrill RM, et al. The diversification of Heliconius butterflies: What have we learned in 150 years? J Evol Biol. 2015;28:1417–1438. doi: 10.1111/jeb.12672. [DOI] [PubMed] [Google Scholar]
- 43.Owen DF, Smith DAS, Gordon IJ, Owiny AM. Polymorphic Müllerian mimicry in a group of African butterflies–A reassessment of the relationship between Daneus chrysippus, Acraea encedon and Acraea encedana (Lepidoptera, Nymphalidae) J Zool. 1994;232:93–108. [Google Scholar]
- 44.Bates D, Maechler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Software. 2015;67:1–48. [Google Scholar]
- 45.R Core Team 2013 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at www.R-project.org/. Accessed April 17, 2017.
- 46.Jiggins CD, Naisbit RE, Coe RL, Mallet J. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411:302–305. doi: 10.1038/35077075. [DOI] [PubMed] [Google Scholar]
- 47.Nadeau NJ, et al. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature. 2016;534:106–110. doi: 10.1038/nature17961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]