Significance
Under certain conditions during transcription, a single base of the template DNA specifies multiple bases in the RNA transcript due to slippage between the transcript and template. This noncanonical form of RNA synthesis is called “reiterative transcription,” and it plays key regulatory roles in bacteria, eukaryotes, and viruses. In this study, we determined the crystal structure of a bacterial RNA polymerase engaged in reiterative transcription. Our study found a completely unexpected RNA extension pathway during reiterative transcription and uncovered an atypical mechanism for the transition from the open promoter complex to the transcription elongation complex. These findings represent a major advancement in understanding the mechanics and flexibility of transcription.
Keywords: reiterative transcription, RNA polymerase, X-ray crystal structure, transcription initiation, transcript slippage
Abstract
Reiterative transcription is a noncanonical form of RNA synthesis in which a nucleotide specified by a single base in the DNA template is repetitively added to the nascent transcript. Here we determined the crystal structure of an RNA polymerase, the bacterial enzyme from Thermus thermophilus, engaged in reiterative transcription during transcription initiation at a promoter resembling the pyrG promoter of Bacillus subtilis. The structure reveals that the reiterative transcript detours from the dedicated RNA exit channel and extends toward the main channel of the enzyme, thereby allowing RNA extension without displacement of the promoter recognition σ-factor. Nascent transcripts containing reiteratively added G residues are eventually extended by nonreiterative transcription, revealing an atypical pathway for the formation of a transcription elongation complex.
In the canonical form of transcription, RNA polymerase (RNAP) reads the sequence of a template strand DNA one base at a time to produce a complementary strand of RNA. However, in some instances of transcription, a single base in the template DNA can specify multiple bases in the RNA product (1–4). This process is due to multiple rounds of upstream slippage of the RNA without translocation of the template DNA within the active site of RNAP (5). This unconventional reaction is called “reiterative transcription,” and it occurs primarily within a homopolymeric tract in the template DNA. Reiterative transcription can involve the repetitive addition of any nucleotide, and it can occur during all phases of the transcription cycle. During initiation, a homopolymeric tract as short as three residues can enable reiterative transcription (6–8), whereas a significantly longer homopolymeric tract is required during elongation and termination (9–12). This difference reflects the length of the obligatory DNA/RNA hybrid that forms within all transcription complexes, which during initiation can be shorter than the ∼9-bp hybrid that forms during elongation and termination (10, 13).
Reiterative transcription plays key roles in gene expression in eukaryotes, viruses, and especially bacteria, where it has been shown to control gene expression through a variety of mechanisms (1, 3). The earliest example was UTP-sensitive regulation of transcription initiation of the pyrBI operon of Escherichia coli, which encodes two subunits of the pyrimidine nucleotide biosynthetic enzyme aspartate transcarbamylase (14). In this case, reiterative transcription occurs within a T3 tract located in the initially transcribed region 5′-AATTTG (nontemplate strand sequence) of the pyrBI promoter. Reiterative transcription produces transcripts with the sequence 5′-AAUUUUn (where n = 1 to >100), essentially all of which are released from the transcription initiation complex. The extent of this nonproductive reiterative transcription is directly proportional to the intracellular level of UTP, so that pyrBI gene expression is reduced when UTP levels are high. Several other regulatory mechanisms were subsequently discovered that were similar to the pyrBI mechanism in that they relied on variable reiterative transcription at TTTn tracts in initially transcribed regions to produce transcripts that were released immediately after repetitive UMP addition, thereby repressing gene expression (2).
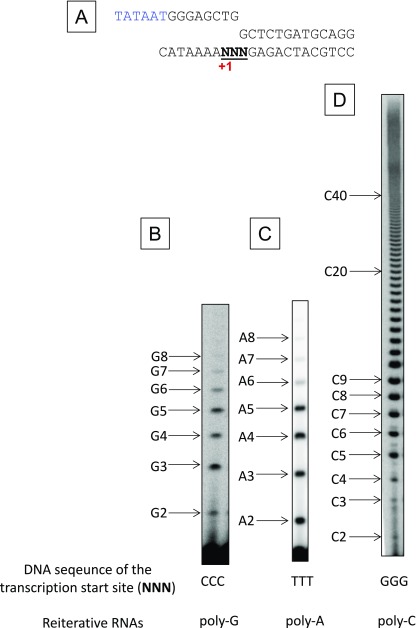
Another well-studied example is CTP-mediated control of pyrG expression in Bacillus subtilis, which produces the pyrimidine biosynthetic enzyme CTP synthetase (15). The nontemplate DNA sequence of the pyrG initially transcribed region is 5′-GGGCT, with transcription initiated predominantly at the first G residue (Fig. 1A). Low intracellular levels of CTP cause transcription pausing at position +4C, which provides time needed for upstream transcript slippage and reiterative transcription to occur at the end of the 5′-CCC tract of the DNA template. This reaction results in the addition of up to nine extra G residues at the 5′ end of the pyrG transcript, after which the transcript is extended by nonreiterative transcription through the pyrG leader region. The extra G tract then forms an antiterminator hairpin with a downstream segment of the nascent transcript that precludes intrinsic transcription termination at an attenuator preceding the pyrG gene. As a result, transcription proceeds through the pyrG gene leading to the production of more CTP synthetase and increased levels of CTP.
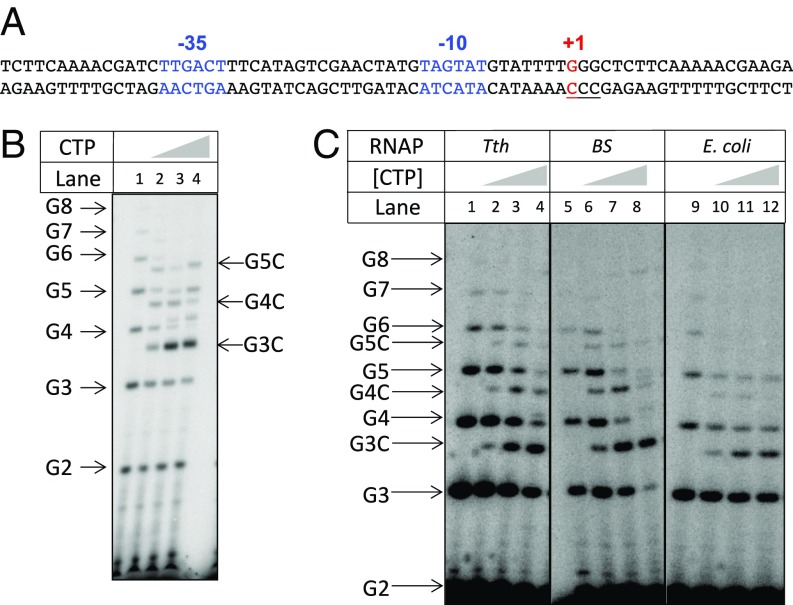
Fig. 1.
Characterization of reiterative transcription by the T. thermophilus RNAP. (A) DNA sequence of the B. subtilis pyrG promoter region. The −35 and −10 regions (blue) and the +1 transcription start site (red) are indicated. The template strand CCC track (from +1 to +3) that permits reiterative transcription is underlined. (B) In vitro reiterative transcription by the T. thermophilus RNAP using a double-stranded DNA containing the pyrG promoter shown in A as template. Transcription assays were performed in the presence of GTP (100 μM GTP plus 1 μCi [γ-32P]GTP) and different concentrations of CTP (0, 1, 10, and 100 μM). The positions of the reiterative poly-G and the poly-G + C products are indicated. Note that poly-G transcripts migrate slower than same-length poly-G + C transcripts. (C) In vitro reiterative transcription with the synthetic scaffold shown in Fig. 2A and different RNAPs. Transcription assays were performed as in B. RNAPs used for these reactions are indicated (Tth, T. thermophilus; BS, B. subtilis).
Although reiterative transcription clearly plays important roles in controlling gene expression, much remains to be learned about the mechanism of this reaction and how it differs from canonical transcription. This is particularly true for the positioning of the nascent RNA inside RNAP. In this study, we have explored this question by preparing a reiterative transcription complex by in crystallo transcription and determining the X-ray crystal structure of a bacterial RNAP in the process of reiterative transcription at a promoter with the same critical features as the pyrG promoter of B. subtilis.
Results
Preparation and Structure of a Reiterative Transcription Complex.
We chose the readily crystallized Thermus thermophilus RNAP σA holoenzyme to examine the structure of a reiterative transcription complex (RTC). To validate reiterative transcription with the T. thermophilus RNAP, we performed in vitro transcription using a double-stranded DNA fragment containing the pyrG promoter of B. subtilis as template (Fig. 1A). The result showed that the T. thermophilus RNAP was capable of producing poly-G transcripts up to eight bases in length in the presence of GTP (Fig. 1B). In the presence of both GTP and CTP, a single C residue was incorporated at the 3′ end of the G3–8 transcripts, and the extent of C-residue incorporation was directly proportional to the concentration of CTP. These results indicate that the T. thermophilus RNAP is proficient at reiterative transcription and that this reaction is comparable to that observed naturally at the pyrG promoter (16).
For the formation of the RTC, we designed a synthetic DNA scaffold corresponding to the transcription bubble and downstream double-stranded DNA of the pyrG promoter (Fig. 2A). To confirm reiterative transcription with the synthetic scaffold as template, it was transcribed in vitro with T. thermophilus RNAP. Under the same reaction conditions as described in Fig. 1B, reiterative transcription was essentially the same as that observed with the natural pyrG promoter (Fig. 1C). Similar results were obtained when the synthetic scaffold was transcribed by the primary RNAPs of B. subtilis and E. coli (Fig. 1C), indicating the universal nature of the reiterative transcription reaction in bacteria.
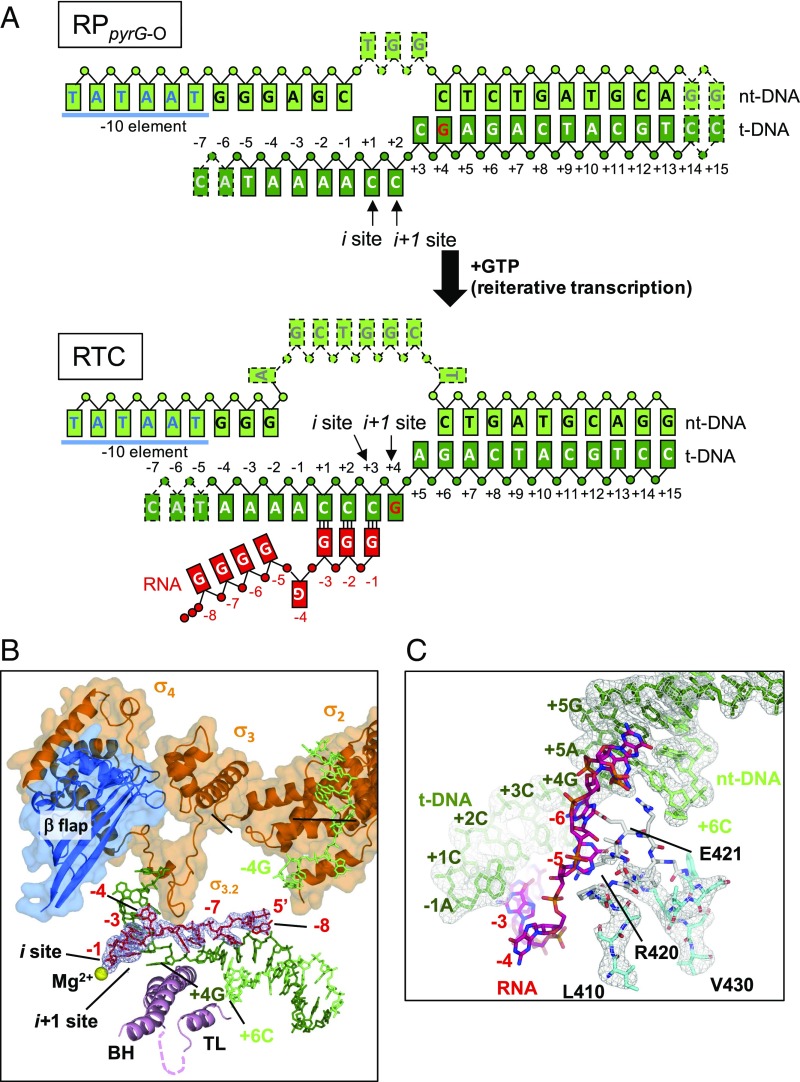
Fig. 2.
Structure of the reiterative transcription complex. (A) Schematic representation of the RTC formed by the RPpyrG-O in the presence of GTP. The DNA scaffold used for crystallization and reiterative transcript are depicted in boxes [template DNA (t-DNA)], dark green; nontemplate DNA (nt-DNA), light green; RNA, red; the same colors are used in all other figures). DNA bases disordered in the structures are indicated by dashed boxes. Positions of DNA and RNA bases are indicated by numbers. (B) Ribbon models of RNAP motifs (BH, bridge helix; TL, trigger loop) are shown with transparent surfaces of the σ-factor and the β-flap domain. RNA and DNA are shown as stick models. The disordered region of the TL is shown as a dashed line. The nucleotide binding sites (i and i+1) are indicated, and the active site catalytic Mg2+ is shown as a yellow sphere. The 2Fo–Fc electron density for RNA is shown (gray mesh, 1.5 σ). The RNAP orientation in this figure is similar to Fig. 3A. (C) Stick models of the DNA, RNA, and fork loop 2 in the RTC are shown with 2Fo–Fc electron densities (gray mesh, 1.5 σ) of the DNA and fork loop 2. The disordered part of fork loop 2 (white stick model) is modeled from fork loop 2 of the RPpyrG-O after superimposing these structures.
Using T. thermophilus RNAP and the synthetic DNA scaffold, we formed and crystallized an open complex (RPpyrG-O) as described in Experimental Procedures. The structure of the RPpyrG-O was determined at 2.8-Å resolution to use as a reference in the structural analysis of the RTC (Table S1, Fig. S1, and see description in SI Text). To verify that the RPpyrG-O remained transcriptionally active in the crystalline state, as observed with other crystals of T. thermophilus RNAP (17, 18), we performed in crystallo reiterative transcription by soaking GTP into the RPpyrG-O crystals. The results showed that the crystals produced and retained poly-G transcripts up to 8 nt in length (Fig. S2), similar to the transcripts produced in solution (Fig. 1C). We then soaked a crystal of RPpyrG-O in GTP to trigger reiterative transcription and determined the structure of the resulting RTC at 3.3-Å resolution (Table S1). The structure shows continuous electron density from the RNAP active site that corresponds to a de novo synthesized poly-G transcript containing eight residues (Fig. 2B; hereafter, RNA residues are counted −1, −2, etc. from the 3′ end), which is the same length as the longest poly-G transcript synthesized in solution (Fig. 1 B and C). The electron density of poly-G transcript is gradually decreased from positions −5 to −8, presumably due to less RNA extension beyond a 5-mer transcript in the RTC crystal (Fig. S2) and/or thermal motion of single-stranded RNA. The 3′ end of the RNA is in a posttranslocated state (i.e., in the i site), forming a base pair with template DNA residue +3C, whereas the +4G base is positioned at the i+1 site, waiting for a molecule of CTP to extend the nascent RNA by nonreiterative transcription (Fig. 2B).
Table S1.
Data collection and refinement statistics
| Complex | RPpyrG-O | RTC |
| PDB code | 5VOI | 5VO8 |
| Data collection | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a (Å) | 183.18 | 186.54 |
| b (Å) | 103.47 | 101.98 |
| c (Å) | 294.44 | 296.11 |
| β (°) | 99.14 | 98.56 |
| Resolution (Å) | 30–2.80 | 30–3.30 |
| Total reflections | 440,744 | 253,251 |
| Unique reflections | 132,496 | 78,332 |
| Redundancy | 3.3 (2.3) | 3.2 (2.7) |
| Completeness (%) | 98.2 (89.5) | 94.2 (84.1) |
| I/σ | 8.6 (1.67) | 9.9 (2.11) |
| Wilson B factor | 47.8 | 81.8 |
| Rsym (%) | 10.3 (66.5) | 8.7 (47.1) |
| CC1/2 | (0.474) | (0.743) |
| Refinement | ||
| Resolution (Å) | 30–2.80 | 30–3.30 |
| Rwork | 0.192 | 0.185 |
| Rfree | 0.236 | 0.246 |
| No. of atoms | 28,432 | 28,563 |
| B factors | 72.3 | 123.6 |
| R.m.s deviations | ||
| Bond length (Å) | 0.014 | 0.013 |
| Bond angles (°) | 1.48 | 1.58 |
| Clashscore | 9.5 | 12.4 |
| Ramachandran favored, % | 98.2 | 96.4 |
| Ramachandran outliers, % | 0.06 | 0.87 |
Datasets were collected at MacCHESS F1 line, Ithaca, NY. Highest resolution shells are shown in parentheses.
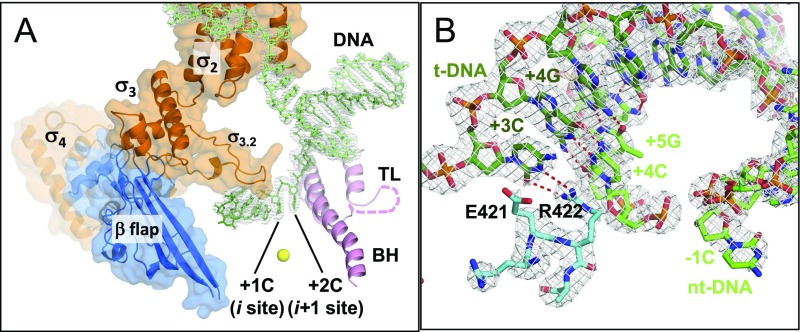
Fig. S1.
Structures of the T. thermophilus RNAP holoenzyme DNA complex mimicking the pyrG promoter open complex. (A) Ribbon models of RNAP domains and motifs are shown with transparent surfaces of the σ-factor and the β-flap domain (TL, trigger loop; BH, bridge helix). DNA strands are shown as stick models (template DNA, dark green; nontemplate DNA, light green). The disordered region of the TL is shown as a dashed line. The nucleotide binding sites (i and i+1) are indicated, and the active site catalytic Mg2+ is shown as a yellow sphere. The gray mesh shows the 2Fo–Fc electron density for the DNA strands contoured at 1.5 σ. (B) Stick models of the downstream DNA and the β-subunit fork loop 2 are shown with their 2Fo–Fc electron densities (gray mesh) contoured at 1.5 σ. Hydrogen bonds between DNA bases and the salt bridge between +3G and R422 are depicted as red dashed lines.
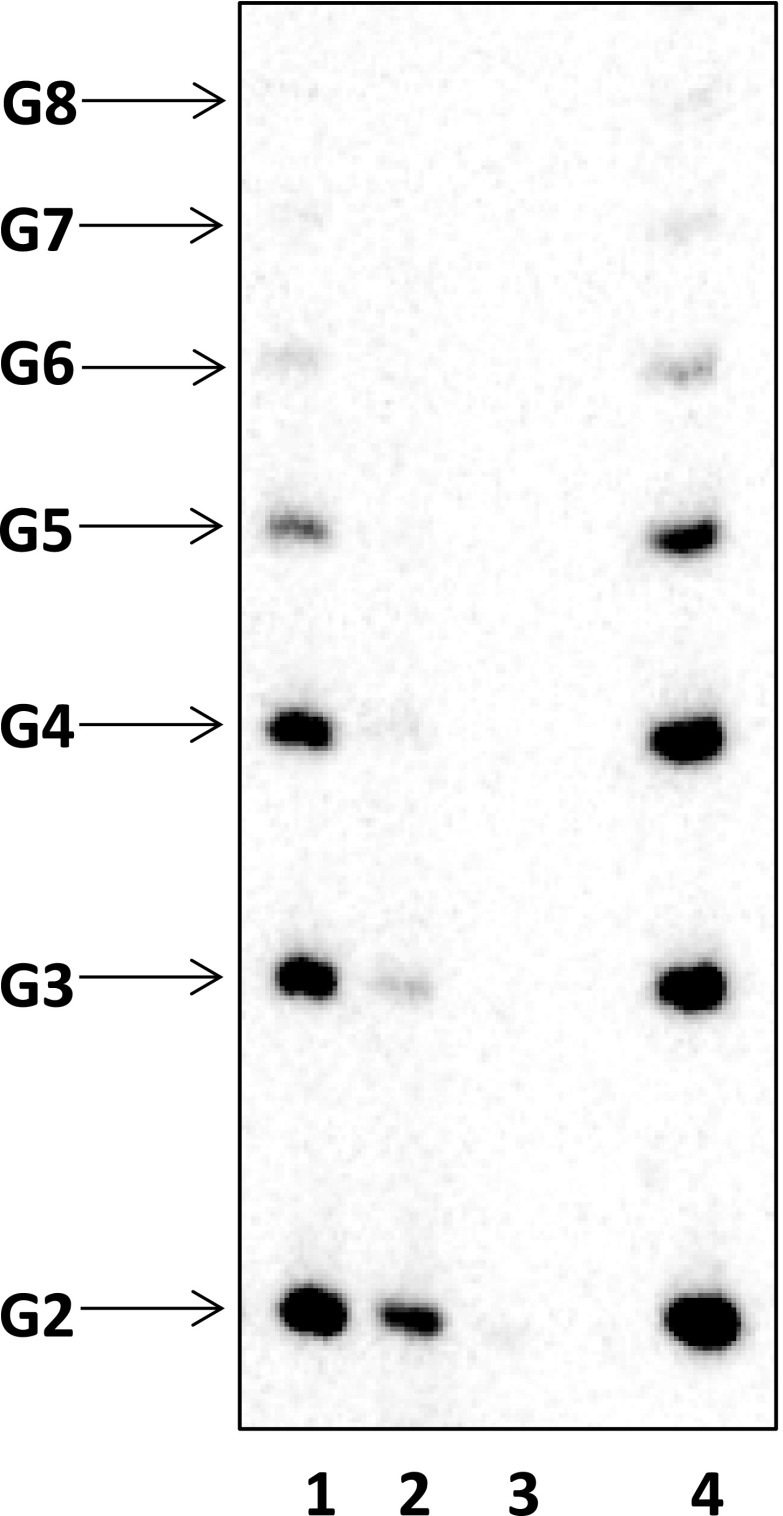
Fig. S2.
Reiterative transcripts in the RTC crystals. In crystallo transcription on pyrG promoter scaffold (Fig. 2A) was performed in the presence of 400 µM GTP and [γ-32P]GTP. (Lane 1) RNAs released from crystals to supernatant in the course of reaction. (Lanes 2 and 3) RNAs released from crystals during two consecutive washes with the crystallization solution without GTP. (Lane 4) RNAs retained in the RTC crystals.
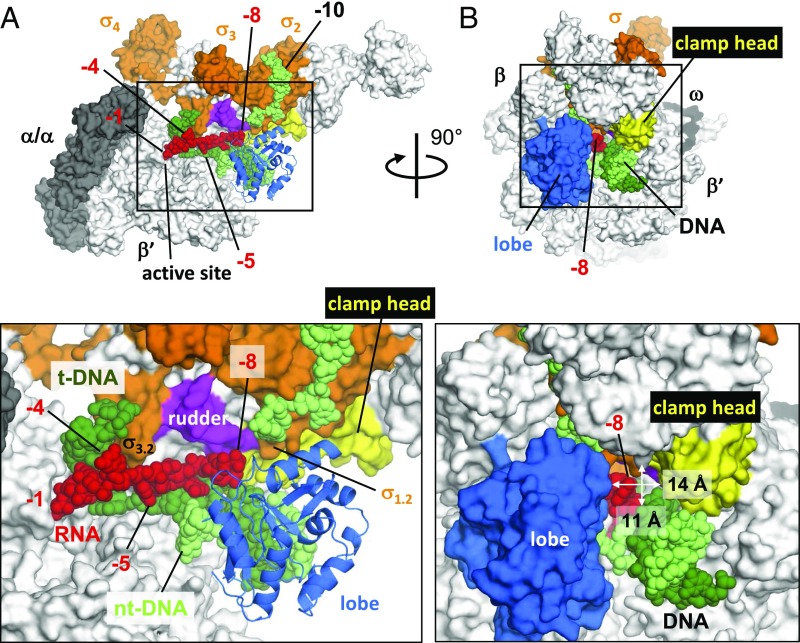
Viewing the overall structure of the RTC reveals the location of the reiterative transcript within RNAP (Fig. 3). Surprisingly, the nascent RNA does not extend toward the dedicated RNA exit channel; instead, it extends toward the main channel of the enzyme. RNA residues −5G and −6G are located near the downstream DNA and fork loop 2 of the β-subunit (residues 414–424), which appears disordered (Fig. 2C). RNA residues pass beneath σ-region 3.2 (σ3.2), and the 5′ end of the RNA approaches the entrance of the RNAP main channel (Fig. 3A). The terminal RNA residue (−8G) is surrounded by the lobe domain (β-subunit), the rudder, and the clamp head (β′-subunit), the N terminus of σ-region 1.2, and the major groove of the downstream double-stranded DNA (Fig. 3 A and B). The opening into the main channel that would allow the RNA to escape from RNAP is only 14 Å wide and 11 Å high (Fig. 3B), which could present a barrier to further RNA extension.
Fig. 3.
Position of the nascent RNA in the reiterative transcription complex. The overall structure of the RTC is shown with RNAP depicted as a molecular surface model (α, gray; β′, white; σ, orange). RNA, t-DNA, and nt-DNA strands are shown as sphere models and labeled. For clarity, all of the β-subunit except a ribbon model of the lobe domain was removed in A. RNAP motifs discussed in the text are highlighted and labeled. The structure in B is the same as that in A, except that it is rotated clockwise 90°. Bottom shows magnified views of the boxed regions at Top.
To test this barrier hypothesis, we performed in vitro transcription using as template DNA two variants of the pyrG-like DNA scaffold in which the initially transcribed sequence was changed from C3 to either T3 or G3 (Fig. S3A). In the presence of only the corresponding initiating nucleotides ATP or CTP, T. thermophilus RNAP efficiently synthesized reiterative transcripts with both promoter variants (Fig. S3 C and D). With the T3 promoter, a ladder of poly-A transcripts up to 8 residues in length was produced, the same maximum length observed with the standard template and poly-G transcripts (Fig. S3 B and C). In contrast, with the G3 promoter, a ladder of poly-C transcripts with lengths easily exceeding 40 residues was produced. These results, and the fact that pyrimidines are smaller than purines, suggest that the maximum length of reiterative transcripts was defined by steric restrictions imposed by the narrow opening into the main channel (Fig. 3B).
Fig. S3.
Composition of the homopolymeric tract determines the maximum length of reiterative transcripts. In vitro transcription assays were performed with the DNA scaffold containing pyrG variants (A) having homopolymeric tract in the initially transcribed template DNA (sequences of the transcription start site of each template and their RNA products are indicated below B–D). Transcription assays were performed in the presence of GTP (B), ATP (C), or CTP (D) {100 μM NTP plus 1 μCi [γ-32P]GTP, [γ-32P]ATP, or [α-32P]CTP}. The positions of the reiterative products are indicated.
An Alternative RNA Extension Pathway During Reiterative Transcription.
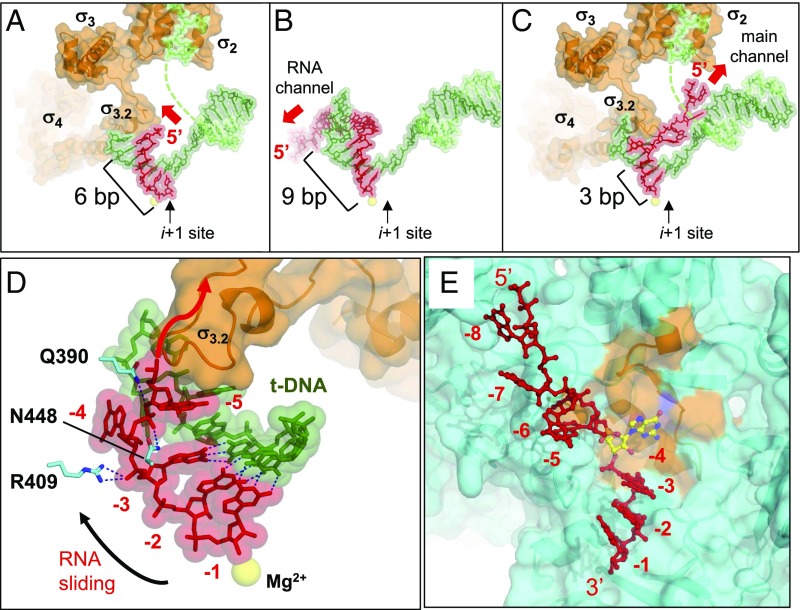
The extension strategy of nascent RNA in the RTC (Fig. 4C) is strikingly different from that exhibited by previously described crystal structures of comparable transcribing complexes containing nonreiterative transcripts (Fig. 4 A and B). In these cases, RNA is directed toward and eventually into a dedicated RNA exit channel located underneath the flap domain (β-subunit). For example, in an initially transcribing complex containing a 6-mer RNA, the 5′ end of RNA abuts σ3.2 [Fig. 4A, Protein Data Bank (PDB): 4G7H] (17). This clash causes the tip of σ3.2 to become disordered, which presumably marks the beginning of σ-release from RNAP. Further RNA extension would completely displace σ3.2, thereby clearing the way for RNA to enter the RNA exit channel (Fig. 4B, PDB: 2O5J) (13, 19). To explore the importance of the 5′-triphosphate group in transcript extension during reiterative transcription, we used the dinucleotide pGpG, which contains a 5′-monophosphate group, to initiate reiterative transcription at the scaffold pyrG promoter. The transcript ladder produced was identical to that obtained by transcription with GTP as sole substrate (Fig. S4).
Fig. 4.
Comparison of RNA paths within different transcription complexes. (A–C) Comparable views of the RNA in (A) initially transcribing complex (PDB: 4G7H), (B) elongation complex (PDB: 2O5J), and (C) reiterative transcription complex (PDB: 5VO8). The σ-factor, DNA, and RNA strands are depicted. The i+1 site is indicated and the catalytic Mg2+ is shown as a yellow sphere. The directions of RNA extension are indicated by red arrows. (D) Another view of the RTC highlights RNA extension without σ-displacement. The DNA, RNA, and σ3.2 are depicted as in C. Hydrogen bonds between the DNA and RNA bases within the 3 bp of the DNA/RNA hybrid and hydrogen bonds/salt bridges between the amino acids and the RNA are shown as blue dashed lines. (E) View of the RTC shows the insertion of RNA base into the rifampin-binding pocket (β-subunit residues 387–413, orange). RNA is shown as a red stick model except −4G (yellow stick). The β-subunit is depicted as a ribbon model with partially transparent surface.
Fig. S4.
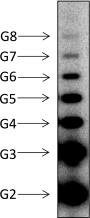
The effect of the phosphate group on reiterative transcription with the synthetic pyrG scaffold shown in Fig. 2A. Transcription assay was performed in the presence of 32P-labeled pGpG primer containing 5′ monophosphate group and 20 μM GTP with T. thermophilus RNAP. The positions of the reiterative poly-G products are indicated.
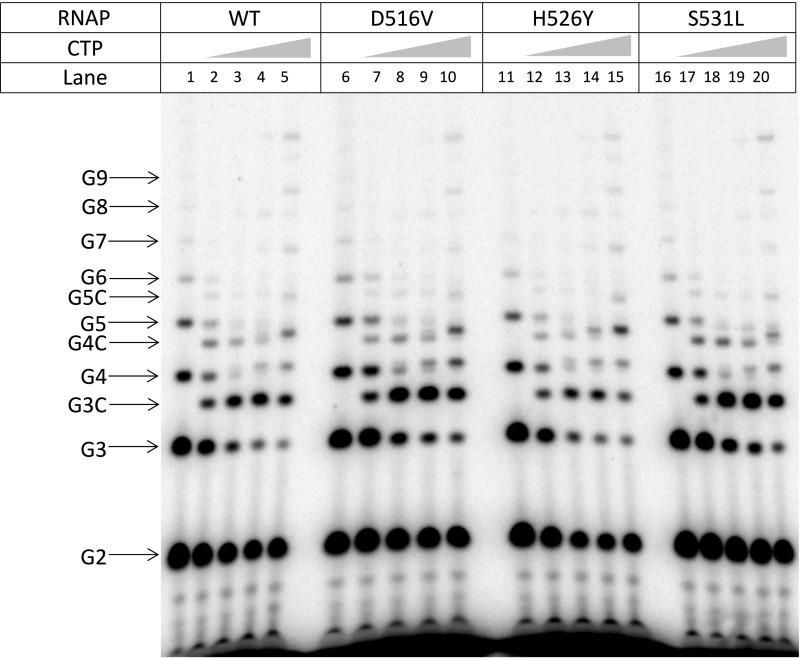
In both the initially transcribing (Fig. 4A) and elongation complexes (Fig. 4B), the 5′ end of RNA is guided toward the RNA exit channel by an extended DNA/RNA hybrid (i.e., 6 bp and an ∼9 bp, respectively). However, in the case of a RTC containing the synthetic pyrG promoter, the DNA/RNA hybrid is only 3 bp long (Fig. 4C). The remaining nonbase-pairing residues of the RNA are turned perpendicular to the DNA/RNA hybrid and directed toward the main channel, without displacement of σ3.2. RNA residue −4G is inserted into a pocket on the surface of the β-subunit known as the rifampin-binding pocket (also known as the rifampin-resistance determining region I) (Fig. 4E). To determine the importance of the interaction between RNA and the rifampin-binding pocket during reiterative transcription, we measured reiterative transcription from the pyrG promoter in solution with three rifampin-resistant mutants of E. coli RNAP having amino acid substitutions either at D516V, H526Y, or S531L of the β-subunit. The D516V and H526Y mutants alter the electrostatic potential and the shape of the rifampin-binding pocket, respectively, whereas the S531L mutant replaces a hydrophilic with a hydrophobic side chain in the rifampin-binding pocket (20). We found that all mutants synthesize equal amounts of reiterative transcripts and that the efficiencies of CMP incorporation to the 3′ end of poly-G transcripts are equally dependent on CTP concentration in mutant and wild-type RNAPs (Fig. S5). These results suggest that the rifampin-binding pocket functions merely to shelter an unpaired RNA base after the transcript changes its direction from the dedicated RNA exit to the main channels of RNAP.
Fig. S5.
Reiterative transcription with the synthetic scaffold and rifampin-resistant RNAPs in vitro. Transcription assays were performed in the presence of GTP {100 μM GTP plus 1 μCi [γ-32P]GTP} and different concentrations of CTP (0, 10, 100, 500, or 1,000 μM). E. coli RNAPs including the wild-type and rifampin-resistant enzymes used for the reactions are indicated. The positions of the reiterative poly-G and the poly-G + C products are indicated. Note that poly-G transcripts migrate slower than same-length poly-G + C transcripts.
The typical transcription elongation complex is optimally stabilized by an ∼9-bp DNA/RNA hybrid (13, 19). In contrast, the RTC at the pyrG promoter demonstrates that a 3-bp hybrid, with a calculated Tm of 17 °C (OligoCalc, in the presence of 100 mM Na+) (21), is sufficient to maintain the RNA in the transcription complex and to allow its extension one base at a time. The 3-bp hybrid does this while still being weak enough to allow the upstream slippage of the RNA, which is a central feature of reiterative transcription. RNA residues from −3G to −5G interact with amino acid residues Q390, R409, and N448 of the β-subunit, which presumably would limit diffusion of the nonbase-paired region of the RNA (Fig. 4D).
SI Text
We crystallized the T. thermophilus RNAP holoenzyme bound to the synthetic DNA scaffold (RNAP–pyrG promoter open complex, RPpyrG-O) and determined its structure at 2.8-Å resolution (Table S1) to use as a reference in the structural analysis of the reiterative transcription complex (RTC). The overall shape of RPpyrG-O is identical to previously reported T. thermophilus RNAP–promoter open complex (RPO) structures (17, 27), having the straight conformation of the bridge helix and the open conformation of the trigger loop with a disordered tip (residues 1,238–1,253) (Fig. S1A). The high-resolution electron density map of the RPpyrG-O resolved individual DNA bases, which unambiguously determined the register of DNA sequence. The +1C and +2C bases of the template DNA are positioned at the i and i+1 sites, respectively, ready to base pair with two molecules of GTP (Fig. 2A and Fig. S1A). The +3C base of the template DNA is located at the i+2 site (Fig. S1B), which is on the opposite side of the bridge helix and at the edge of downstream dsDNA. In the previously described RPO structures, the +3 bases of the template and nontemplate strands were base paired. However, in the RPpyrG-O, the +3 base of the nontemplate DNA is unwound and disordered (which is also observed with the +1 and +2 bases of the nontemplate DNA). The R422 side chain of fork loop 2 is located in front of +3C of the template DNA and forms Watson–Crick-like hydrogen bonds and a cation–π interaction with +4G of the nontemplate DNA, which facilitates positioning of downstream DNA (Fig. S1B). The significance of an unwound +3 position and the interaction between R422 and +3C in terms of reiterative transcription remains to be elucidated.
Discussion
This study reports the structure of a bacterial RNAP in the process of reiterative transcription, revealing a unique mechanism for extending RNA within the transcription initiation complex. This process includes an atypical RNA extension pathway, a stably maintained 3-bp DNA/RNA hybrid, and extension of the nascent RNA without displacement of σ3.2.
Elucidating the mechanism of reiterative transcription at the pyrG promoter in solution is difficult because reiterative transcripts are quickly released from the initiation complex, only a minor fraction of transcription complexes contain the longest 8-mer poly-G transcript at any given time (Fig. 1 B and C), and the stochastic nature of RNA slippage prevents control of reiterative transcript length. However, using in crystallo transcription, we were able to prepare a stable reiterative transcription complex containing poly-G transcripts up to 8 nt in length. Direct analysis of the composition of the RNA transcripts in crystals shows a mixed population varying in length from 2 to 8 nt (Fig. S2), similar to transcripts produced during reiterative transcription in solution (Fig. 1C). Although this heterogeneity results in a gradual decrease of the RNA electron density from residues −5 to −8, all transcripts appear to follow the atypical exit pathway as evidenced by the lack of any electron density along the canonical RNA exit pathway. Thus, the summation of RNA electron densities from somewhat heterogeneous reiterative transcription complexes in a single crystal provides a means for establishing the location of an otherwise unstable reiterative RNA. The crystalline state of T. thermophilus RNAP–promoter DNA complex has a high solvent content (∼60%), is fully hydrated to allow nucleotide access to the enzyme active site, and can produce either a canonical RNA transcript with 6-bp DNA/RNA hybrid (17) or a reiterative transcript with a 3-bp DNA/RNA hybrid (this study) depending on only the initially transcribed region of the DNA template and the nucleotide(s) soaked into the crystal. The unique, ideal, and near physiological environment of RNAP in crystallo captures a hitherto undetected and unstable intermediate by freezing crystals, the structure of which reveals an atypical RNA extension pathway at atomic resolution.
Alternative Pathway for Transcription Elongation Complex Formation.
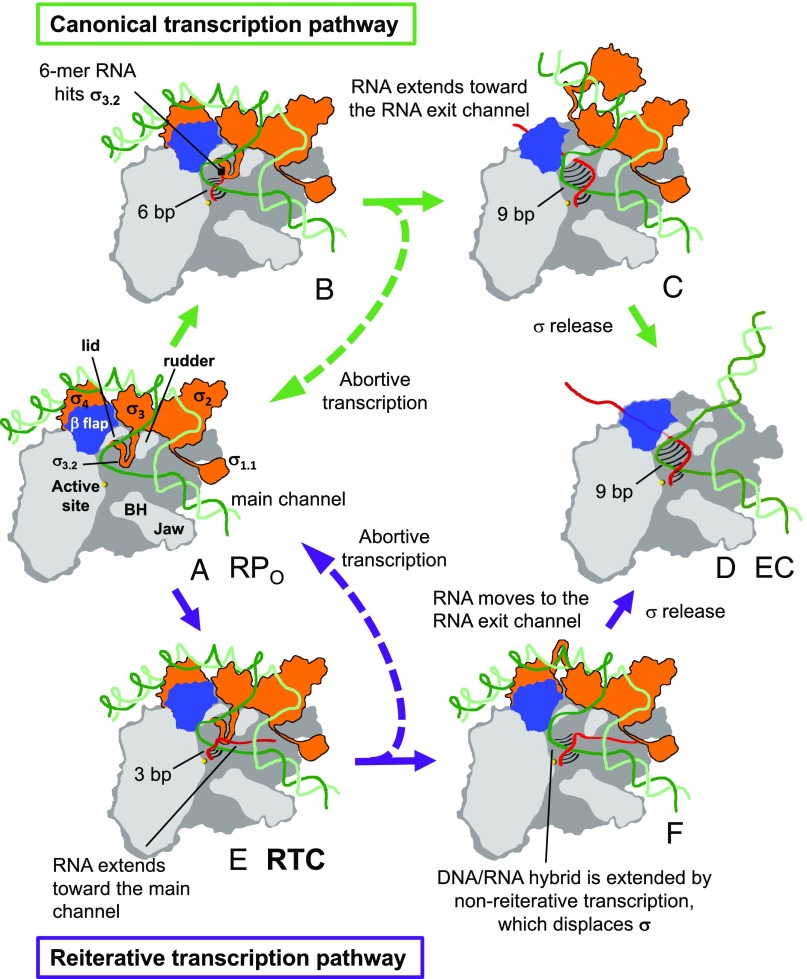
Because pyrG transcripts containing extra G residues at their 5′ ends are eventually fully extended by canonical transcription in B. subtilis, our results reveal an alternative pathway for the formation of a transcription elongation complex (Fig. 5). During reiterative transcription at the pyrG promoter, only a 3-bp DNA/RNA hybrid forms, and the first 5′-end RNA base not included in the DNA/RNA hybrid fills the rifampin-binding pocket (Fig. 5E). The 5′-end RNA is redirected away from σ3.2 and toward the main channel. At some point during further RNA extension up to approximately eight bases, there can be a switch to nonreiterative transcription (Fig. 5F). This switch is accompanied by an increase in the length of the DNA/RNA hybrid, which would displace σ3.2 to expose the dedicated RNA exit channel of the core enzyme (Fig. 5F). Possibly, the 5′ end of the nascent RNA transcript is then redirected toward the RNA exit channel. We speculate that reiterative transcription pauses when the 5′-end RNA reaches the narrow opening of the main channel of RNAP (Fig. 3C and Fig. S3), providing time for CTP to be incorporated at the 3′ end of RNA and the switch from reiterative to canonical transcriptions. Increasing the length of the DNA/RNA hybrid triggers the eventual displacement of σ from core enzyme (Fig. 5F). Without σ-factor, the main and RNA exit channels are separated only by a weak interaction between the lid region (β′-subunit) and the inner surface of the flap domain (β-subunit) (22), which can be disrupted by opening of the RNAP clamp and/or flap (23). Therefore, disruption of this interaction during the transition from reiterative to nonreiterative transcription might be involved in redirecting the RNA toward the dedicated RNA exit channel.
Fig. 5.
Alternative pathways for transcription elongation complex formation. Two distinct pathways are shown for elongation complex formation following canonical transcription (B and C) or reiterative transcription (E and F) during initiation at the pyrG promoter. Cross-sectional views of the RNAP holoenzyme (β-flap, blue; σ, orange; rest of RNAP, gray; catalytic Mg2+, yellow sphere), promoter DNA (template strand, dark green; nontemplate strand, light green) and the RNA transcript (red) are shown. Base pairs between the DNA and RNA are depicted as black lines. (A and D) Open and the elongation complexes, respectively. In the canonical transcription pathway (Top), the initially transcribing complex (B) and the complex at the stage of promoter clearance (C) are shown. In the reiterative transcription pathway, the RTC (E) and the transcription complex switched right after from the reiterative to nonreiterative transcriptions (F) are depicted.
Our studies also suggest that the maximum length of reiterative pyrG transcripts is established when an 8-mer RNA collides with a narrow opening into the main channel of RNAP (Fig. 3 and Fig. S3). Apparently, this opening is too small for further extension of poly-G (and poly-A) transcripts, but not for poly-C transcripts. Establishing the general nature of this restriction will require structural studies of RTCs, including other promoters and RNAPs.
Although our study provides a detailed view of reiterative transcription at the molecular level, much remains to be known about the mechanism of RNA slippage. In the future, we intend to apply time-dependent soak–trigger–freeze X-ray crystallography (24) to visualize RNA slippage during reiterative transcription. It is also important to note that the fates of reiterative transcripts from other promoters are fundamentally different from that of the pyrG transcript. Most notably, during transcription initiation at some bacterial promoters (e.g., the pyrBI promoter of E. coli), once extra nucleotides are added to a nascent RNA by reiterative transcription, there is no switch to canonical transcription (2). As a consequence, these reiterative transcripts are released from the transcription initiation complex, resulting in reduced gene expression.
Interestingly, in all known examples of reiterative transcription leading to release of transcripts during initiation, the promoters involved differ from the pyrG promoter in a similar way. All of these promoters include an initially transcribed region in which the homopolymeric tract at which reiterative transcription occurs is located one or two bases downstream from the transcription start site (2). Thus, at the start of reiterative transcription, the DNA/RNA hybrid will be one or two base pairs longer than that observed at the pyrG promoter. This small difference might be sufficient to prevent a switch from reiterative to canonical transcription. Although the RNAP structural features controlling these alternative fates are not known, it is possible that the presence of the 5′ end base(s) preceding the homopolymeric tract causes nascent transcripts to be directed into a pathway within RNAP that precludes transcribing downstream DNA sequence. Confirming such a model will require structural comparisons of reiterative transcription complexes that differ in transcript fate.
Experimental Procedures
Preparation and Purification of T. thermophilus, E. coli, and B. subtilis RNAPs.
T. thermophilus, E. coli, and B. subtilis RNAP holoenzymes were purified as described previously (25–27). The rifampin-resistant E. coli RNAPs were purified as described (20).
Preparation of pyrG Promoter DNA Scaffold for the Crystallization.
The promoter DNA scaffold that resembles B. subtilis pyrG promoter region was constructed by using two oligodeoxynucleotides and used for the crystallization. The sequences of the nontemplate strand and template strand were 5′-TATAATGGGAGCTGGCTCTGATGCAGG -3′ and 5′- CCTGCATCAGAGCCCAAAATAC-3′, respectively. The two oligonucleotides were annealed in 40 μL containing 5 mM Tris⋅HCl (pH 7.7), 200 mM NaCl, and 10 mM MgCl2 to the final concentration of 0.5 mM. The solution was heated at 95 °C for 10 min, and the temperature was gradually decreased to 22 °C.
Crystallization of the T. thermophilus RNAP Promoter DNA Complex.
The crystals were prepared by mixing the purified T. thermophilus RNAP holoenzyme (18 μM) and the pyrG promoter DNA scaffold (27 μM) in the crystallization buffer [20 mM Tris⋅HCl (pH 8 at 4 °C), 100 mM NaCl, 0.1 mM EDTA, 1% glycerol, 1 mM DTT]. The full-sized crystal in the hanging drop containing 0.1 M Tris⋅HCl (pH 8 at 22 °C), 0.2 M KCl, 50 mM MgCl2, 9.5% PEG4000 was harvested and cryoprotected by stepwise transferring in the solutions 0.1 M Tris⋅HCl (pH 8 at 22 °C), 0.2 M KCl, 50 mM MgCl2, 15% PEG4000, and 15% butanediol. The crystals were frozen by liquid nitrogen. To prepare the crystals of the RTC, the RNAP and pyrG promoter complex crystals were transferred to the cryosolution containing 2 mM GTP and incubated for 30 min at room temperature.
X-Ray Data Collections and Structure Determinations.
The X-ray dataset was collected and structure was determined as previously described (17).
Reiterative in Vitro Transcription Assay with CTP Concentration Titration.
The same DNA scaffold used in crystal formation was used as template DNA. In vitro transcription assays were performed in 10 μL containing 250 nM RNAP holoenzyme, 250 nM DNA scaffold, 40 mM Tris⋅HCl (pH 8.0 at 25 °C), 30 mM KCl, 10 mM MgCl2, 15 μM acetylated BSA, 1 mM DTT, 100 μM GTP, 1 μCi (1 Ci = 37 GBq) [γ-32P]GTP, and various concentrations of CTP. The samples were incubated for 10 min at 37 °C (B. subtilis and E. coli) or 55 °C (T. thermophilus), and the reactions were stopped by adding 10 μL of 2× stop buffer (90% formamide, 50 mM EDTA, xylene cyanol, and bromophenol blue). The reaction products were electrophoretically separated on a denaturing 24% polyacrylamide/7 M urea gel and visualized with a phosphorimager (Typhoon 9410; GE Healthcare).
Acknowledgments
We thank Dr. Ritwika Basu for the T. thermophilus RNAP preparation, Dr. Shoko Murakami for critical advice for crystallographic data collection, Dr. Masaya Fujita for providing the B. subtilis strain for the RNAP preparation, Dr. Paul Babitzke for critically reading the manuscript, and the staff at the Cornell High Energy Synchrotron Source (CHESS)/Macromolecular Diffraction Facility at CHESS for support of crystallographic data collection. This work was supported by NIH Grants GM087350 (to K.S.M.) and GM94466 (to C.L.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5VOI and 5VO8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702741114/-/DCSupplemental.
References
- 1.Turnbough CL., Jr Regulation of gene expression by reiterative transcription. Curr Opin Microbiol. 2011;14:142–147. doi: 10.1016/j.mib.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: Repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacques JP, Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991;5:707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlin M, Berg P. Deoxyribonucleic acid-directed synthesis of ribonucleic acid by an enzyme from Escherichia coli. Proc Natl Acad Sci USA. 1962;48:81–94. doi: 10.1073/pnas.48.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anikin M, Molodtsov V, Temiakov D, McAllister WT. Transcript Slippage and Recoding. Nucleic Acids and Molecular Biology. Vol 24. Springer; New York: 2010. pp. 409–432. [Google Scholar]
- 6.Cheng Y, Dylla SM, Turnbough CL., Jr A long T. A tract in the upp initially transcribed region is required for regulation of upp expression by UTP-dependent reiterative transcription in Escherichia coli. J Bacteriol. 2001;183:221–228. doi: 10.1128/JB.183.1.221-228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo HC, Roberts JW. Heterogeneous initiation due to slippage at the bacteriophage 82 late gene promoter in vitro. Biochemistry. 1990;29:10702–10709. doi: 10.1021/bi00499a019. [DOI] [PubMed] [Google Scholar]
- 8.Xiong XF, Reznikoff WS. Transcriptional slippage during the transcription initiation process at a mutant lac promoter in vivo. J Mol Biol. 1993;231:569–580. doi: 10.1006/jmbi.1993.1310. [DOI] [PubMed] [Google Scholar]
- 9.Barr JN, Wertz GW. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: Implications for the mechanism of transcription termination. J Virol. 2001;75:6901–6913. doi: 10.1128/JVI.75.15.6901-6913.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou YN, et al. Isolation and characterization of RNA polymerase rpoB mutations that alter transcription slippage during elongation in Escherichia coli. J Biol Chem. 2013;288:2700–2710. doi: 10.1074/jbc.M112.429464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molodtsov V, Anikin M, McAllister WT. The presence of an RNA:DNA hybrid that is prone to slippage promotes termination by T7 RNA polymerase. J Mol Biol. 2014;426:3095–3107. doi: 10.1016/j.jmb.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18:3529–3535. doi: 10.1093/nar/18.12.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Heath LS, Turnbough CL., Jr Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 1994;8:2904–2912. doi: 10.1101/gad.8.23.2904. [DOI] [PubMed] [Google Scholar]
- 15.Meng Q, Turnbough CL, Jr, Switzer RL. Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc Natl Acad Sci USA. 2004;101:10943–10948. doi: 10.1073/pnas.0403755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen-MacAllister IE, Meng Q, Switzer RL. Regulation of pyrG expression in Bacillus subtilis: CTP-regulated antitermination and reiterative transcription with pyrG templates in vitro. Mol Microbiol. 2007;63:1440–1452. doi: 10.1111/j.1365-2958.2007.05595.x. [DOI] [PubMed] [Google Scholar]
- 17.Basu RS, et al. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J Biol Chem. 2014;289:24549–24559. doi: 10.1074/jbc.M114.584037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird JG, et al. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature. 2016;535:444–447. doi: 10.1038/nature18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korzheva N, et al. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 20.Molodtsov V, Scharf NT, Stefan MA, Garcia GA, Murakami KS. Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis. Mol Microbiol. 2017;103:1034–1045. doi: 10.1111/mmi.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kibbe WA. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 23.Darst SA, et al. Conformational flexibility of bacterial RNA polymerase. Proc Natl Acad Sci USA. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basu RS, Murakami KS. Watching the bacteriophage N4 RNA polymerase transcription by time-dependent soak-trigger-freeze X-ray crystallography. J Biol Chem. 2013;288:3305–3311. doi: 10.1074/jbc.M112.387712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami KS. X-ray crystal structure of Escherichia coli RNA polymerase σ70 holoenzyme. J Biol Chem. 2013;288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita M, Sadaie Y. Rapid isolation of RNA polymerase from sporulating cells of Bacillus subtilis. Gene. 1998;221:185–190. doi: 10.1016/s0378-1119(98)00452-1. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]