Significance
We describe the phage FLiP, an ssDNA virus with an icosahedral capsid and an internal lipid membrane. The FLiP genome shows limited similarity to known sequences, although an ssDNA replication mechanism was implied by genome analysis. However, because the capsid protein fold indicates relatedness with the dsDNA viruses of the PRD1–adenovirus lineage, FLiP exhibits a unique combination of structural and replication modules. It is suggested that the capsid protein structure could be used to complement the sequence data when classifying viruses and in detecting their deep evolutionary relationships, especially in the absence of sequence similarities. Furthermore, these findings demonstrate the value of characterizing unknown viruses from diverse environmental sources to understand the diversity of the microbial world.
Keywords: cryo-electron microscopy, Flavobacterium, lipids, genome, structure
Abstract
Viruses have impacted the biosphere in numerous ways since the dawn of life. However, the evolution, genetic, structural, and taxonomic diversity of viruses remain poorly understood, in part because sparse sampling of the virosphere has concentrated mostly on exploring the abundance and diversity of dsDNA viruses. Furthermore, viral genomes are highly diverse, and using only the current sequence-based methods for classifying viruses and studying their phylogeny is complicated. Here we describe a virus, FLiP (Flavobacterium-infecting, lipid-containing phage), with a circular ssDNA genome and an internal lipid membrane enclosed in the icosahedral capsid. The 9,174-nt-long genome showed limited sequence similarity to other known viruses. The genetic data imply that this virus might use replication mechanisms similar to those found in other ssDNA replicons. However, the structure of the viral major capsid protein, elucidated at near-atomic resolution using cryo-electron microscopy, is strikingly similar to that observed in dsDNA viruses of the PRD1–adenovirus lineage, characterized by a major capsid protein bearing two β-barrels. The strong similarity between FLiP and another member of the structural lineage, bacteriophage PM2, extends to the capsid organization (pseudo T = 21 dextro) despite the difference in the genetic material packaged and the lack of significant sequence similarity.
Although conservative calculations estimate that the number of viruses in the biome exceed 1031 virions (1), this enormous group of biological entities is largely unexplored because only a fraction of viruses have been studied in detail. In recent years, the enormous diversity of the viral world (2) has been revealed by high-throughput sequencing of viral genomes. Although the traditionally used virus detection methods (epifluorescence microscopy, pulse-field gel electrophoresis, and early metagenomic studies) have been selective for dsDNA viruses (3–5), recent metagenomic studies have revealed that the number of ssDNA viruses in nature has been grossly underestimated. In fact, ssDNA viruses are widespread and may predominate in certain habitats, including the oceans and arctic freshwaters (6–10). However, only a few ssDNA viruses have been cultivated in laboratory conditions, and therefore detailed structural and biochemical analyses of distinct ssDNA viruses are mostly lacking. Detailed characterization of novel virus isolates is essential to expand our understanding of evolutionary relationships among viruses and their role in microbial communities (11).
Using solely sequence-based methods for virus classification and phylogeny is complicated because of the enormous genetic diversity among viruses (11, 12). However, essential viral structures and functions are often strongly conserved (13, 14). There seem to be only a limited number of folds a functional protein may adopt, and stringent structural constraints further decrease the number of folds suitable for forming a capsid protein. These constraints make the serendipitous invention of new architectures very unlikely, so that only a very limited number of lineages of virion architecture have been observed (15, 16). Therefore, it has been suggested, that the major capsid protein (MCP) fold and overall virion structure could be used to classify viruses and track down their deeper evolutionary relationships (17).
We describe here an icosahedral virus with an ssDNA genome and an internal lipid membrane designated “FLiP” (Flavobacterium-infecting, lipid-containing phage). The 9,174-nt-long genome of FLiP has limited sequence similarities with previously identified viruses. Based on cryo-electron microscopy (cryo-EM) at 4-Å resolution, the virion structure of FLiP displays the pseudo triangulation number (T) = 21 dextro organization previously shown only for the marine dsDNA phage PM2. The MCP consists of two β-barrels with jellyroll topology and closely resembles dsDNA viruses of the PRD1–adenovirus lineage. Thus, these structural data indicate evolutionary relatedness between some ssDNA and dsDNA viruses and suggest that it would be beneficial to complement traditional sequence-based systems with structure-based approaches.
Results
FLiP Is a Phage That Infects Flavobacterium.
The virus designated FLiP was isolated together with the host bacterium from a boreal freshwater habitat in Central Finland in September, 2010. The host bacterium was identified as a member of genus Flavobacterium (Bacteroidetes) by 16S rRNA gene sequencing. Flavobacterium species are important members of freshwater bacterial communities in the boreal regions (18, 19). The greatest similarity to the obtained 1,422-bp-long 16S rRNA gene sequence was to several Flavobacterium sp. strains (99%). Accordingly, the host bacterium strain of FLiP was designated as Flavobacterium sp. B330.
FLiP virions were collected for purification from plate lysate. Phage particles were concentrated from the filtered lysate with PEG-NaCl precipitation and then were purified by rate zonal and equilibrium centrifugation to near homogeneity. The rate zonal centrifugation was optimized using different gradient materials (iodixanol, glycerol, and sucrose). The phage yield was significantly higher in the sucrose gradient; therefore this gradient material was used routinely for purification in both rate zonal and equilibrium centrifugation. This method yielded highly purified particles with specific infectivity of 1.28 × 1012 pfu/mg of protein. The typical recovery and specific infectivity for each purification step are presented in Table S1.
Table S1.
FLiP purification
| Purification step | Recovery of infectivity, pfu | Recovery of infectivity, % |
| Plate lysate | 7.8 × 1013 | |
| PEG precipitate | 1.6 × 1013 | 20.6 |
| Purified by rate zonal centrifugation* | 4.2 × 1012 | 5.4 |
| Purified by equilibrium centrifugation* | 9.6 × 1011 | 1.2 |
| Concentrated virus | 1.6 × 1011 | 0.2 |
In sucrose.
The FLiP Genome Is a Circular ssDNA Molecule with 16 Putative ORFs.
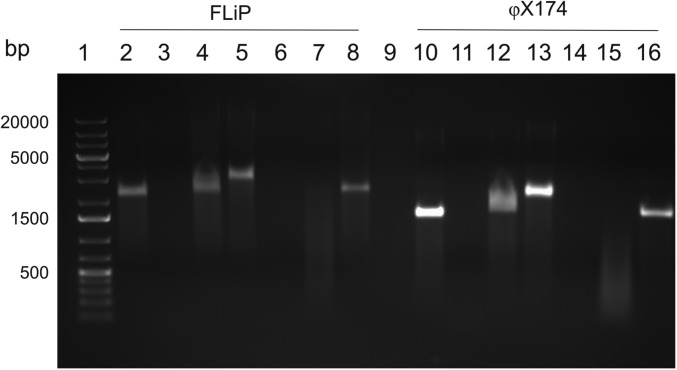
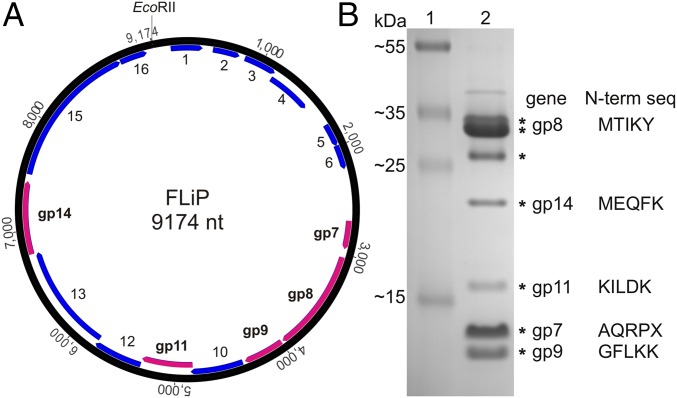
FLiP genomic nucleic acid was extracted from purified virions. Nuclease treatments (Fig. S1) revealed that the genome is an ssDNA molecule. Genome sequencing resulted in a 9,174-nt-long molecule, with an overall guanine–cytosine (GC) content of 34% (Fig. 1A). A total of 16 putative ORFs were identified from the genome using the programs Glimmer (20) and GeneMark (21). All the ORFs were oriented in the same direction. Most of the ORFs showed limited similarity to other sequences in the public databases (Table S2), as is common for environmental phage isolates.
Fig. S1.
Analysis of the ssDNA genome of FLiP. Agarose gel electrophoresis (AGE, 1% agarose) analysis of FLiP (lanes 2–9) and φX174 genomes (lanes 10–16). Lanes 2 and 10: untreated samples; lanes 3 and 11: samples treated with DNase I; lanes 4 and 12, samples treated with RNase A; lanes 5 and 13: samples treated with RNase I; lanes 6 and 14, samples treated with S1 nuclease; lanes 7 and 15: samples treated with Mung bean nuclease; lanes 8 and 16: lanes treated with EcoRI. The DNA ladder (Fermentas #R0491) is in lane 1.
Fig. 1.
Circular ssDNA genome of FLiP. (A) A graphic representation of the genome (9,174 nt, numbering starting from a unique EcoRII restriction site); predicted ORFs are labeled and shown in blue. Those identified by proteomics to encode for structural proteins are indicated in purple. (B) SDS/PAGE (16% acrylamide) analysis of the structural proteins. Mass standards (Fermentas, 26619) are shown in lane 1. Major protein bands (lane 2) that were subjected to mass spectrometry are indicated by an asterisk. N-terminal sequences determined by N-terminal protein sequencing are indicated.
Table S2.
Sequence analysis of FLiP genome and ORFs
| ORF or gene | Direction | Nucleotide coordinates, start–stop | GC, % | Length, aa | Calculated. mass, kDa | Calculated isoelectric point | Protein | Function | Transmembrane helix predictions, n | Blast (E-value) | HHpred (% probable) |
| 1 | F | 155–445 | 31.96 | 97 | 11.2 | 9.70 | |||||
| 2 | F | 540–782 | 32.51 | 81 | 9.6 | 4.33 | |||||
| 3 | F | 836–1,138 | 33.66 | 101 | 11.8 | 6.22 | 1 | ||||
| 4 | F | 1,138–1,557 | 34.52 | 140 | 15.8 | 4.57 | Helix-turn-helix motif (98.6) | ||||
| 5 | F | 1,799–2,008 | 39.52 | 70 | 7.1 | 10.5 | |||||
| 6 | F | 2,011–2,229 | 31.05 | 73 | 9.0 | 9.80 | |||||
| 7 | F | 2,703–2,957 | 33.73 | 85 | 10.3 | 12.92 | gp7 | S | 1 | ||
| 8 | F | 3,036–3,971 | 37.07 | 312 | 34.6 | 5.07 | gp8 | S, MCP | Cellulophaga phage φ48:2 (0.020) | ||
| 9 | F | 3,992–4,363 | 43.01 | 124 | 12.9 | 10.77 | gp9 | S | 1 | Cellulophaga phage φ12a:1 (5e-04) | |
| 10 | F | 4,385–4,861 | 35.01 | 159 | 18.5 | 6.83 | 1 | ||||
| 11 | F | 4,845–5,321 | 37.32 | 159 | 17.9 | 9.42 | gp11 | S | 1 | ||
| 12 | F | 5,324–5,770 | 33.56 | 149 | 17.0 | 4.63 | |||||
| 13 | F | 5,770–6,780 | 35.61 | 337 | 38.4 | 4.31 | |||||
| 14 | F | 6,780–7,445 | 37.39 | 222 | 25.2 | 9.61 | gp14 | S, lytic enzyme | 1 | Lytic enzyme (4e-28) | Lytic enzyme (99.8) |
| 15 | F | 7,510–8,853 | 31.99 | 448 | 53.5 | 9.06 | Cellulophaga phage φ48:2 (3e-12) | Replication initiation protein (97.0) | |||
| 16 | F | 8,865–9,110 | 28.86 | 82 | 9.3 | 8.46 | Coiled-coiled protein (92.1) |
ORF or gene as in Fig. 1; nucleotide coordinates refer to FLiP genome; gene functions as predicted by BlastP, Psi-Blast, and HHpred programs. The E-value of the most relevant Blast match is indicated. The most relevant HHpred predictions are listed. F, forward; gp, confirmed by proteomics (see Table S3); R, reverse; S, structural protein.
Identification of FLiP Structural Proteins.
Purified virions were subjected to SDS/PAGE analysis. The most abundant protein species detected were ∼35 kDa in size (Fig. 1B). Resolved structural proteins were identified by mass spectrometry, and N-terminal sequencing was performed for the most abundant protein species (Table S3). According to these analyses, five ORFs (7–9, 11, 14) were designated as genes because their protein products were identified (Fig. 1B). Because of its high expression level, gene 8 was predicted to encode the MCP. Translated gene 14 showed a high sequence similarity to several lytic transglycosylases and contained the conserved transglycosylase domain (E value 4.97 × 10−4), indicating that a protein with lytic activity is present in the virion structure.
Table S3.
FLiP structural proteins identified by MS and N-terminal sequencing
| Protein | MS | N-terminal sequence |
| ORF7 | ND | AQRPX |
| ORF8 | Identified | MTIKY |
| ORF9 | Identified | GFLKK* |
| ORF11 | Identified | KILDK |
| ORF14 | Identified | MEQFK |
ND, not determined.
Last determined amino acid is uncertain (X/K/N/M).
FLiP Obtains Lipids Selectively from the Host.
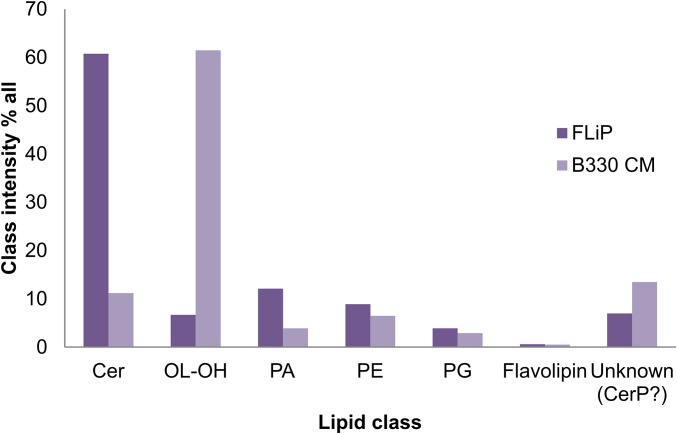
Low buoyant density (1.21 g/cm3 and 1.18 g/cm3 in CsCl2 and sucrose gradients, respectively) and chloroform sensitivity indicated the presence of a lipid membrane in the virion. The lipid class and molecular species composition of the virion and the host bacterium inner membrane were analyzed from the extracted lipids by mass spectrometry [triple quadrupole electrospray ionization (ESI)-MS/MS] (Fig. S2). FLiP contains significantly more ceramide (60% of the total lipid composition) than the host cytoplasmic membrane. Ornithine lipids are enriched in the host membrane but are present only in a negligible amount (∼7%) in the phage. The differing lipid compositions indicate that the phage obtains the lipids from the host selectively.
Fig. S2.
Comparison of lipid class intensities between the host cytoplasmic membrane (CM) and the FLiP membrane. Class intensity percentage of each lipid class identified by mass spectrometry from Flavobacterium sp strain B330 cytoplasmic membrane compared with that of the FLiP membrane. Cer, ceramide; CerP, ceramide 1-phosphate; OL-OH, ornithine lipid; PA, phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphatidylglycerol.
Cryo-EM Reconstruction of FLiP Virion and MCP Structure.
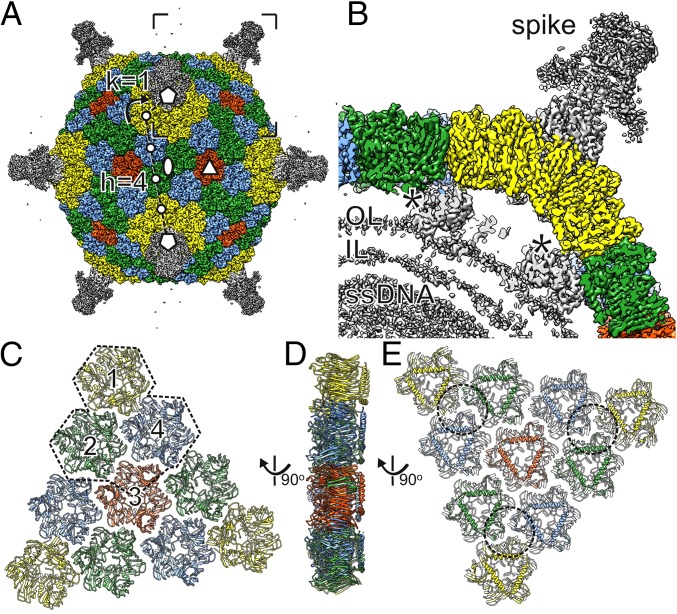
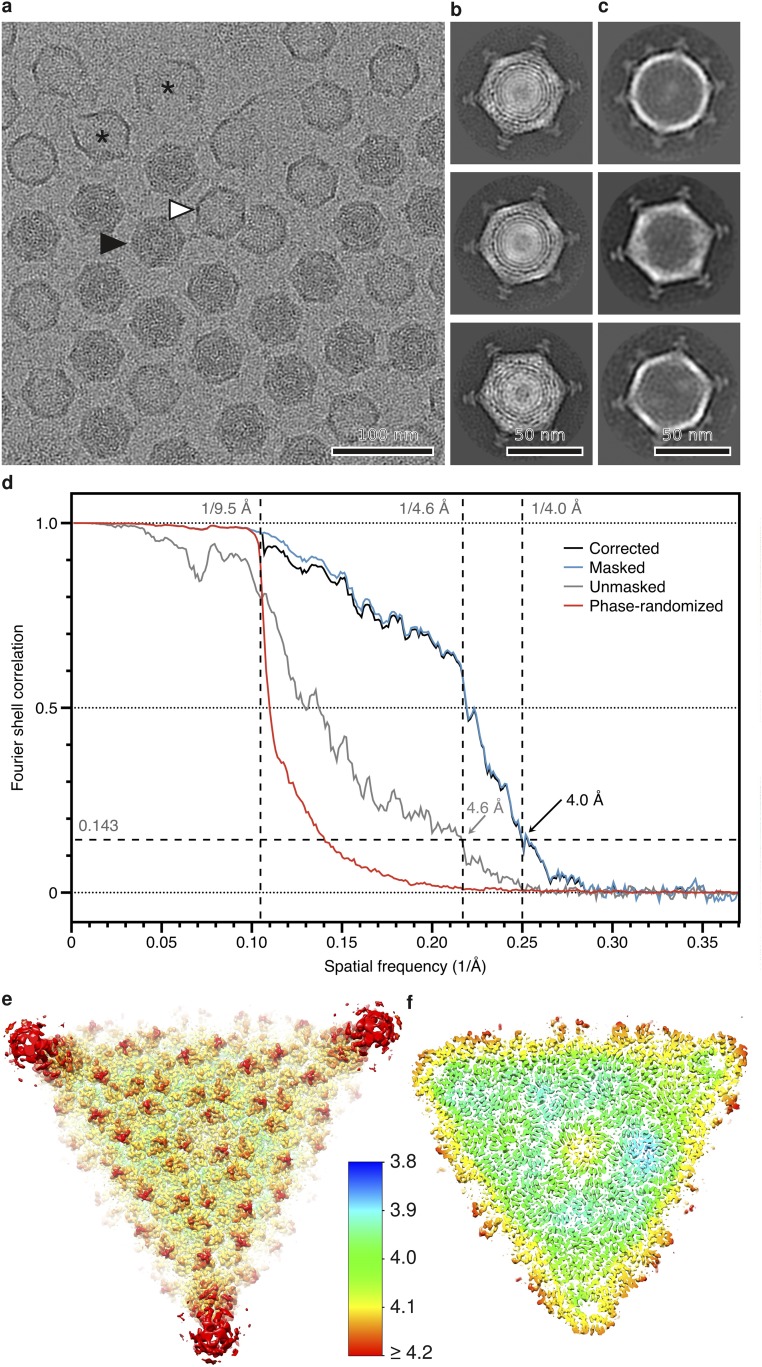
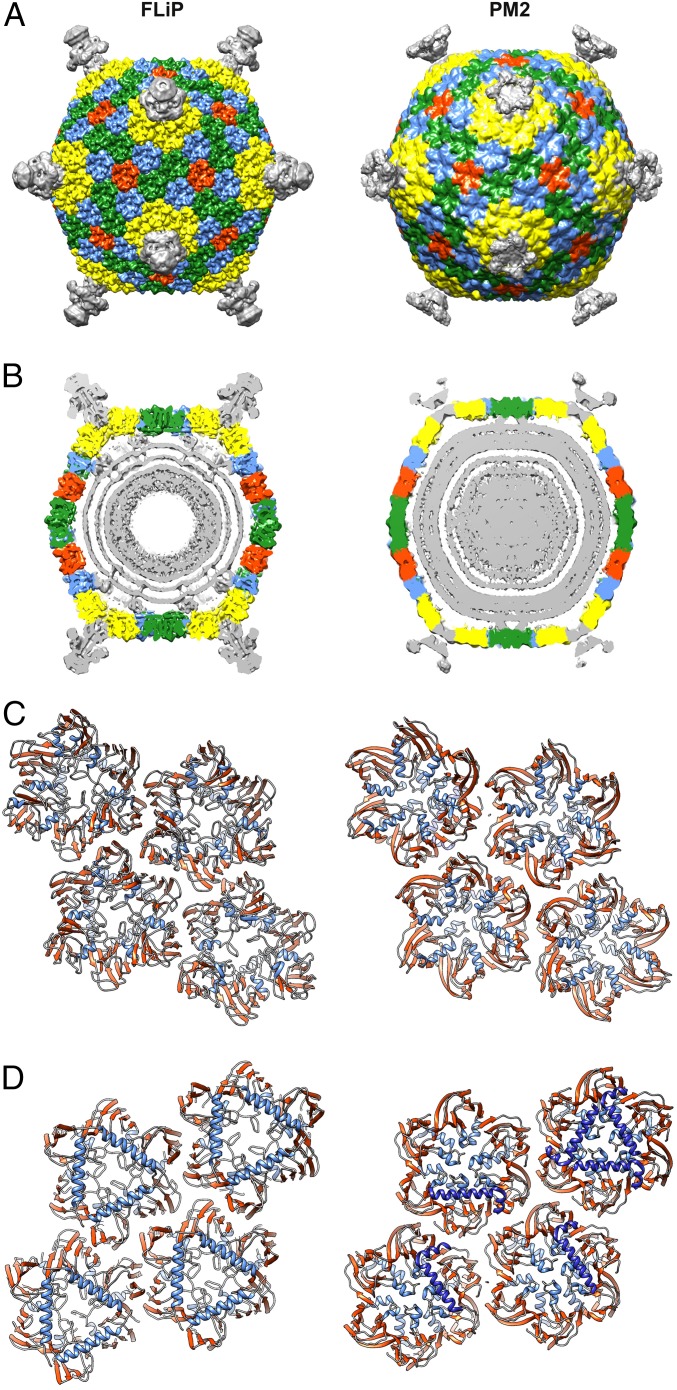
We solved the structure of the FLiP virion using cryo-EM (Fig. 2A). Micrographs of purified virions revealed spherical particles that were ∼60 nm in diameter (Fig. S3 A and B). Some of the particles appeared hollow, suggesting they were empty, lacking the DNA genome and possibly also the internal lipids (Fig. S3C). Single-particle analysis of 2,203 full particles and gold-standard 3D structure refinement (22) yielded an icosahedrally symmetric reconstruction of the FLiP virion solved from a final set of 934 particles (Fig. 2A and Table S4). The resolution of the viral protein capsid was 4.0 Å on average, as estimated by Fourier shell correlation (FSC) at a threshold of 0.143 (Fig. S3D). Some local areas of the viral surface were less resolved, but resolution in the capsid interior approached 3.9-Å resolution (Fig. S3 E and F). Reaching such resolution from a relatively small number of particles has been reported before, for example in the case of deformed wing virus (EMD-3570) (23). In the FLiP reconstruction, pentameric spikes (12 nm high) protrude from the 12 icosahedral vertices (Fig. 2 A and B). The MCPs forming the outer protein shell follow a pseudo T = 21 dextro icosahedral capsid organization (Fig. 2A) also observed in the dsDNA phage PM2 (24). The shape of the shell is highly faceted, with facet-to-facet and edge-to-edge distances of 53 nm and 55 nm, respectively, compared with a vertex-to-vertex distance of 59 nm as measured from the base of the spike. The outer protein shell covers a 5-nm-thick lipid bilayer membrane (Fig. 2B). Minor structural components, located between the outer protein shell and the membrane, bridge the two together (Fig. 2B).
Fig. 2.
3D cryo-EM reconstruction of the FLiP virion. (A) Surface rendering of the FLiP virion. The MCPs are colored yellow, green, blue, and red. Other structural components are colored gray. The icosahedral twofold (ellipse), threefold (triangle), and fivefold (pentagons) axes of symmetry are indicated. The triangulation number T = 21 of the icosahedral lattice of the capsid is calculated as T = h2 + h × k + k2 where lattice indices are h = 4 and k = 1 as indicated. The lattice is right-handed (dextro) as indicated by the curved arrow. (B) A section of virion density is shown from the area indicated in A. Spike structure, minor proteins (asterisks), the outer leaflet (OL) and inner leaflet (IL) of the lipid bilayer, and density corresponding to the ssDNA are indicated. (C) The capsid is composed of 20 faces, each consisting of a group of 10 pseudohexameric MCP trimers (ribbons). Each face is divided in three asymmetric units (one is outlined), each consisting of three MCP trimers (1, 2, 4) and one chain from MCP trimer 3 (in addition to the base domain of the spike). (D and E) Group-of-ten seen from the side (D) and below (E). Approximate footprints of the minor capsid proteins indicated in B are outlined.
Fig. S3.
Cryo-EM of FLiP. (A) A representative cryo-EM micrograph of purified FLiP particles. Full (black arrowhead) and empty (white arrowhead) particles are indicated. Broken particles are labeled with an asterisk. (Scale bar, 100 nm.) (B and C) Representative 2D class averages of full (B) and empty (C) particles. Both revealed an outer protein capsid with spikes at the vertices. The hollow particles were empty, lacking both the membrane and DNA. (Scale bars, 50 nm.) (D) FSC calculated between two half-maps as a function of spatial frequency is shown. FSC is shown for the original, unmasked half-maps (gray), masked half-maps that had most of the membrane and genome density removed (blue), and phase-randomized half-maps (red) in which phases were randomized at frequencies higher than 1/9.5 Å. The phase-randomized FSC drops sharply at the cutoff frequency below the noise threshold (0.143), as expected. The phase-randomization test was used to take the effect of masking on the half-maps into account before calculating the final, corrected FSC curve (black). Good agreement between the masked and corrected curves indicated that masking did not cause overestimation of resolution. The corrected curve drops below the noise threshold at 1/4.0 Å indicating a resolution of 4.0 Å in the reconstruction. (E and F) Two slabs of density are shown from the capsid surface (E) and capsid interior (F) along the threefold axis of symmetry. The surface was colored based on local resolution as indicated in the color bar.
Table S4.
FLiP data collection and processing
| Parameter | Value |
| Cryo-EM data collection | |
| Voltage, kV | 300 |
| Magnification, × | 37,037 |
| Defocus, μm | 0.7–2.5 |
| Dose rate, electrons per pixels per second | 8 |
| Frames | 22 |
| Frame length, s | 0.2 |
| Total dose, e−/Å2 | 22 |
| Micrographs | 235 |
| Cryo-EM data processing | |
| Particles | 934 |
| Box size, pixels | 700 |
| Pixel size, Å | 1.35 |
| Symmetry | I1 |
| Resolution, Å | 4.0 |
| Map sharpening B-factor, Å2 | −100 |
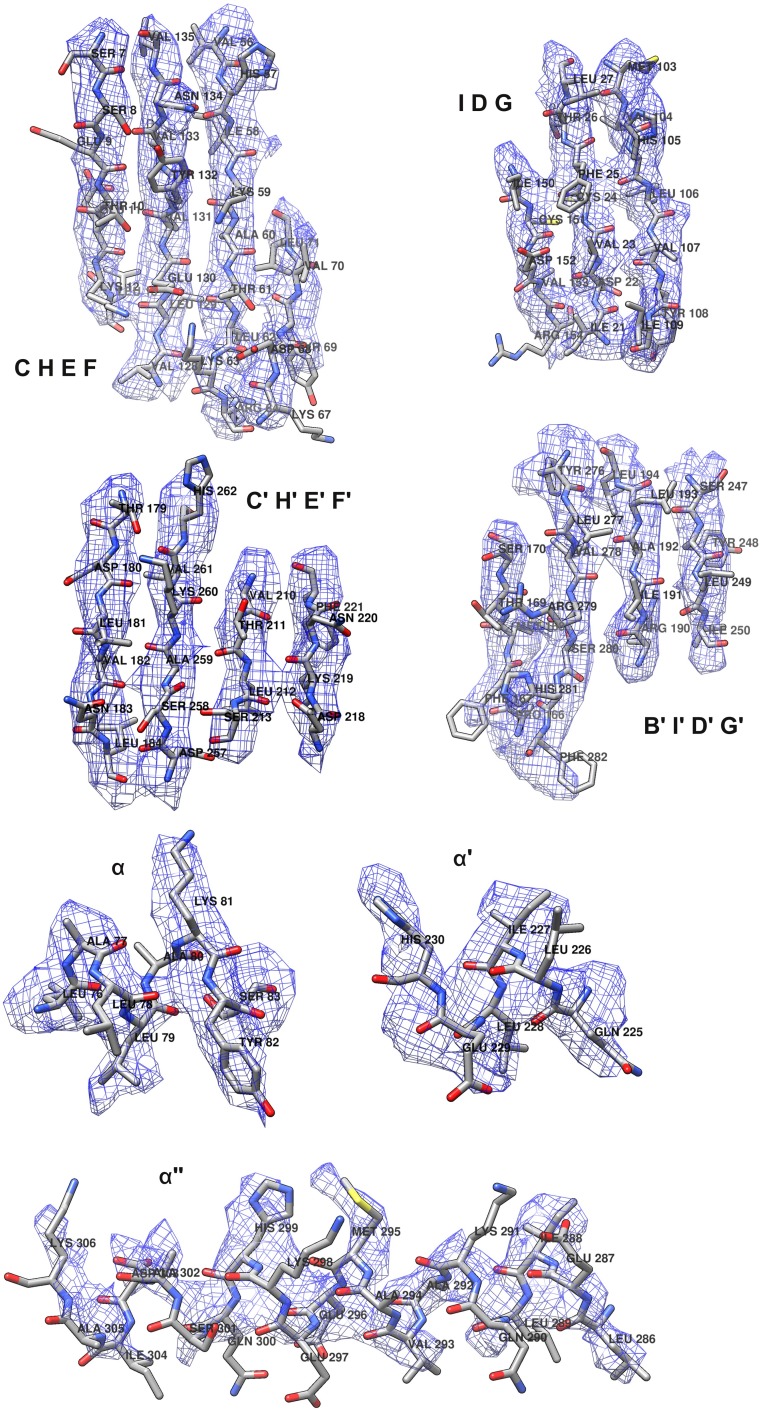
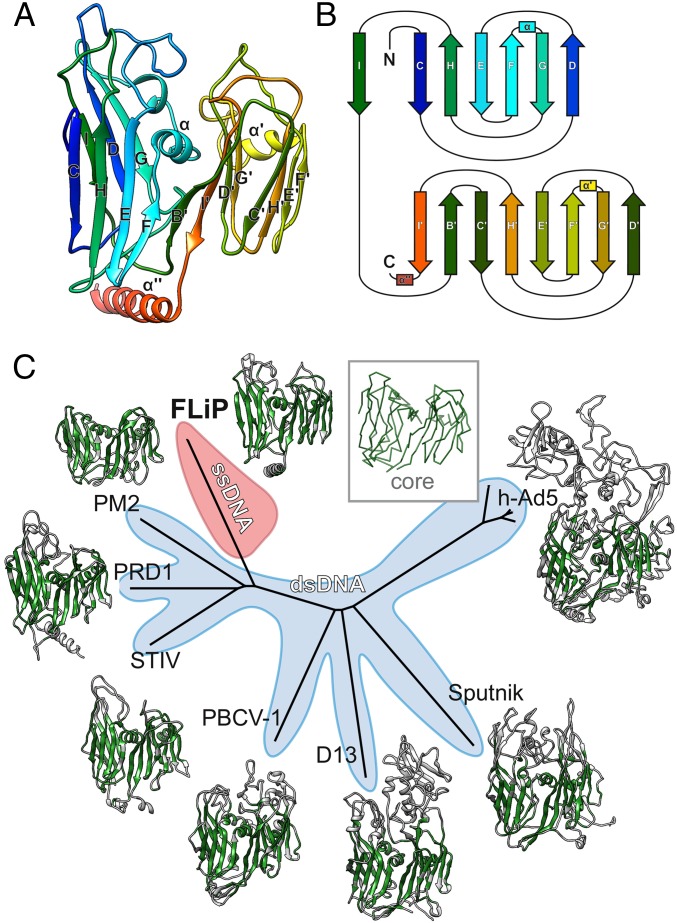
The near-atomic resolution of the FLiP cryo-EM reconstruction facilitated building an atomic model of the MCP de novo (Fig. S4 and Table S5). Minor structural proteins that were resolved in the reconstructions, such as the pentameric spikes, were not built in the absence of sequence assignments. The MCP consists of two β-barrels/sandwiches, each of which is composed of two antiparallel β-sheets (Fig. 3A). Both barrels have a jellyroll topology (Fig. 3B). Strands C, H, E, and F contribute to the sheets facing the surface of the trimer, whereas strands B, I, D, and G contribute to the sheets facing toward the center of the trimer (Fig. 3 A and B). Strand B is absent in the N-terminal β-barrel. The two β-barrels are decorated by loops in addition to α-helices embedded between strands F and G. A long C-terminal α-helix resides horizontally at the base of the MCP. Trimers of the double β-barrel MCP are formed with the six β-barrels of the MCP subunit arranged to form a pseudohexameric molecule (Fig. 2).
Fig. S4.
Atomic model of FLiP MCP. Atomic models and cryo-EM density corresponding to the FLiP MCP secondary structure elements are shown. Secondary structure elements are labeled as in Fig. 4.
Table S5.
MCP structure refinement and model validation (full particle)
| Parameter | Value |
| Refinement | |
| Resolution, Å | 3.9 |
| Number of atoms | 1,453,200 |
| FSC, overall | 0.789 |
| FSC, around masked atoms | 0.811 |
| CC, overall | 0.781 |
| CC, around masked atoms | 0.757 |
| Rmsd | |
| Bonds, Å | 0.006 |
| Angles, ° | 0.956 |
| Average B-factor, Å2 | 84.31 |
| EMRinger score | 2.32 |
| MolProbity | |
| Clash score, all atoms | 5.66 |
| MolProbity score | 1.94 |
| Rotamer outliers, % | 0.00 |
| Ramachandran, % favored | 86.55 |
| Ramachandran, % allowed | 13.15 |
| Ramachandran, % outliers | 0.29 |
CC, cross correlation.
Fig. 3.
FLiP MCP fold and structure-based phylogeny. (A) The structure of the FLiP MCP is shown as a ribbon representation, colored from the N terminus (blue) to the C terminus (red). The β-strands in the N-terminal β-barrel are labeled C–I, and the β-strands in the C-terminal β-barrel are labeled B′–I′. α-Helices are labeled α, α′, and α′′. (B) A schematic diagram illustrating the topology of the double β-barrel jellyroll fold. The secondary structure elements are labeled and colored as in A. C, C terminus; N, N terminus. (C) Structure-based phylogenetic tree derived from alignment of MCPs (30) from FLiP, bacteriophage PM2 (PDB code: 2WOC), bacteriophage PRD1 (PDB ID code: 1HX6), Sulfolobus turreted icosahedral virus (STIV; PDB ID code: 2BBD), Paramecium bursaria chlorella virus 1 (PBCV-1; PDB ID code: 1M3Y), Vaccinia virus Western Reserve D13 (D13; PDB ID code: 2YGB), Sputnik virophage (PDB ID code: 3J26), and human adenovirus 5 (h-Ad5; PDB ID code 1P30). The region of each protein used in the structure-based alignment is colored in green. A generalized common core of the fold is shown as a C-αtrace in the Inset. Branches corresponding to viruses with dsDNA genomes are shaded light blue. The branch corresponding to FLiP, the only characterized member the lineage with an ssDNA genome, is highlighted in red.
The FLiP MCP Fold Suggests the Virus Belongs to the PRD1–Adenovirus Lineage.
The overall virion architecture and details of the MCP fold (including the topology and disposition of the β-barrels and the position of the embedded α-helices) closely resemble those observed in members of the PRD1–adenovirus lineage (Figs. 3C and 4) (14). This lineage of icosahedral viruses until now has comprised solely dsDNA viruses, although the viruses infect organisms ranging from bacteria [e.g., PRD1 (25) and PM2 (26)] through archaea [Sulfolobus turreted icosahedral virus (27)] to green algae [Paramecium bursaria Chlorella virus 1 (28)] and higher eukaryotes [e.g., adenovirus (29)]. We calculated a structure-based phylogenetic tree by aligning MCPs of different members of this lineage against FLiP using the Homologous Structure Finder program (Fig. 3C) (30). FLiP branches from the bacterial/archaeal arm of the tree, being in no sense an outlier, and structurally is most closely related to the bacteriophage PM2. For these two viruses, the structural homology extends beyond the conserved double β-barrel MCP fold. Notably, FLiP and PM2 share the same pseudo T = 21 dextro capsid organization, although PM2 is slightly larger in size than FLiP (vertex-to-vertex distance 63 vs. 59 nm) (Fig. 4) (24). The two viruses also differ in how the capsid is bridged to the internal lipid bilayer. In both viruses, minor membrane proteins interact with the MCP inner surface via α-helices that are parallel to the capsid (Fig. 4D). In FLiP, these parallel helices correspond to the C terminus of the MCP (Figs. 2E and 3A, α′′, Ser285–Lys360), whereas in PM2 the helices belong to another minor protein, P3 (26).
Fig. 4.
Comparison of FLiP and PM2 capsid architecture. (A) Surface-rendered views of FLiP (this study) and PM2 (EMDB accession no. 1082) virion structures. Both maps have been low-pass filtered to 8-Å resolution. MCP trimers are colored yellow, blue, green, and red, depending on their location in the icosahedral capsid. Other densities, including the spikes in the icosahedral vertices, are colored gray. (B) A central section of density is shown for the two virions. Coloring is as in A. Capsid-to-membrane contacts are indicated by arrowheads. In FLiP, these densities have not been assigned. In PM2, they correspond to minor protein P3 (26). (C and D) A ribbon representation of four different types of MCPs seen from outside (C) and inside (D) the virion. In addition to the MCP, α-helices of the PM2 minor protein P3 interacting with the MCP (26) are shown in dark blue.
Discussion
Here we describe a virus, FLiP, which infects a Flavobacterium host. The FLiP virion consists of an icosahedrally symmetric protein capsid enclosing a circular ssDNA genome 9,174 nt in length (Fig. 1). The inner surface of the capsid is covered by a lipid membrane, making FLiP unique among previously described ssDNA phages. Even outside those ssDNA genomes, few bacterial viruses with internal lipid membranes have been characterized (31). The presence of the membrane evidently facilitates virus–host interactions (31), and, as does FLiP, some viruses have been shown to derive lipids selectively from the host (32, 33). FLiP contained significantly more ceramide than the host cytoplasmic membrane, possibly because the cone-shaped structure of ceramide might favor the formation of the highly curved viral membrane.
The FLiP genome is considerably larger than that of the previously isolated nontailed ssDNA phages except for the Cellulophaga phage phi48:2 (34). FLiP shows limited sequence similarity to any known sequences in the database (Table S2), as is typical for environmental phage isolates, probably reflecting the limited number of cultured virus isolates. Similarities were detected to Flavobacterium and Zunongwangia profunda as well as to ssDNA phages infecting Cellulophaga (34, 35). Z. profunda and Cellulophaga belong to the Bacteroidetes phylum, as does the FLiP host Flavobacterium. Moreover, ORF4 contains the conserved DNA-binding helix-turn-helix domain of the Xre family. Interestingly, ORF15 has resemblance to rolling circle replication initiator proteins from Staphylococcus aureus and Geobacillus stearothermophilus, suggesting that FLiP might use replication mechanisms similar to those found in other ssDNA replicons.
However, despite the unique combination of the ssDNA genome and the lipid membrane and the limited sequence similarities, the further structural characterization of FLiP implies intriguing connections to previously identified dsDNA viruses in the PRD1–adenovirus lineage. Although the number of available high-resolution structures of viral MCPs is increasing, most viruses seem to fit into just a few structure-based lineages, to the extent that certain DNA viruses (e.g., ssDNA phage phiX174 and papilloma and polyomaviruses with dsDNA genomes) have been assigned to the same lineage together with some positive-sense ssRNA viruses (15). The overall virion structure of FLiP suggests a relationship with the members of the PRD1–adenovirus lineage and with PM2 in particular. In addition, the MCP of PM2 is the closest homolog of the FLiP MCP, strongly suggesting that FLiP is a member of the PRD1–adenovirus lineage, even though the genomes of other members of that lineage are dsDNA as opposed to the circular ssDNA of FLiP (Fig. 3C). This notion contrasts with the traditional classification of viruses accepted by the International Committee on the Taxonomy of Viruses in which genome type (ssDNA, dsDNA, ssRNA, and dsRNA) is used as the top level of classification into virus groups. The data we present here together with previous observations (15, 36) suggest that it might be appropriate to place more emphasis on the virion architecture and the structure of the capsid proteins when classifying viruses and assigning distant viruses to higher taxonomic levels. To gain a more comprehensive view of the evolution of the virus world, it would seem beneficial to complement the genomic analyses with structural data.
Pervasive shuffling of genes and gene modules is undoubtedly a key feature in virus evolution. Viral genomes commonly encompass structural and replication modules, which may have different evolutionary provenances (37, 38). The recombination of these modules may provide adaptive advantage for viruses and give rise to novel virus types (37). The genetic resemblance of FLiP ORF15 to rolling circle replication initiation proteins indicates that this virus may display a fascinating combination of a replication mechanism typical of ssDNA viruses and structural characteristics of dsDNA viruses. However, the replication mechanism of FLiP is yet to be confirmed. Interestingly, PM2-like prophages with differing replication machineries seem to be common in aquatic bacteria, although their structural proteins have remained conserved (39).
It is evident that bacterial viruses represent an enormous reservoir of genetic diversity. However, only a small fraction of them have been structurally and biochemically characterized. The finding of FLiP exemplifies the importance of characterizing novel viruses from diverse environments in detail to obtain deeper understanding of the microbial world. This characterization is especially important for ssDNA viruses, whose diversity and ecological impact have remained poorly understood despite their abundance.
Methods
Isolation of the Bacteriophage and Host Bacterial Strains.
The bacteriophage FLiP and its host bacterium Flavobacterium sp. B330 were isolated from a freshwater sample taken from lake Jyväsjärvi in Central Finland (62°13.840′ N 025°44.510′ E) as described elsewhere (40). Afterward B330 was routinely grown in Shieh medium (41) at room temperature with constant agitation of 110 rpm. FLiP virions were isolated from a filtered (pore size 0.45 µm; Micropore) water sample by enriching the host bacterium in LB liquid medium diluted 1:5 with the filtrated water sample at room temperature for 2 d (110 rpm). The enrichment culture was mixed with soft agar [0.7% (wt/vol)] in 1:5 LB medium and was plated on solid agar. The plate was incubated at room temperature for 2 d; then single plaques were picked, suspended in 500 µL of 1:5 LB medium, and stored at 4 °C. Three rounds of plaque purification were performed. The bacteriophage was propagated using the standard double agar layer technique (42). In brief, 100 µL of the bacteriophage suspension in different dilutions was added to 3 mL of molten soft agar [0.7% (wt/vol)] together with 200 µL of the host cell culture. The mixture was poured onto the surface of Shieh agar plates [1% (wt/vol)] and incubated overnight at room temperature.
16S rRNA Gene Sequencing of the Host Bacterium.
Genomic DNA of B330 was extracted with the GeneJET Genomic DNA Purification Kit (Fermentas) according to the manufacturer’s instructions. The gene encoding the 16S rRNA was amplified by PCR using primers forward D1 and reverse D1 (43). The PCR products were purified with the QIAquick PCR Purification Kit (Thermo Scientific) and sequenced using the BigDye Terminator v3.1 Kit and an ABI Prism Genetic Analyzer 3100 (Life Technologies). The sequence data were analyzed in June 2015 using BlastN (44).
Amplification, Purification, and Stability of FLiP-Virions.
For the preparation of high-titer phage lysates, plates showing confluent lysis were used. Shieh (5 mL) was added onto each plate, and the suspension was incubated at a cold (∼6 °C) temperature for 3 h with agitation. The medium was collected from the plates, and the resulting supernatant was filtrated through a membrane filter (pore size 0.45 µm; PALL Life Sciences). The viral titer of the filtrated supernatant was determined by the plaque assay, and the phage stock was stored at 4 °C and for longer periods at −80 °C with 20% glycerol.
For phage purification, the lysate was collected from several hundred confluent plates. The lysate was filtered, and virus particles were precipitated with 10% (wt/vol) polyethylene glycol 6000 and 0.5 M NaCl. Phage precipitate was collected by centrifugation (Sorvall SLA3000) at 11,000 × g for 30 min at 4 °C and was suspended in 20 mM potassium phosphate buffer (PPB) (pH 7.2). Three different gradient materials (all in 20-mM PPB) were tested for optimal purification: glycerol (5–30%), OptiPrep (5–20%), and sucrose (5–20%). Sucrose was used for subsequent experiments. The suspension was layered on top of a 5–20% (wt/vol) sucrose gradient (in 20-mM PPB, pH 7.2) for subsequent rate zonal centrifugation using a Beckman Optima L-K90 ultracentrifuge with an SW28 rotor at 104,000 × g for 1 h at 15 °C. The light-scattering zone containing the virus was collected and further purified by equilibrium centrifugation in 20–70% (wt/vol) sucrose gradient (in 20 mM PPB, pH 7.2) using a Beckman Optima L-K90 centrifuge with an SW41 rotor at 175,000 × g for 19 h at 5 °C. Light-scattering zones were collected; then the phage particles were pelleted by centrifugation (Beckman Optima L-K90, 70Ti rotor, 112,000 × g, 3 h, 5 °C), suspended in 20 mM PPB (pH 7.2), and stored at −20 °C or −80 °C.
Analysis of Phage Proteins.
The protein concentrations of purified phage solutions were measured by the Bradford assay (45) using BSA as a standard. Structural proteins of the purified virions were resolved by 16% SDS/PAGE. The gel was stained with Coomassie brilliant blue to detect the proteins. For structural protein identification, protein bands were cut from the gel.
Structural proteins were identified by nanoHPLC-ESI-MS/MS in Proteome Factory (Proteome Factory AG). The LC-MS system included an Agilent 1100 nanoHPLC system (Agilent), a PicoTip electrospray emitter (New Objective), and an LTQ-FT or an Orbitrap XL Ultra mass spectrometer (Thermo Fisher). For LC-MS/MS analysis SDS/PAGE-separated proteins were excised from the gels and in-gel digested by trypsin. Peptides were first trapped and then desalted on the enrichment column (Zorbax SB C18, 0.3 × 5 mm; Agilent) for 5 min (solvent: 2.5% acetonitrile/0.5% formic acid), after which they were separated on a Zorbax 300SB-C18, 75 μm × 150 mm column (Agilent) using a linear gradient from 10 to 32% B (solvent A: 5% acetonitrile in water, solvent B: acetonitrile, both with 0.1% formic acid). Ions of interest were data-dependently subjected to MS/MS according to the expected charge state distribution of peptide ions. Proteins were identified by database search against a database containing the RefSeq entries of Flavobacterium johnsoniae UW101 (National Center for Biotechnology Information, NCBI) and FLiP sequences using MS/MS ion search of the Mascot search engine (Matrix Science). Only peptides matches with a score of 20 or higher were accepted.
Gel Electrophoresis and Edman Degradation.
Gel electrophoresis and Edman degradation were conducted in Proteome Factory (Proteome Factory AG). The sample (5–23 µg) was loaded onto 15% SDS/PAGE under reducing conditions. After electrophoresis, proteins were transferred onto an Immobilon-P PVDF membrane (pore size 0.45 mm; Millipore) using a Trans-Blot SD Semi-Dry Transfer Cell [Bio-Rad; 5 V; 16 h, 4 °C; blot-buffer: 40 mM borat (pH 9), 20% methanol] and visualized by Coomassie blue staining. Afterwards an ABI Procise 491 Protein Sequencer (Applied Biosystems) was used for automated N-terminal sequencing according to the manufacturer’s protocol.
Isolation, Sequencing, and Bioinformatics of the Phage Genome.
To isolate phage DNA, purified phage particles were disrupted by treatment with 2% SDS and 1.2 µg/mL proteinase K (37 °C, 45 min), followed by phenol-ether extraction and precipitation with sodium acetate and ethanol. The purified phage genome was digested with different nucleases [DNase I (Fermentas), RNase A (Sigma-Aldrich), RNase I (Thermo Scientific), S1 nuclease (Thermo Scientific), Mung bean nuclease (Promega), and EcoRI (Fermentas)] according to the manufacturers’ instructions. The same treatments were performed for the single-stranded DNA genome of φX174 (Thermo Scientific) as control. Digestion products were analyzed by agarose gel electrophoresis (1% agarose in 1× Tris-EDTA buffer).
Complementary strands were synthesized for the FLiP ssDNA genome using random hexamer primers and the Klenow fragment. Subsequently, the double-stranded cDNA was amplified by the Illustra GenomiPhi V2 DNA Amplification Kit and sequenced using Roche 454 GS FLX+ sequencer at the Institute of Biotechnology, University of Helsinki, Helsinki. These initial sequence data were used to design specific oligonucleotides for primer walking to fill in the sequence gaps between six contigs. The Klenow fragment was used again in these complementary PCR reactions. The resulting PCR products were first amplified by the Illustra GenomiPhi V2 DNA Amplification Kit and then were sequenced using conventional Sanger sequencing with the BigDye Terminator v3.1 Cycle Sequencing Kit and ABI Prism Genetic Analyzer 3100 (Life Technologies) to obtain the complete genome sequence of FLiP. Finally, the whole genome was covered by primer walking as described above.
The GC content of the genome was calculated using the Infoseq program from the EMBOSS package. Putative protein-encoding ORFs were predicted from the genome using the algorithms of GeneMark (21) and Glimmer (20). Proteins homologous to translated FLiP ORFs were found using the NCBI programs BlastP and PSI-Blast (46) against the nonredundant GenBank protein database. NCBI Conserved Domain Search was used to search for protein motifs. Translated ORFs were characterized by molecular mass and isoelectric point using programs included in the ExPASy Proteomics tools. Transmembrane domains were predicted using the TMHMM program (47).
Extraction of Cytoplasmic Membrane and Lipid Analysis.
Extraction of cytoplasmic membrane from the cells of Flavobacterium sp strain B330 was done according to Laurinavičius et al. (32) using a floatation gradient to separate membranes. Lipid extraction and analysis of the cytoplasmic membrane and purified phage was done commercially by the Lipidomics Unit at the University of Helsinki, using triple quadrupole ESI-MS/MS.
Cryo-EM.
An aliquot (3 µL) of purified particles was applied on a glow-discharged EM grid (C-flat; Protochips) and plunge-frozen in liquid ethane using a vitrification apparatus (Vitrobot mark IV; FEI) operated at room temperature and above 80% relative humidity. Data were acquired using a 300-kV transmission electron microscope (Tecnai F30 Polara; FEI) operated at liquid nitrogen temperature and equipped with an energy filter (GIF Quantum LS; Gatan; zero-loss mode with 20-eV slit width) and a direct electron detector (K2 Summit; Gatan). Movies (22 frames, total electron dose 22 electrons/Å2) were collected in electron-counting mode using a dose rate of eight electrons per pixel per second. Frames were aligned in MotionCor2 to compensate for specimen drift and electron beam-induced damage and were averaged together (48). Particles were picked from averaged images automatically using ETHAN (49). The picked particles were divided manually into two datasets, consisting of full and empty particles. Contrast transfer function (CTF) parameters were estimated locally using GCTF (50). The 3D structure of full particles was determined in Relion using established protocols for image classification and gold-standard structure refinement (23). As a starting model, the previously published structure of PM2 [Electron Microscopy Data Bank (EMDB) accession code EMD-1082] was used and was filtered to low resolution (40 Å) to avoid bias. The resolution of the reconstruction was estimated by FSC using a threshold of 0.143. The map was sharpened by applying an inverse B-factor of −100 Å2, and local resolution was estimated in Relion.
Model Building and Refinement.
The PM2 MCP [Protein Data Bank (PDB) ID code: 2VVF] was used to generate the FLiP asymmetric unit; 10 protein chains were fitted in the FLiP cryo-EM density using Chimera (51). Subsequently the density corresponding to the asymmetric unit was extracted with Phenix suite phenix.map_box (52) and CCP4 suite (53). To facilitate the sequence assignment, the map features were improved by B-factor sharpening in REFMAC (54) and density modification in PHENIX (52). The atomic model of a single MCP protomer was traced manually in the density map filtered to 3.9 Å using COOT (55). The initial model was rebuilt and refined in Rosetta release version 2016.32.58837 using protocols optimized for cryo-EM maps (56). The best-scoring model as estimated by density fit and geometry was selected and used in COOT to guide further model building and optimization. The final model was refined with Phenix suite phenix.real_space_refine using icosahedral constraints, secondary structure restraints, and reciprocal space B-factor refinement. The model was validated by calculating model-to-map real-space cross-correlation and FSC with and without masking the map around the model. Side-chains were validated by EMRinger (57) and model geometry was validated by MolProbity (58). Structural alignments of FLiP MCP and other viral MCPs with a double β-barrel fold and a phylogenetic tree were calculated using Homologous Structure Finder software (30). Figures were generated with Chimera (51).
Acknowledgments
We thank Professor Jaana Bamford for discussions and support; Ms. Heidi Pirttinen for assistance in the laboratory; Mr. Petri Papponen for help with transmission electron microscopy (TEM); Dr. Alistair Siebert for cryo-TEM support; and Dr. Pietro Roversi, Professor Axel Brünger, and Professor Frank DiMaio for helpful discussions on model building and refinement in EM maps. This work was supported by the Academy of Finland Center of Excellence Program in Biological Interactions 2012–2017 Grant 252411, by Academy of Finland Grant 266879 (to L.-R.S.), by the Jane and Aatos Erkko Foundation, by Medical Research Council Grant MR/N00065X/1 (to D.I.S.), and by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme Grant 649053 (to J.T.H.). A.G. is supported by Wellcome Trust 4-y PhD Studentship 106274/Z/14/Z. The Oxford Particle Imaging Centre was founded by Wellcome Trust Joint Infrastructure Fund Award 060208/Z/00/Z and is supported by Welcome Trust Equipment Grant 093305/Z/10/Z. The Wellcome Trust Centre for Human Genetics is supported by Wellcome Trust Centre Grant 090532/Z/09/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome sequence of FLiP has been submitted to GenBank (accession no. MF361639). The cryo-electron microscopy 3D reconstruction map of FLiP has been deposited with the Electron Microscopy Data Bank (EMDB) (accession code EMD-3771). The modeled structure of the virus MCP shell has been deposited with the Protein Data Bank (PDB) (accession code 5OAC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703834114/-/DCSupplemental.
References
- 1.Suttle CA. Marine viruses–Major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 2.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomaru Y, Nagasaki K. Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine eukaryotic microalgae. J Oceanogr. 2007;63:215–221. [Google Scholar]
- 4.Steward GF. Marine Microbiology, Methods in Microbiology. Elsevier; St. Petersburg, FL: 2001. Fingerprinting viral assemblages by Pulsed Field Gel Electrophoresis (PFGE) pp. 85–103. [Google Scholar]
- 5.Breitbart M, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angly FE, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 8.Desnues C, et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- 9.Tucker KP, Parsons R, Symonds EM, Breitbart M. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J. 2011;5:822–830. doi: 10.1038/ismej.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguirre de Cárcer D, López-Bueno A, Pearce DA, Alcamí A. Biodiversity and distribution of polar freshwater DNA viruses. Sci Adv. 2015;1:e1400127. doi: 10.1126/sciadv.1400127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krupovic M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr Opin Virol. 2013;3:578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Hatfull GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bamford DH, Burnett RM, Stuart DI. Evolution of viral structure. Theor Popul Biol. 2002;61:461–470. doi: 10.1006/tpbi.2002.1591. [DOI] [PubMed] [Google Scholar]
- 14.Bamford DH. Do viruses form lineages across different domains of life? Res Microbiol. 2003;154:231–236. doi: 10.1016/S0923-2508(03)00065-2. [DOI] [PubMed] [Google Scholar]
- 15.Abrescia NGA, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 16.Oksanen HM, et al. Viruses: Essential Agents of Life. Springer; Dordrecht, The Netherlands: 2012. Virus universe: Can it be constructed from a limited number of viral architectures; pp. 83–105. [Google Scholar]
- 17.Krupovič M, Bamford DH. Order to the viral universe. J Virol. 2010;84:12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eiler A, Bertilsson S. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ Microbiol. 2004;6:1228–1243. doi: 10.1111/j.1462-2920.2004.00657.x. [DOI] [PubMed] [Google Scholar]
- 19.Eiler A, Bertilsson S. Flavobacteria blooms in four eutrophic lakes: Linking population dynamics of freshwater bacterioplankton to resource availability. Appl Environ Microbiol. 2007;73:3511–3518. doi: 10.1128/AEM.02534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Škubník K, et al. Structure of deformed wing virus, a major honey bee pathogen. Proc Natl Acad Sci USA. 2017;114:3210–3215. doi: 10.1073/pnas.1615695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huiskonen JT, Kivelä HM, Bamford DH, Butcher SJ. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat Struct Mol Biol. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 25.Benson SD, Bamford JKH, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 26.Abrescia NGA, et al. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol Cell. 2008;31:749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Khayat R, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts MM, White JL, Grütter MG, Burnett RM. Three-dimensional structure of the adenovirus major coat protein hexon. Science. 1986;232:1148–1151. doi: 10.1126/science.3704642. [DOI] [PubMed] [Google Scholar]
- 30.Ravantti J, Bamford D, Stuart DI. Automatic comparison and classification of protein structures. J Struct Biol. 2013;183:47–56. doi: 10.1016/j.jsb.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Atanasova NS, et al. Advances in Virus Research. Vol 92. Academic; Waltham, MA: 2015. Comparison of lipid-containing bacterial and archaeal viruses; pp. 1–61. [DOI] [PubMed] [Google Scholar]
- 32.Laurinavičius S, Käkelä R, Somerharju P, Bamford DH. Phospholipid molecular species profiles of tectiviruses infecting Gram-negative and Gram-positive hosts. Virology. 2004;322:328–336. doi: 10.1016/j.virol.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Laurinavičius S, Bamford DH, Somerharju P. Transbilayer distribution of phospholipids in bacteriophage membranes. Biochim Biophys Acta. 2007;1768:2568–2577. doi: 10.1016/j.bbamem.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Holmfeldt K, et al. Twelve previously unknown phage genera are ubiquitous in global oceans. Proc Natl Acad Sci USA. 2013;110:12798–12803. doi: 10.1073/pnas.1305956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmfeldt K, Odić D, Sullivan MB, Middelboe M, Riemann L. Cultivated single-stranded DNA phages that infect marine Bacteroidetes prove difficult to detect with DNA-binding stains. Appl Environ Microbiol. 2012;78:892–894. doi: 10.1128/AEM.06580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietilä MK, Roine E, Sencilo A, Bamford DH, Oksanen HM. Pleolipoviridae, a newly proposed family comprising archaeal pleomorphic viruses with single-stranded or double-stranded DNA genomes. Arch Virol. 2016;161:249–256. doi: 10.1007/s00705-015-2613-x. [DOI] [PubMed] [Google Scholar]
- 37.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krupovic M, Koonin EV. Multiple origins of viral capsid proteins from cellular ancestors. Proc Natl Acad Sci USA. 2017;114:E2401–E2410. doi: 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krupovic M, Bamford DH. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics. 2007;8:236. doi: 10.1186/1471-2164-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laanto E, Sundberg L-R, Bamford JKH. Phage specificity of the freshwater fish pathogen Flavobacterium columnare. Appl Environ Microbiol. 2011;77:7868–7872. doi: 10.1128/AEM.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song YL, Fryer JL, Rohovec JS. Comparison of six media for the cultivation of Flexibacter columnaris. Fish Pathol. 1988;23:81–94. [Google Scholar]
- 42.Adams MH. Bacteriophages. John Wiley & Sons, Ltd; New York: 1959. [Google Scholar]
- 43.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 45.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Sonnhammer E. Protein family databases for automated protein domain identification. Database. 1998;9:68–78. [Google Scholar]
- 48.Zheng S, Palovcak E, Armache J-P, Cheng Y, Agard D. Anisotropic correction of beam-induced motion for improved single-particle electron cryo-microscopy. Nat Meth. 2016;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. Local average intensity-based method for identifying spherical particles in electron micrographs. J Struct Biol. 2000;131:126–134. doi: 10.1006/jsbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 50.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.DiMaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barad BA, et al. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]