Significance
Life-threatening susceptibility to common respiratory infections in previously healthy children can be indicative of pathogen-specific primary immunodeficiencies due to rare deleterious variants in key genes and pathways of the immune system. These findings have implications for prevention and treatment of susceptible children.
Keywords: respiratory syncytial virus, rhinovirus, IFIH1, RIG-I-like receptor family, severe pediatric infectious disease
Abstract
Viral respiratory infections are usually mild and self-limiting; still they exceptionally result in life-threatening infections in previously healthy children. To investigate a potential genetic cause, we recruited 120 previously healthy children requiring support in intensive care because of a severe illness caused by a respiratory virus. Using exome and transcriptome sequencing, we identified and characterized three rare loss-of-function variants in IFIH1, which encodes an RIG-I-like receptor involved in the sensing of viral RNA. Functional testing of the variants IFIH1 alleles demonstrated that the resulting proteins are unable to induce IFN-β, are intrinsically less stable than wild-type IFIH1, and lack ATPase activity. In vitro assays showed that IFIH1 effectively restricts replication of human respiratory syncytial virus and rhinoviruses. We conclude that IFIH1 deficiency causes a primary immunodeficiency manifested in extreme susceptibility to common respiratory RNA viruses.
Viral respiratory tract infections are the most common childhood infections worldwide, with close to 100% of children being infected during the first years of life. Whereas the vast majority of viral respiratory infections are mild and self-limiting, more severe disease leads to the hospitalization of about 3% of individuals in each birth cohort (1). In-hospital mortality rates are limited to <1% with intensive care support; still these infections account for 21% of childhood mortality worldwide (2, 3). The main viral pathogens causing lower respiratory tract infections are human respiratory syncytial virus (RSV), enteroviruses [including human rhinoviruses (HRV)], adenoviruses, human metapneumovirus, coronavirus, influenza, and parainfluenza viruses, with RSV being responsible for the majority of the hospitalized pediatric cases (4, 5).
A number of risk factors including socioeconomic and environmental influences, preterm birth, chronic diseases, and immunosuppression are associated with more severe clinical presentation (6). However, ∼1 out of 1,000 children without any known risk factor will require intensive care support due to life-threatening manifestations of common viral respiratory infections. In the absence of established differences in pathogen virulence, we hypothesized that human genetic variation contributes to unusual susceptibility to severe disease due to common viruses. Supporting evidence is provided by a recent study, which showed that rare variants in IRF7 resulted in life-threatening influenza in an otherwise healthy child (7).
We combined exome sequencing, transcriptomic analysis, and in vitro functional testing to identify and characterize potentially causal genetic variants in a prospective cohort of previously healthy children requiring intensive care support for common respiratory viral infections. We report the identification of a pathogen-restricted immunodeficiency due to loss-of-function variants in IFIH1, which result in defective innate recognition of RNA viruses, preventing the activation of an efficient antiviral IFN response.
Results
Study Participants.
We enrolled 120 previously healthy children admitted to pediatric intensive care units (PICUs) with respiratory failure due to a common viral respiratory infection. The most common clinical presentation was bronchiolitis (n = 105, 88%) and the median age was 78 d (interquartile range, IQR: 37–769). RSV was the most common pathogen, identified in 67 (56%) of the cases, followed by HRV in 31 (26%) of the cases (Table 1).
Table 1.
Baseline characteristics of the 120 study participants
| Parameter | Variable | Median (IQR) or N (%) |
| Age, d | 78 (37–269) | |
| Weight, kg | 5.9 (4.4–9.8) | |
| Country of recruitment | Australia | 100 (83%) |
| Switzerland | 20 (17%) | |
| Ethnicity | Caucasian | 90 (78%) |
| African | 4 (4%) | |
| Asian | 4 (4%) | |
| Australian aboriginal | 6 (5%) | |
| Pacific Islander | 11 (10%) | |
| Sex | Male | 70 (58%) |
| Female | 50 (42%) | |
| Clinical phenotype | Bronchiolitis | 105 (88%) |
| Pneumonia | 8 (7%) | |
| Laryngotracheobronchitis | 5 (4%) | |
| Reactive airway disease | 2 (2%) | |
| Virus identified in respiratory sample | RSV | 67 (56%) |
| Entero/rhinovirus | 31 (26%) | |
| Adenovirus | 17 (14%) | |
| Human bocavirus | 9 (8%) | |
| Influenza | 2 (2%) | |
| Parainfluenza | 6 (5%) | |
| Human metapneumovirus | 3 (2%) | |
| CoV-HKU1 | 3 (2%) | |
| CoV-NL63 | 1 (1%) | |
| Respiratory support | HFNC | 99 (83%) |
| CPAP or BiPAP | 30 (25%) | |
| Invasive ventilation | 31 (26%) | |
| HFOV | 6 (5%) | |
| Length of PICU stay, d | 2.7 (1.6–5.0) | |
| Expected mortality | Pediatric index of mortality 2 (%) | 0.02 (0.01–0.7) |
| Observed mortality | Pediatric index of mortality 2 (%) | No fatal case |
CoV-HKU1, coronavirus HkU1; CoV-NL63, coronavirus NL63.
Exome Sequencing and Analysis.
DNA samples were sequenced to a mean coverage of 70×, with 96% of exonic bases achieving at least 10× coverage and 78% achieving at least 30× coverage. The final set of variants included 2,793 stop-gained single-nucleotide variants (SNVs), 297 splice-site SNVs, and 951 frame-shift indels. Among these putative loss-of-function variants (LoFs), we searched for variants that were homozygous in at least one study participant, and with a higher minor allele frequency in our cohort than in the genome Aggregation Database (gnomAD) (8) and in an in-house collection of 485 exomes. Six putative LoFs fitting these criteria were identified (SI Appendix, Table S3). This set included a variant in the IFN induced with helicase C domain 1 (IFIH1) gene, which encodes an RIG-I-like cytoplasmic sensor of long double-stranded RNA (dsRNA) and plays a major role in innate immune recognition of RNA viruses (9).
LoF Variants in IFIH1.
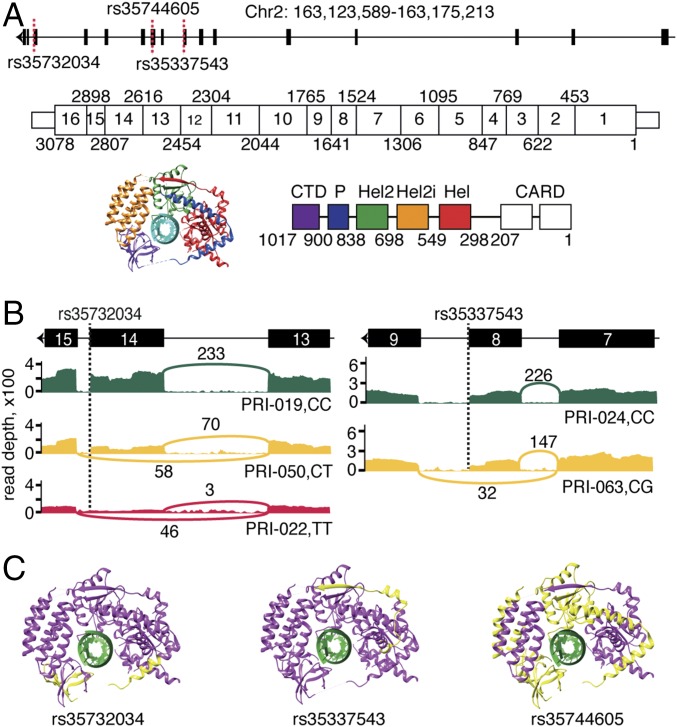
In total, eight study participants carried putative LoF variants in IFIH1 (Table 2). Four study participants carried a rare [gnomAD minor allele frequency (MAF) = 0.64%] splicing variant, rs35732034 (Fig. 1A): one in homozygous and three in heterozygous form. We used RNA sequencing to characterize the transcriptomic impact of this variant. We observed that the minor allele T causes skipping of exon 14 (IFIH1-Δ14) (Fig. 1B), which results in a frame shift and an early stop codon in exon 15. The resulting protein lacks the final 153 amino acids of wild-type IFIH1, including the C-terminal regulatory domain (CTD), which is essential for binding to viral dsRNA (10) (Fig. 1C). Western blot analyses of peripheral blood mononuclear cells from the homozygous patient and her heterozygous parents demonstrated that the IFIH1-Δ14 protein is expressed upon in vitro RSV infection (SI Appendix, Fig. S1). We identified two additional rare LoF variants in IFIH1, present in heterozygous form in a total of four study participants (Fig. 1A): the splicing variant rs35337543 (n = 3, gnomAD MAF = 0.67%) and the stop-gained variant rs35744605 (n = 1, gnomAD MAF = 0.32%). RNA sequencing showed that the minor allele G at rs35337543 causes skipping of exon 8 (IFIH1-Δ8) (Fig. 1B), which removes 39 amino acids at the end of the helicase 1 domain and in the linker part between helicase 1 and helicase 2, but does not result in a frame shift. rs35744605 is a stop-gained SNV in exon 10 that leads to the loss of 399 amino acids from the C-terminal end of IFIH1 (IFIH1-ΔCTD) (Fig. 1C).
Table 2.
Characteristics of the eight study participants carrying an IFIH1 loss-of-function variant
| Patient ID | Zygosity | Sex | Age at admission, d | Ethnicity | Virus | Ventilation required | Clinical presentation | Parental allele | Variant ID | Nucleotide change | Amino acid change | gnomAD AC (#hom), MAF |
| PRI_022 | hom | F | 493 | White | RSV | I | Bronchiolitis pneumonia | het in both parents | rs35732034 | 2:163124596 C/T | p.Ile872Ter | 1673 (7), 0.006 |
| PRI_050 | het | M | 34 | Aboriginal | HRV | NI | Recurrent bronchiolitis | NA | ||||
| PRI_061 | het | M | 41 | White | RSV | NI | Bronchiolitis | het in father | ||||
| PRI_122 | het | F | 48 | White | RSV | NI | Bronchiolitis | het in mother | ||||
| PRI-063 | het | M | 22 | White | RSV | NI | Bronchiolitis | NA | rs35337543 | 2:163136505 C/G | p.Leu509_Glu547del | 1847 (8), 0.006 |
| PRI-065 | het | M | 38 | White | RSV | NI | Bronchiolitis | NA | ||||
| PRI-116 | het | M | 479 | White | RSV | NI | Bronchiolitis | NA | ||||
| PRI-080 | het | M | 343 | White | HRV | I | Bronchiolitis pneumonia | NA | rs35744605 | 2:163134090 C/A | p.Glu627Ter | 887 (1), 0.003 |
NA, not available; het, heterozygous; hom, homozygous; I, invasive; NI, noninvasive; AC, allele count.
Fig. 1.
LoF variants identified in IFIH1. Related to SI Appendix, Fig. S1. (A) Schematic representation of IFIH1 DNA, mRNA, and protein. The identified variants are indicated with dashed red lines. Exon boundaries are marked with nucleotide coordinates. Protein domain boundaries are marked with amino acid coordinates. (B) Alternative splicing of IFIH1 associated with rs35732034 and rs35337543 genotypes, as seen in RNA sequencing data. The T allele at rs35732034 leads to skipping of exon 14. The G allele at rs35337543 leads to skipping of exon 8. The Sashimi plots illustrate the genotype-dependent abundance of splice junctions. The number of observed reads spanning the respective splice junctions is indicated on the Bezier curves, which connect exons. (C) Schematic 3D representation of IFIH1 (Protein Data Bank ID code: 4GL2, image produced using UCSC Chimera). The parts of the protein that are predicted to be missing due to rs35732034, rs35744605 and rs35337543 variants are indicated in yellow. Hel, helicase domain; P, pincer.
Description of Study Participants Carrying IFIH1 LoF Variants.
A 16-mo-old girl was homozygous for rs35732034. She presented with respiratory failure due to RSV infection requiring invasive ventilation. The disease course was complicated by a pulmonary superinfection with Staphylococcus aureus. She had a full recovery and did not develop any other severe infection up to the age of 3. Her phenotype and history was otherwise unremarkable. In particular, she did not develop any complication after live vaccine administration. Full blood count, Ig levels and IgG subclasses, and lymphocyte subclasses were within normal limits. Three infants requiring noninvasive respiratory support for bronchiolitis were heterozygous for rs35732034. One of these had recurrent severe viral lower respiratory tract infections leading to repeated PICU admissions during childhood. Three children were heterozygous for rs35337543 and required noninvasive ventilatory support for RSV bronchiolitis. One infant was heterozygous for rs35744605 and required invasive ventilatory support for HRV-positive bronchiolitis. Parental DNA was available for three of the eight children. Targeted genotyping confirmed that the relevant IFIH1 variant was present in heterozygous form in one of the parents for two heterozygous individuals, and in both parents for the homozygous patient. Parental medical history was unremarkable in all cases.
Functional Characterization of IFIH1 Variants.
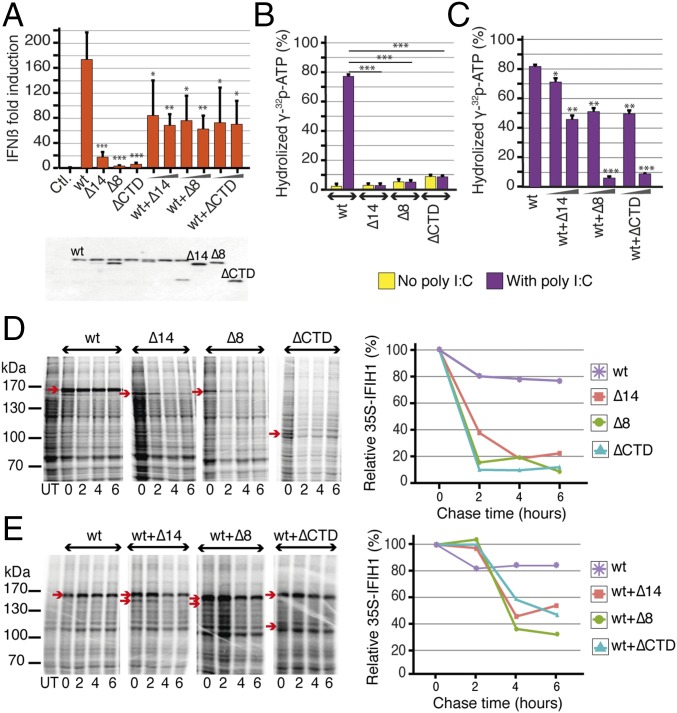
To functionally characterize the identified variants, we first measured the ability of wild-type (IFIH1-wt) and mutant IFIH1 isoforms to induce IFN β (IFNβ) in vitro. We transfected plasmids carrying IFIH1-wt, IFIH1-Δ8, IFIH1-Δ14, and IFIH1-ΔCTD into 293T cells. Overexpression of IFIH1-wt, but not of any of the mutant IFIH1 isoforms, led to IFNβ induction. Cotransfection of IFIH1-wt with each of the mutant IFIH1 isoforms showed interference with IFIH1-wt-induced IFNβ production (P < 0.05, Fig. 2A). We then tested the ATPase activity of recombinant IFIH1-wt and mutant IFIH1 isoforms. IFIH1-wt was able to hydrolyze ATP and showed a typical dsRNA-dependent increase in enzymatic activity, whereas the mutant isoforms had no detectable ATPase activity, even upon stimulation with polyinosinic:polycytidylic acid (polyI:C), a synthetic analog of dsRNA (Fig. 2B). Furthermore, all mutant isoforms decreased IFIH1-wt ATPase activity in a dose-dependent manner, an interference that was specific to mutant IFIH1 isoforms, as demonstrated by the absence of any effect of BSA on IFIH1-wt ATPase activity (P < 0.05, Fig. 2C and SI Appendix, Fig. S2 A and B). Finally, we checked the stability of the various IFIH1 protein isoforms by performing pulse-chase experiments in transfected 293T cells. The three mutant isoforms were less stable than IFIH1-wt (Fig. 2D) and had a negative impact on the stability of the wild-type isoform when cotransfected (Fig. 2E). Jointly, these experiments demonstrate that the three putative LoF variants identified in our study population lead to severe disruption of IFIH1 signaling function, enzymatic activity, and protein stability in vitro. In addition, the observations that the mutant IFIH1 isoforms interfere with the wild-type protein in terms of IFNβ induction, and enzymatic activity and protein stability suggest a dominant negative role for heterozygous LoF variants in IFIH1.
Fig. 2.
Functional characterization of the IFIH1 variants. Related to SI Appendix, Fig. S2. (A) Transfection with IFIH1-wt plasmid (20 ng) results in strong IFNβ induction in 293T cells, whereas transfection of plasmids harboring any of the IFIH1 variants cannot induce IFNβ (240 ng, n = 4). Cotransfection experiments demonstrate an interference of the mutant plasmids with IFIH1-wt on IFNβ induction (20-ng wt plasmid, 20- and 120-ng mutant plasmids, n = 4). Expression levels of the IFIH1 isoforms are shown under the plot in the Western blot gel. *P < 0.05, **P < 0.01, ***P < 0.001. (B) RNA-induced ATPase activity of purified IFIH1-wt protein and alternate IFIH1 isoforms; IFIH1-wt can hydrolyze ATP in the presence of polyI:C, whereas IFIH1-Δ14, IFIH1-Δ8, and IFIH1-ΔCTD lack ATPase activity, with (purple) or without (yellow) polyI:C stimulation. (C) ATPase activity of IFIH1-wt is reduced upon coincubation with the alternate isoforms in a dose-dependent manner (300-ng wt protein, 300- or 600-ng of each alternate isoform, 10-ng polyI:C, n = 2). *P < 0.05, **P < 0.01, ***P < 0.001. (D and E) Protein stability followed by pulse chase in 293T cells expressing IFIH1-wt, IFIH1-Δ14, IFIH1-Δ8, or IFIH1-ΔCTD. Each protein is marked on the gel by an arrow, and relative amounts of proteins are shown in the graphs. IFIH1-wt is more stable than the alternate IFIH1 isoforms, and the stability of IFIH1-wt is reduced upon coexpression with any of the alternate isoforms. Molecular mass markers are shown on the left of each gel (kilodaltons) and the bands corresponding to IFIH1-wt, IFIH1-Δ14, IFIH1-Δ8, or IFIH1-ΔCTD are indicated using a red arrow (2-μg plasmid expressing wt protein, 2 μg of each mutant plasmid, n = 1). Data are represented as mean ± SD; polyI:C, polyinosinic:polycytidylic acid.
Role of IFIH1 in RSV and HRV Replication.
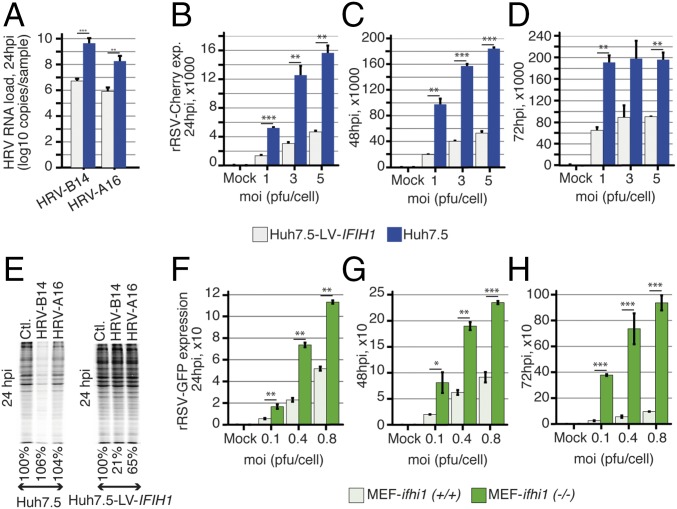
Viral testing of respiratory samples showed that six of the patients harboring IFIH1 LoF alleles were infected with RSV and two with HRV. To study the effect of IFIH1 on RSV and HRV replication, we used Huh7.5 cells, which lack endogenous expression of IFIH1 and express a mutated, inactive form of RIG-I, and thus are completely unreactive to the RNA pathogen-associated molecular patterns that normally activate these pathways (11). The cells were transduced with an IFIH1-wt-expressing lentiviral vector, which made them highly responsive to polyI:C stimulation (SI Appendix, Fig. S3A) without causing any nonspecific or constitutive activation of the IFN system (SI Appendix, Fig. S3B). We observed a much higher level of viral replication in native than in IFIH1-wt-transduced Huh7.5 cells upon infection with HRV-B14, HRV-A16, and RSV (Fig. 3 A–D). Furthermore, RSV replication level was higher in cells transduced with the mutant forms of IFIH1 than in IFIH1-wt-transduced cells (P < 0.05, SI Appendix, Fig. S4 A and B). The role of IFIH1 in HRV restriction was further demonstrated by 35S labeling of infected cells, which showed a stronger shutoff of cellular protein synthesis in native than in IFIH1-transduced Huh7.5 cells, due to higher replication of the virus in the absence of IFIH1 (Fig. 3E). We also measured RSV replication in mouse embryonic fibroblasts (MEFs), ifih1(+/+), and in IFIH1-knockout MEFs, ifih1(−/−), and obtained similar results (Fig. 3 F–H). Together, these results affirm the central role of IFIH1 in innate immune recognition of RSV and HRV (12, 13). Therefore, LoF variants in IFIH1 can be reasonably expected to increase susceptibility to these viruses.
Fig. 3.
IFIH1 restricts HRV and RSV replication. Related to SI Appendix, Figs. S3 and S4. (A) Real-time PCR of HRV-B14 and HRV-A16 RNA in Huh7.5 and Huh7.5 cells transduced with a lentiviral vector expressing IFIH1 (Huh7.5-LV-IFIH1). HRV-B14 and HRV-A16 replicate more efficiently in the absence of IFIH1 at 24 h postinfection (hpi) (n = 5 for HRV-B14 and n = 2 for HRV-B16). *P < 0.05, **P < 0.01, ***P < 0.001. (B–D) FACS analyses of mCherry expressing recombinant RSV (rRSV-Cherry) in Huh7.5 and Huh7.5-LV-IFIH1 transduced cells at 24, 48, and 72 hpi show that RSV replicates more efficiently in the absence of IFIH1 (n = 2). (E) Cellular proteins labeled with 35S at 24 hpi with both HRV-B14, HRV-A16 showing a much stronger shutoff of cellular protein synthesis in Huh7.5 cells than in Huh7.5-LV-IFIH1 transduced cells, due to higher viral replication. (F–H) FACS analyses of GFP expressing recombinant RSV (rRSV-GFP) in ifih1 knockout MEFs [MEF-ifih1(−/−)] and ifih1 knockout MEFs transduced with a lentiviral vector expressing IFIH1 (MEF-LV-IFIH1). RSV replicates more efficiently in the absence of IFIH1 at 24, 48, and 72 hpi (n = 2). *P < 0.05, **P < 0.01, ***P < 0.001. Data are represented as mean ± SD. GFP, green florescent protein; MEF, mouse embryonic fibroblast; moi, multiplicity of infection; pfu, plaque-forming unit.
Discussion
We hypothesized that extreme susceptibility to common viral respiratory infection in previously healthy children––a rare, potentially lethal phenotype––could reflect an underlying primary immunodeficiency. Using an unbiased exomewide approach in a prospective cohort of carefully selected individuals requiring intensive care support, we identified a rare monogenic defect predisposing to severe clinical presentations of RSV and HRV infections.
Three deleterious variants were observed in IFIH1, which encodes a cytoplasmic receptor critical for viral RNA sensing. It has been shown previously that IFIH1, alone or in combination with RIG-I, recognizes and limits the replication of many RNA viruses including: positive single-stranded RNA (ssRNA) viruses like picornaviruses (14–16), negative ssRNA viruses like paramyxoviruses (17–19), and dsRNA viruses like reoviruses (20).
IFIH1 recognizes viral RNA via interaction of its CTD and helicase domains with long dsRNA molecules. This is an ATP-dependent reaction that leads to polymerization of IFIH1 molecules into a filament and assembly of IFIH1 caspase activation recruitment domains (CARDs) (21, 22). This in turn initiates a signaling cascade that results in type 1 IFN production and activation of antiviral genes (23). Our transfection and transduction analyses show that this process is disrupted in the presence of any of the IFIH1 rare variants found in our study population. Our exome and RNA sequencing data predict that the loss of IFIH1 function is due to loss of the CTD (rs35732034 and rs35744605) or to partial loss of the helicase domain (rs35337543).
We observed interference between IFIH1-wt and the three mutant proteins in terms of stability, ATPase activity, and capacity to induce IFNβ production, suggesting a dominant negative effect, which provides a rationale for the unusual susceptibility to respiratory viruses observed in heterozygous individuals. The exact interfering mechanism is not known but could involve physical interaction between IFIH1-wt and mutant proteins, preventing the formation of normal, multimeric IFIH1 filaments.
While the revised version of this paper was under review, an independent study showed an association between IFIH1 deficiency and life-threatening infections with HRV and other respiratory viruses in a child carrying another homozygous missense IFIH1 variant (24). This observation further supports a causal role for IFIH1 deficiency in extreme susceptibility to common respiratory viruses.
The three IFIH1 variants described in this study have allele frequencies of 0.32–0.67% in gnomAD. The cumulative frequency of all putative LoF alleles is 1.89% in the same database, which is significantly less than the 3.75% cumulative frequency observed in our study population (P = 0.037, Fisher’s exact test). Nevertheless, the presence of alleles of potentially devastating consequences at such frequency in the general population is intriguing, as they are expected to be removed by purifying selection. Two nonexclusive mechanisms can explain this observation: balancing selection and incomplete penetrance. We here show that some IFIH1 alleles increase susceptibility to viral respiratory infections, but the same LoF variants are known to be protective against type 1 diabetes and other autoimmune diseases (25–30), strongly suggesting a role for balancing selection in their maintenance. In a comparable example, rare nonsynonymous variants in TYK2, a known primary immunodeficiency gene, were shown to be protective against rheumatoid arthritis (31). Incomplete penetrance, on the other hand, could be due to modulating effects of environmental or genetic factors, like compensatory mutations, or to functional redundancy in innate immune response to RNA viruses (32, 33). This hypothesis is in line with several recent publications (34–37), which suggest that incomplete penetrance and genetic heterogeneity are likely to be the rule rather than the exception in severe clinical presentations of infectious diseases.
On top of their associations with autoimmunity, more common IFIH1 variants have also been associated with hepatitis C virus clearance (38). Additionally, rare gain-of-function mutations in IFIH1 dramatically up-regulate type I IFN production, resulting in Aicardi–Goutières syndrome or Singleton–Merten syndrome (39–41). At the functional level, Gorman et al. recently studied the effects on viral sensing and autoimmune pathogenesis of rs1990760, a missense IFIH1 variant that is associated with multiple autoimmune diseases (30). They showed that the allele providing better defense against viral infection also bolsters autoimmune responses against self-RNA (42). Together, these results underscore the pivotal role of innate immune recognition and activation in the intricate balance between host defense, inflammation, and autoimmunity.
Our study demonstrates the power of using an unbiased, exome sequencing approach to variant discovery in prospective cohorts of extreme infectious disease phenotypes. Nevertheless, LoF variants in IFIH1 were only found in a minority (n = 8, 6.2%) of the 120 children enrolled in our study, suggesting that other genetic or nongenetic risk factors remain to be discovered. Larger sample sizes will be required to delineate the relevance of other rare potentially causal alleles. Whole genome sequencing will also be needed to obtain a more complete coverage of exonic regions (43), and to explore noncoding and large-scale structural variation.
RSV and rhinovirus infections are the two most common viral respiratory infections in children. The elucidation of the human genetic basis of extreme susceptibility to these viruses provides insight into pathogenesis and innate immune response. An immediate practical implication is the possibility to develop diagnostic assays to identify susceptible individuals who could benefit from specific preventive and interventional measures. By highlighting the genes and pathways that play an essential role in host–pathogen interaction, genetic discovery in individuals with extreme phenotypes also provides the opportunity to design new therapeutic strategies that could be useful for the vast majority of patients with milder clinical presentation.
Materials and Methods
Between December 2010 and October 2013, we prospectively enrolled previously healthy children below 4 y of age suffering from severe lower respiratory tract infection and requiring invasive or noninvasive respiratory support in five specialized PICUs from Australia and Switzerland. The study was approved by the respective institutional Human Research Ethics Committees. Written informed consent was obtained from parents or legal guardians.
Exclusion criteria were the presence of any significant underlying disease or comorbidity, including prematurity, congenital cardiac disease, chronic lung disease, sickle cell disease, hepatic, renal, or neurologic chronic conditions, solid and hematological malignancies, and known primary immunodeficiency. Respiratory support was defined as noninvasive ventilation including high-flow nasal cannulae (HFNC) and continuous or bilevel positive airway pressure (CPAP and BiPAP), or invasive ventilation including conventional and high-frequency oscillation ventilation (HFOV). The following demographic and clinical information was collected: age, gender, weight, ethnicity, type of ventilation, length of ventilation in days, clinical outcome, microbiological diagnostic procedures and results including rapid antigenic test for RSV and influenza, respiratory virus PCR panel, and viral cultures. For each study participant, we obtained a nasopharyngeal aspirate or endotracheal tube aspirate, 1 mL EDTA blood in vacutainer tubes, and 2.5 mL blood in PAXgene blood RNA tubes. Samples were immediately frozen at −70 °C until shipment, and then analyzed in batch.
We generated high-coverage exome sequencing data for all study participants (SI Appendix, Table S1). We then used a combination of three variant calling methods (GATK, Platypus, and SAMtools) and only kept SNVs and small insertion and deletions (SI Appendix, Table S2). Assuming that causal genetic variants are likely to be highly deleterious, we focused on rare gene knockout events, defined as homozygous, putative LoF variants (stop-gained and splice-site SNVs, frame-shift indels) with an MAF of <1% in the ExAC (SI Appendix, Table S3). We performed RNA sequencing to assess the transcriptomic impact of candidate DNA variants, and characterized the infecting viruses using multiplex PCR assays. We then used in vitro functional testing to demonstrate the biological relevance of candidate variants and tested in vivo expression of the affected protein (SI Appendix).
Supplementary Material
Acknowledgments
We express our sincere appreciation to all study participants and their families for their participation and support. We thank Deon Venter and Marc Vipond, Mater Pathology, Mater Health Services, Brisbane, as well as Patricia Keith, Institute for Health and Biomedical Innovation, Queensland University of Technology, Brisbane, Australia, for their help in sample processing and sampling logistics; Mark Peeples (Columbus, OH) for sharing rRSV-GFP, and Jean-François Eléouët (Institut National de la Recherche Agronomique, Jouy-en-Josas, France) for sharing rRSV- mCherry. mCherry. This study was funded by Swiss National Science Foundation Grants PP00P3_133703 and PP00P3_157529 (to J.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704259114/-/DCSupplemental.
References
- 1.Meissner HC. Viral bronchiolitis in children. N Engl J Med. 2016;374:62–72. doi: 10.1056/NEJMra1413456. [DOI] [PubMed] [Google Scholar]
- 2.Nair H, et al. Severe Acute Lower Respiratory Infections Working Group Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: A systematic analysis. Lancet. 2013;381:1380–1390. doi: 10.1016/S0140-6736(12)61901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: An updated systematic analysis. Lancet. 2015;385:430–440. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 4.Jain S, et al. CDC EPIC Study Team Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372:835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debiaggi M, Canducci F, Ceresola ER, Clementi M. The role of infections and coinfections with newly identified and emerging respiratory viruses in children. Virol J. 2012;9:247. doi: 10.1186/1743-422X-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez DA, et al. Predictors of severity and mortality in children hospitalized with respiratory syncytial virus infection in a tropical region. Pediatr Pulmonol. 2014;49:269–276. doi: 10.1002/ppul.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciancanelli MJ, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 10.Takahasi K, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: Identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q, Langereis MA, van Kuppeveld FJ. Induction and suppression of innate antiviral responses by picornaviruses. Cytokine Growth Factor Rev. 2014;25:577–585. doi: 10.1016/j.cytogfr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grandvaux N, et al. Sustained activation of interferon regulatory factor 3 during infection by paramyxoviruses requires MDA5. J Innate Immun. 2014;6:650–662. doi: 10.1159/000360764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barral PM, et al. MDA-5 is cleaved in poliovirus-infected cells. J Virol. 2007;81:3677–3684. doi: 10.1128/JVI.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slater L, et al. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. 2010;6:e1001178. doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gitlin L, et al. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6:e1000734. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shingai M, et al. Differential type I IFN-inducing abilities of wild-type versus vaccine strains of measles virus. J Immunol. 2007;179:6123–6133. doi: 10.4049/jimmunol.179.9.6123. [DOI] [PubMed] [Google Scholar]
- 19.Baños-Lara Mdel R, Ghosh A, Guerrero-Plata A. Critical role of MDA5 in the interferon response induced by human metapneumovirus infection in dendritic cells and in vivo. J Virol. 2013;87:1242–1251. doi: 10.1128/JVI.01213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loo Y-MM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci USA. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawling DC, Pyle AM. Parts, assembly and operation of the RIG-I family of motors. Curr Opin Struct Biol. 2014;25:25–33. doi: 10.1016/j.sbi.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loo Y-MM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamborn IT, et al. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J Exp Med. 2017;214:1949–1972. doi: 10.1084/jem.20161759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chistiakov DA, Voronova NV, Savost’Anov KV, Turakulov RI. Loss-of-function mutations E6 27X and I923V of IFIH1 are associated with lower poly(I:C)-induced interferon-β production in peripheral blood mononuclear cells of type 1 diabetes patients. Hum Immunol. 2010;71:1128–1134. doi: 10.1016/j.humimm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 28.Yang H, et al. IFIH1 gene polymorphisms in type 1 diabetes: Genetic association analysis and genotype-phenotype correlation in Chinese Han population. Autoimmunity. 2012;45:226–232. doi: 10.3109/08916934.2011.633134. [DOI] [PubMed] [Google Scholar]
- 29.Cen H, et al. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity. 2013;46:455–462. doi: 10.3109/08916934.2013.796937. [DOI] [PubMed] [Google Scholar]
- 30.Chistiakov DA. Interferon induced with helicase C domain 1 (IFIH1) and virus-induced autoimmunity: A review. Viral Immunol. 2010;23:3–15. doi: 10.1089/vim.2009.0071. [DOI] [PubMed] [Google Scholar]
- 31.Diogo D, et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS One. 2015;10:e0122271. doi: 10.1371/journal.pone.0122271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen S, Thomsen AR. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, et al. Loss of RIG-I leads to a functional replacement with MDA5 in the Chinese tree shrew. Proc Natl Acad Sci USA. 2016;113:10950–10955. doi: 10.1073/pnas.1604939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong X-FF, et al. Haploinsufficiency at the human IFNGR2 locus contributes to mycobacterial disease. Hum Mol Genet. 2013;22:769–781. doi: 10.1093/hmg/dds484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim HK, et al. TLR3 deficiency in herpes simplex encephalitis: High allelic heterogeneity and recurrence risk. Neurology. 2014;83:1888–1897. doi: 10.1212/WNL.0000000000000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conley ME, Casanova J-LL. Discovery of single-gene inborn errors of immunity by next generation sequencing. Curr Opin Immunol. 2014;30:17–23. doi: 10.1016/j.coi.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann FS, et al. Polymorphisms in melanoma differentiation-associated gene 5 link protein function to clearance of hepatitis C virus. Hepatology. 2015;61:460–470. doi: 10.1002/hep.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice GI, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oda H, et al. Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am J Hum Genet. 2014;95:121–125. doi: 10.1016/j.ajhg.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutsch F, et al. A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. Am J Hum Genet. 2015;96:275–282. doi: 10.1016/j.ajhg.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorman JA, et al. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol. 2017;18:744–752. doi: 10.1038/ni.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilissen C, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.