Significance
Conformational properties of unfolded and intrinsically disordered proteins (IDPs) under native conditions are important for understanding the details of protein folding and the functions of IDPs. The average dimensions of these systems are quantified using the mean radius of gyration and mean end-to-end distance, measured by small-angle X-ray scattering (SAXS) and single-molecule Förster resonance energy transfer (smFRET), respectively, although systematic discrepancies emerge from these measurements. Through holistic sets of studies, we find that the disagreements arise from chemical heterogeneity that is inherent to heteropolymeric systems. This engenders a decoupling between different measures of overall sizes and shapes, thus leading to discrepant inferences based on SAXS vs. smFRET. Our findings point the way forward to obtaining comprehensive descriptions of ensembles of heterogeneous systems.
Keywords: single-molecule FRET, intrinsically disordered proteins, denatured-state ensemble, protein folding, polymer theory
Abstract
Unfolded states of proteins and native states of intrinsically disordered proteins (IDPs) populate heterogeneous conformational ensembles in solution. The average sizes of these heterogeneous systems, quantified by the radius of gyration (RG), can be measured by small-angle X-ray scattering (SAXS). Another parameter, the mean dye-to-dye distance (RE) for proteins with fluorescently labeled termini, can be estimated using single-molecule Förster resonance energy transfer (smFRET). A number of studies have reported inconsistencies in inferences drawn from the two sets of measurements for the dimensions of unfolded proteins and IDPs in the absence of chemical denaturants. These differences are typically attributed to the influence of fluorescent labels used in smFRET and to the impact of high concentrations and averaging features of SAXS. By measuring the dimensions of a collection of labeled and unlabeled polypeptides using smFRET and SAXS, we directly assessed the contributions of dyes to the experimental values RG and RE. For chemically denatured proteins we obtain mutual consistency in our inferences based on RG and RE, whereas for IDPs under native conditions, we find substantial deviations. Using computations, we show that discrepant inferences are neither due to methodological shortcomings of specific measurements nor due to artifacts of dyes. Instead, our analysis suggests that chemical heterogeneity in heteropolymeric systems leads to a decoupling between RE and RG that is amplified in the absence of denaturants. Therefore, joint assessments of RG and RE combined with measurements of polymer shapes should provide a consistent and complete picture of the underlying ensembles.
Quantitative characterizations of the sizes, shapes, and amplitudes of conformational fluctuations of unfolded proteins under denaturing and native conditions are directly relevant to advancing our understanding of the collapse transition during protein folding. These types of studies are also relevant to furthering our understanding of the functions and interactions of intrinsically disordered proteins (IDPs) in physiologically relevant conditions (1). Polymer physics theories provide the conceptual foundations for analyzing conformationally heterogeneous systems such as IDPs and unfolded ensembles of autonomously foldable proteins (2–4). Specifically, order parameters in theories of coil-to-globule transitions and analytical descriptions of conformational ensembles (5, 6) are based on ensemble-averaged values of radii of gyration (RG) and amplitudes of fluctuations measured by end-to-end distances (RE).
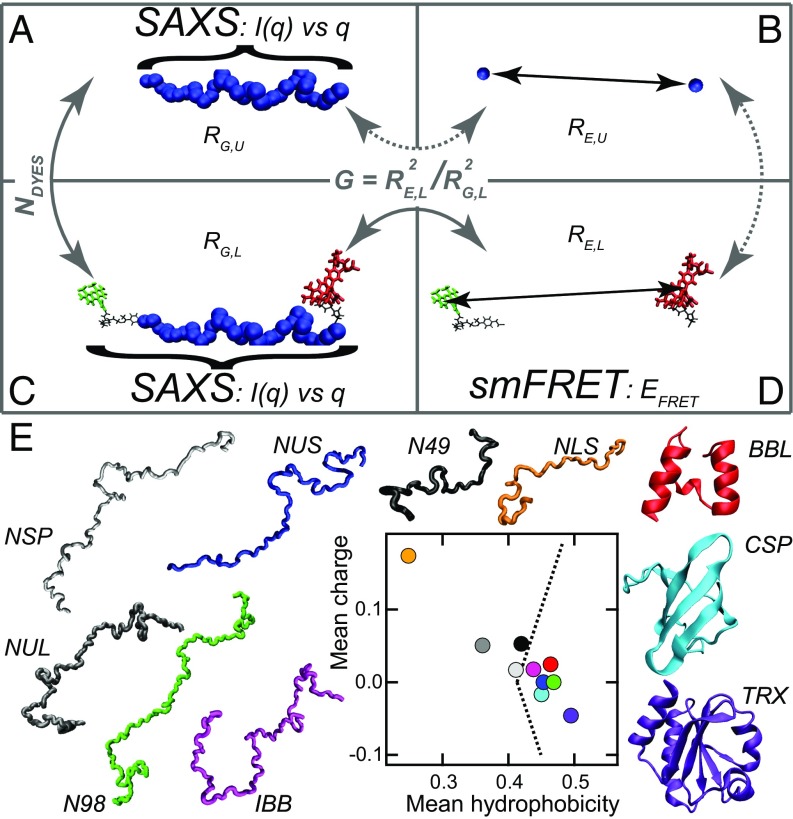
Estimates of RG are accessible through small-angle X-ray scattering (SAXS) measurements because scattering intensities are directly related to the global protein size (Fig. 1) (7, 8). At finite concentrations, assuming the absence of intermolecular interactions, RG is proportional to the square root of the mean square of interatomic distances within individual molecules averaged over the conformations of all molecules in solution (see SI Appendix, Table S1 for details). Estimates of RE can be made from single-molecule Förster resonance energy transfer (smFRET) experiments. Here, donor and acceptor fluorophores are covalently attached to N- and C-terminal ends of the protein of interest and the measured mean FRET efficiencies () are used to infer the mean distances between dyes (RE,L) (Fig. 1D). This serves as a useful proxy for estimating RE although it requires the assumption of an a priori functional form for the distribution of interdye distances, which is often based on the Gaussian chain model (9–13). Because dyes are attached to the protein sidechain via flexible linkers, RE,L is different from the actual end-to-end distance RE, which we denote as RE,U (Fig. 1B). Similarly, the RG of an unlabeled protein, RG,U, should be numerically different from the RG of a labeled protein, RG,L (compare Fig. 1 A and C).
Fig. 1.
The combined SAXS/smFRET approach. Proteins are depicted as a chain of beads (blue), where each bead represents an amino acid residue. Donor dye (Alexa488), acceptor dye (Alexa594), and their linkers are shown in green, red, and black traces, respectively. (A) The radius of gyration of an unlabeled protein, RG,U, can be estimated from a SAXS profile were the intensity of scattered X-rays is recorded as a function of the scattering vector q. (B) The end-to-end distance of the polymer, RE,U, is not directly accessible by smFRET. (C) The radius of gyration of a labeled protein, RG,L, can also be measured by SAXS. (D) The donor-to-acceptor distance, RE,L, can be estimated via the FRET efficiencies (EFRET) measured by smFRET upon assumption of a model. RE,L and RG,L can be related to each other via the G ratio (RE2/RG2). (E) Mean charge of the 10 proteins used in this study plotted against their mean hydrophobicity. Dashed lines show the theoretical prediction separating IDPs (Left) from folded proteins (Right).
Proteins that fold autonomously under physiological conditions can be denatured in high concentrations of urea or guanidinium hydrochloride (14). Upon dilution of denaturants, proteins collapse and fold to form compact structures. An unresolved issue is the nature of the collapse transition (2, 4, 13, 15, 16). Inferences from smFRET measurements suggest that proteins, including IDPs, undergo continuous contraction as the denaturant concentration is decreased (4, 16, 17). The implication for protein folding is that the acquisition of persistent local and nonlocal contacts might follow barrierless collapse that leads to the formation of globules. Inferences from SAXS measurements provide a discrepant view of the collapse transition for protein folding and for IDPs (15, 18, 19). In these experiments, the measured RG values are shown to change minimally over a wide range of denaturant concentrations. Therefore, one might conclude that the collapse transition is virtually nonexistent for IDPs and abrupt and concomitant with the rate-limiting folding transition for autonomously folding proteins. The discrepancies in interpretations regarding the collapse transition for protein folding and for IDPs have led to numerous debates (4, 9, 15, 20–23).
Why do SAXS and smFRET lead to apparently conflicting inferences regarding the collapse transition and the nature of heterogeneous ensembles, especially under physiologically relevant conditions and away from high concentrations of denaturants? Both techniques have distinct strengths and weaknesses (15, 20–23). Strengths of smFRET measurements include the ultralow protein concentrations, at which experiments can be conducted, the ability to resolve distinct conformational populations, and the advantage of following motions across timescales that range from the nanosecond to the millisecond regimes. Weaknesses of smFRET experiments derive from the possibility that fluorescent dyes, tethered via flexible linkers to protein sidechains, could engender nontrivial alterations to the dimensions of unfolded proteins and IDPs. Also, with typical dye pairs, smFRET affords accurate estimates of distances that are limited to the range of ∼2 nm to ∼10 nm. In contrast, SAXS measurements do not require the attachment of labels and the measured scattering intensities are weighted averages over all of the protein molecules in solution, thus enabling direct investigations of chain dimensions. However, SAXS experiments require higher protein concentrations and the averaging over the conformations of all molecules in solutions makes it difficult to obtain assessments of conformational populations and insights regarding fluctuations that are smaller than the global dimensions of the protein. Here, we ask whether the discrepancies between inferences drawn from SAXS vs. smFRET measurements are due to the perceived weaknesses of the methods themselves or because the two methods provide complementary insights that have to be analyzed jointly to obtain a robust quantitative assessment of conformational features of heterogeneous systems.
We performed SAXS measurements on labeled and unlabeled IDPs as well as chemically denatured proteins. Inferences from these measurements were compared with those from smFRET measurements of labeled molecules. Atomistic Monte Carlo simulations based on the ABSINTH (self-assembly of biomolecules studied by an implicit, novel, and tunable Hamiltonian) implicit solvation model (24) were used to generate quantitative insights and to aid in the joint analysis of SAXS and smFRET data. We made rigorous comparisons between RE,L, calculated from smFRET measurements and atomistic simulations of dye-labeled proteins, and the values of RG,U and RG,L obtained from SAXS measurements. We find that the dyes do not significantly influence the SAXS measurements, under either native conditions or denatured conditions. Instead, estimates of RG and RE yield different inferences because these quantities interrogate distinct length scales and are influenced by very different types of averaging. For finite-sized heteropolymeric sequences, we show that large changes in RE are compatible with negligible changes in RG (22, 25). We discuss that such differences are minimized in long homopolymers and long block copolymers that are characterized by the chemical similarity of the interacting units (25). Accordingly, the estimates of RG and RE lead to mutually consistent inferences regarding conformational preferences and the physics of coil-to-globule transitions for long homopolymers (26). A similar robustness prevails for proteins in highly denaturing environments where preferential interactions between denaturants and chain units appear to have a homogenizing effect on the pattern of intrachain interactions (3, 23, 27–29), in line with the observations we report from the different methods.
Therefore, at a minimum, it becomes important to measure both RG and RE if we are to obtain a reliable description of global chain density through RG, amplitudes of fluctuations through RE, and deviations from uniform expansion/contraction by assessments of the overall shape that can be estimated by quantifying the ratio G = (RE2/RG2). Alternatively, we show that a more rigorous assessment of overall shapes and the decoupling between shape and size fluctuations can be derived from analysis of the entire SAXS profile. This provides a more complete description compared with extracting estimates of RG alone. However, if intermolecular interactions at high protein concentrations required for SAXS are an issue, then global analysis of data from multiple, independent smFRET measurements performed using constructs distinguished by different linear separations between dye pairs would be a promising route to pursue (3).
Results
The Protein Set, Labeling Scheme, and Experimental Design.
We selected a set of 10 protein sequences with lengths between 38 residues and 178 residues, covering different amino acid compositions and physicochemical properties (Fig. 1E and SI Appendix, Table S2 and Note S1). Three of the 10 proteins fold to form stable structures under native conditions whereas the other 7 are IDPs that remain disordered in the absence of denaturant. To avoid potential uncertainties that can (30), but must not (31) arise from random labeling of proteins, we exploited the advantages of site-specific, unambiguous dual labeling. Specifically, the donor dye Alexa488 (SI Appendix, Fig. S1A) was attached via oxime ligation to the unnatural amino acid p-acetylphenylalanine, engineered at the penultimate position of the polypeptide chain using amber suppression technology (32). The acceptor fluorophore Alexa594 (SI Appendix, Fig. S1B) was reacted with a cysteine residue located at the second position via maleimide chemistry. Single-molecule measurements were made using the doubly labeled proteins under strongly denaturing conditions (6 M urea) and in (near)-native conditions with urea virtually absent (see buffer details in SI Appendix, Note S2 and experimental smFRET details in SI Appendix, Note S3).
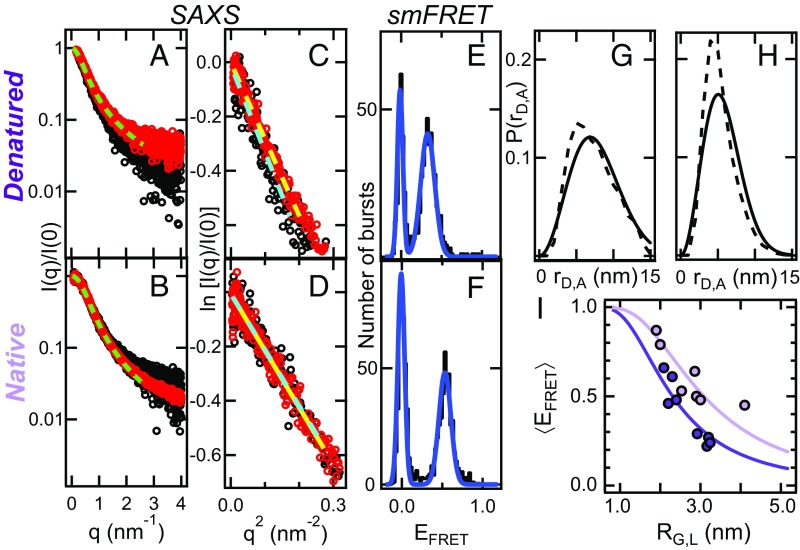
SAXS measurements were performed using unlabeled and labeled samples (see experimental SAXS details in SI Appendix, Note S4). As an example of experimental results, we show the SAXS profiles (Fig. 2 A and B) and Guinier fits (Fig. 2 C and D) for the IDP NUS, under denaturing (Fig. 2 A and C) and native conditions (Fig. 2 B and D). The RG is typically calculated from a plot of the SAXS intensity I(q) vs. the momentum transfer q, using the Guinier approximation (SI Appendix, Note S4):
| [1] |
Fig. 2.
Representative experimental results and estimating G. The data correspond to the IDP NUS. (A) SAXS profile of unlabeled (black line) and labeled NUS (red line) measured in denaturing buffer. (B) SAXS profile of unlabeled (black line) and labeled NUS (red line) measured in native buffer. In A and B the dashed green lines are fits to a mass fractal dimension (for labeled proteins only) to obtain the parameter ν, related to the scaling of internal distances (Eq. 4). (C) Guinier fits of the SAXS profiles shown in A. RG values are directly proportional to the slope of such plots (Eq. 1). (D) Guinier fits to the SAXS profiles shown in B. In A and C the dashed green lines are fits to a mass fractal dimension (for labeled proteins only) to obtain the parameter ν, related to the scaling of internal distances (Eq. 4). In C and D dashed cyan lines and dashed yellow lines represent the Guinier fits of unlabeled and labeled proteins, respectively. (E) EFRET histogram of NUS measured in denaturing buffer. (F) EFRET histogram of NUS measured in native buffer. In E and F the blue lines are fits using a double Gaussian function to get mean FRET values (<EFRET>). (G) Probability distribution functions used to infer RE,L from the mean EFRET shown in E, using Eq. 2. (H) Probability distribution functions used to infer RE,L from the mean EFRET shown in F. In G and H the models are as follows: Gaussian chain (solid line) and CAMPARI simulations reweighted to match <EFRET> and RG,U2 (dashed line). (I) <EFRET> as a function of RG,L under denaturing (dark violet circles) and native conditions (light violet circles). Each circle corresponds to exactly the same protein (i.e., double labeled) measured by smFRET and SAXS. Fits to a distribution of distances according to a Gaussian chain model (with SI Appendix, Eq. S15 and Eq. 2) are shown as dark violet lines (G = 7.1 ± 0.5, proteins denatured in urea) and light violet lines (G = 4.3 ± 0.4, IDPs in native buffer).
Alternatively, RG can be estimated from the pair–distance distribution function (SI Appendix, Note S4). RG,U and RG,L calculated from either the Guinier approximation or the pair–distance distribution function were found to be similar to one another (values in SI Appendix, Table S3). Fig. 2 also shows the smFRET histograms (Fig. 2 E and F) and the most common distance distribution functions used to infer RE,L from (Fig. 2 G and H) corresponding to the same protein (NUS) under denaturing (Fig. 2 E and G) and native (Fig. 2 F and H) conditions. The peak at EFRET near zero in the smFRET histograms arises from donor-only species (33), whereas the second population, originating from molecules containing an active donor–acceptor pair, appears at EFRET ∼ 0.55 for native NUS. The parameter RE,L quantifies the ensemble-averaged root mean-squared distance between the donor and acceptor dyes and it is related to via
| [2] |
Here, R0 or the Förster distance (the distance at which FRET efficiency is 50%) depends on the specific dye pair and it is usually around 5 nm (our measured values are in SI Appendix, Table S4); P(rD,A; RE,L) is a probability distribution function that quantifies the likelihood of realizing values of interdye distances, within an interval rD,A and rD,A + drD,A given a mean donor-to-acceptor distance of RE,L. The form for P(rD,A; RE,L) is unknown a priori and is usually chosen from a list of polymer models that includes the Gaussian chain model, the self-avoiding random walk (SARW) model, or a distribution of points inside a sphere of fixed diameter (34) (SI Appendix, Notes S3 and S8). These models are parameterized in terms of RE,L, which reflects the contribution of the first (mean) and second (variance) moments of the distribution P(rD,A; RE,L) (35).
Fig. 2 shows illustrative datasets from smFRET and SAXS measurements. The complete sets of data from smFRET measurements for all proteins and conditions are shown in SI Appendix, Table S4 (FRET parameters); SI Appendix, Table S5 (anisotropies); SI Appendix, Fig. S2 (gamma and quantum yields); and SI Appendix, Fig. S3 (FRET efficiencies). Similarly, the complete SAXS data are shown in SI Appendix, Fig. S4 A–D (SAXS profiles, Guinier plots, Kratky plots, and pair distance distribution function, respectively). Importantly, to deal with the fact that smFRET and SAXS measurements were performed at very different concentrations, we carried out additional experiments to ensure that the large differences in concentration are not the source of discrepancies in inferences drawn from these measurements (SI Appendix, Note S5 and Fig. S5). Analyses of the datasets, which include information regarding (originating from smFRET), RG,L (measured by SAXS), and RG,U (also from SAXS), are presented in the following sections, first for denatured proteins and then for IDPs under native conditions.
Measurements of RG and Estimates of RE from Measurements of Yield Mutually Consistent Inferences for Denatured Proteins.
We performed SAXS experiments using labeled and unlabeled molecules to quantify the impact of fluorescent dyes on the global dimensions of flexible polymers. SI Appendix, Fig. S6A shows RG,U,D (yellow points) and RG,L,D (red points) calculated from the Guinier approximation as a function of the number of residues (NRES) for eight proteins denatured in 6 M urea. Here, the letters L and U in the subscripts refer to labeled vs. unlabeled molecules and D refers to denaturing conditions (and N refers to native). Our dataset includes five IDPs and three proteins that fold autonomously. The differences between RG,L,D and RG,U,D were generally small, with a root mean-squared deviation (rmsd) of ∼0.3 nm between both datasets. For flexible polymers, a scaling law governs the value of RG whereby
| [3] |
Here, NRES is the number of residues in the chain. The exponent ν quantifies the correlation length and is governed by the solvent quality. In good, theta (indifferent), and poor solvents the values of ν for long homopolymers are 0.59, 0.5, and 0.33, respectively (26). Scattering data for a given protein can be analyzed within an intermediate q range to quantify ν (SI Appendix, Fig. S7A) because
| [4] |
For reference, the full form factor is shown in SI Appendix, Eqs. S19 and S20 (36). An example of the fitting of SI Appendix, Eq. 4 to the experimental SAXS profile is shown in Fig. 2A for denatured NUS (all proteins can be found in SI Appendix, Fig. S4A). In 6 M urea, we find that ν = 0.55 ± 0.04 for unlabeled proteins. Within error, this value is similar to the value for labeled samples, ν = 0.58 ± 0.03 (SI Appendix, Table S6). These findings suggest that the dyes do not fundamentally alter the balance of chain–chain and chain–solvent interactions (SI Appendix, Fig. S7B), thus leaving the solvent quality unchanged. For the analysis that follows, we used an average value of νD = 0.57 ± 0.03 for proteins in 6 M urea. This value for ν is in line with the expected value for the SARW model and the analysis of larger datasets from previous measurements (37, 38), which suggest that high concentrations of denaturants are good solvents for generic protein sequences (3, 23). To test whether smFRET measurements yield similar inferences regarding solvent quality, we calculated the values of G = RE2/RG2. For chains in a good solvent G ∼ 7 (26), and obtaining such a value would require accurate estimates of RE from the smFRET data. In Fig. 2I we plot against RG,L,D, which is extracted from SAXS using exactly the same labeled proteins. The data were analyzed using a Gaussian chain model for the distribution of interdye distances (9–12), with G as the fitting parameter (SI Appendix, Eq. S15 and Note S3). For denatured proteins we obtained GD = 7.1 ± 0.5. This value is in line with theoretical expectations for a swollen chain in good solvent (39) and is larger than the value of 6 expected for random coils (40) in theta solvents (RE,L values in SI Appendix, Table S7 and G values in SI Appendix, Table S8). Taken together, our analyses of SAXS and smFRET data yield mutually consistent inferences regarding solvent quality for denatured proteins in 6 M urea. Importantly, our data establish that the dyes do not materially impact the analysis of chain dimensions of denatured proteins.
SAXS and smFRET Yield Discrepant Inferences Regarding IDP Dimensions in Native Conditions.
We applied the analyses described above to the set of seven IDPs under native conditions to calculate νN and GN. Analysis of SAXS profiles for each of the labeled and unlabeled IDPs yielded similar values for νN (SI Appendix, Fig. S4A), suggesting that dyes do not have a major impact on the dimensions of IDPs under native conditions. The mean value of νN = 0.50 ± 0.04 (SI Appendix, Table S6) is in line with values reported for IDPs with similar compositional biases (3, 7, 41). This suggests that for a class of IDP sequences, the effects of chain–chain and chain–solvent interactions are, on average, mutually compensatory, thus unmasking statistics that are similar to those of chains in theta solvents (29, 41)—a result that has previously been described for unfolded protein ensembles under folding conditions (3). For G, we obtained a mean value of GN = 4.3 ± 0.4, and this is different from the value of 6 that is expected for chains in theta solvents (35, 39, 40). To test whether the anomalous value of G reflects differences in the changes of RG vs. RE, we quantified the swelling ratios that compare the dimensions in 6 M urea vs. native conditions. The swelling ratios are defined as
| [5] |
The inferred values of RE,L of denatured IDPs (RE,L,D) are considerably larger than those of native IDPs (RE,L,N). However, the values of RG,L for denatured IDPs (RG,L,D) are only moderately yet systematically different from those of native IDPs (RG,L,N). This is evidenced by larger values of α(RE,L) vs. smaller values of α(RG,L) (on average 2.02 ± 0.18 vs. 1.27 ± 0.12, respectively; individual values given in SI Appendix, Table S9). These findings are concordant with previous results, which point to disagreements between inferences from SAXS/small-angle neutron scattering (SAXS/SANS) and smFRET measurements at low denaturant concentrations (15, 21). SAXS measurements of labeled vs. unlabeled molecules rule out the dyes as the source of the discrepancy. Once we rule out specific errors with the smFRET measurements, which are presented in Discussion, we are left with three other possible sources for the observed discrepancies: (i) the nature of the averaging that goes into the calculation of RG is likely to make this quantity relatively insensitive to small changes in solvent quality (20), especially for heteropolymers that transition between coil-like ensembles corresponding to ν ∼ 0.59 and ν ∼ 0.5 (42); (ii) because RE quantifies the average distance between a pair of residues, as opposed to an average over all interresidue distances, it is possible that this quantity is more sensitive to fluctuations due the dangling ends of chains (43); and (iii) it is also possible that the inferred values of RE are subject to errors due to assumptions of a Gaussian chain model for the distribution of interdye distances. Each of these factors contributes to the discrepancies between inferences drawn from analysis of SAXS vs. smFRET data. We demonstrate this by analyzing conformational distributions extracted from atomistic simulations that accounted for the presence of fluorescent dyes.
Source of the Discrepant Inferences Regarding the Extent of Collapse Observed Using SAXS vs. smFRET.
We performed all-atom Metropolis Monte Carlo thermal replica exchange simulations for five of the IDPs, using the ABSINTH implicit solvation model and force-field paradigm (24). This combination has proved to be useful for the analysis of conformationally heterogeneous IDPs (42, 44). Details of the simulations are described in SI Appendix, Note S6. For each sequence, we used the measured values of in native conditions to generate reweighted ensembles that match the experimental data. Then, we selected the ensemble corresponding to the lowest simulation temperature (SI Appendix, Table S10) that best matched the experimental observable of interest (more details in SI Appendix, Note S6). To calculate we incorporated atomistic descriptions of rotamers of fluorescent dyes into the simulated ensembles. For each conformation of a specific sequence, we placed roughly 103 distinct dye rotamers in different mutual orientations and distances and calculated FRET efficiencies for each conformation. This process was repeated across the entire ensemble to calculate across the ensemble. Conformations were reweighted based on the agreement between the measured and calculated values of The reweighting of ensembles based on experimental data was performed using COPER (45), which is a maximum-entropy reweighting method that attempts to give conformations similar weights while simultaneously attempting to match an experimental observable or a set of experimental observables.
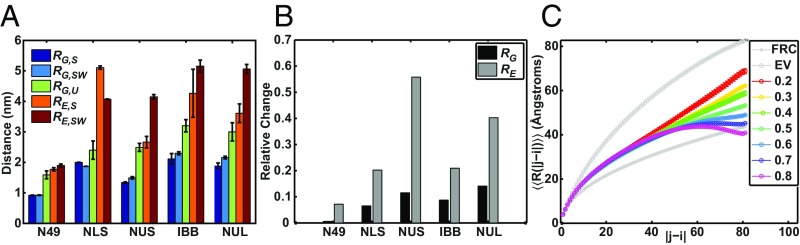
Fig. 3A shows the values of RE and RG that were extracted from the unbiased ensembles (denoted as RE,S and RG,S) and the ensembles reweighted to match (RE,SW and RG,SW) corresponding to native conditions. The subscript S refers to values obtained from simulations and W refers to cases where the simulation values were weighted to match an experimental observable. Here, RE was calculated as the distance between the Cα atoms of the first and last residues and RG was calculated only over the protein atoms. The reweighting procedure reveals an interesting decoupling between the values of RG and RE. Ensembles that were reweighted to match showed minimal changes between RG,S and RG,SW and large changes between RE,S and RE,SW (Fig. 3B). This is consistent with the idea that large changes to and hence RE are compatible with minimal changes to RG. If true, then the discrepant inferences between SAXS and smFRET measurements must originate in the ability to decouple measures of specific pairwise distances such as RE from the averaging over the square of all pairwise distances, which is the case with RG2.
Fig. 3.
Simulated ensembles reweighted to match <EFRET> suggest decoupling between RG and RE. (A) RG and RE values extracted from unbiased (RG,S and RE,S) and reweighted (RG,SW and RE,SW) ensembles for N49, NLS, NUS, IBB, and NUL. Here, reweighted ensembles refer to the ensembles generated by reweighting to <EFRET> values under native conditions. Error bars indicate the SEM over three independent simulations. The experimental RG,U values determined under native conditions are plotted for reference. RG,U is used as a reference given that for the simulated ensembles RG is calculated only over the protein. (B) The relative change in RG and RE between unbiased and reweighted ensembles calculated as |RG,SW − RG,S|/RG,S and |RE,SW − RE,S|/RE,S, respectively. (C) Internal scaling plots for NUS simulated ensembles reweighted to match RG,U2 and one of the following <EFRET> values: [0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8]. For every pair of residues at a given sequence separation (|j–i|) the average through-space distance for that given sequence separation (<<R(|j–i|)>>) is plotted. Here, i and j are the residue positions. FRC denotes the internal scaling profile of the Flory random coil (Gaussian chain) reference and EV denotes the internal scaling profile of the excluded volume coil reference.
To put the proposed decoupling between RG and RE on a quantitative footing, we reweighted the NUS ensembles at 360 K to match the experimentally derived and one of the following target values for mean FRET efficiencies: = [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]. Here, in the simulations is the weighted mean square of the RG values calculated over the protein atoms alone. If RG and RE can be decoupled, then ensembles should be generated that satisfy a single value of and a range of values of Indeed, we find that with the exception of the most extreme values (0.1 and 0.9), NUS ensembles can be generated that match and a given value with minimal changes to the force field (SI Appendix, Fig. S8 A and B and Note S6). This suggests that, under certain conditions, an entire spectrum of and therefore multiple RE values are consistent with a given RG value (22). This result is consistent with the finding that large differences in G are virtually indistinguishable by SAXS (SI Appendix, Fig. S7C). Such a result emerges from the combination of two effects: (i) at low to intermediate values of RG small changes in RE (∼1 nm) can lead to large changes in G (SI Appendix, Fig. S9A) and (ii) large, potentially informative fluctuations at the ends of chains have little effect on the global conformational properties measured by SAXS (SI Appendix, Fig. S9 C and D).
The preceding findings do not imply that the ensembles generated to match different values have the same conformational properties. To make this point, we characterized the overall shapes of polymers and scaling of internal distances for ensembles of NUS that match the experimentally derived and one of the following target values for mean FRET efficiencies: = [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]. We quantified overall shape preferences by calculating conformation-specific and ensemble averaged values of asphericity, δ*, that is given in terms of the eigenvalues, λ1, λ2, and λ3 of conformation-specific gyration tensors (46, 47). Here,
| [6] |
For rod-like conformations δ* ∼ 1 and for a perfect sphere δ* ∼ 0 (26, 47). Distributions of δ*SW\ (SI Appendix, Fig. S8D) show that δ*SW decreases as increases, whereas distributions of RG,SW are similar for all values (SI Appendix, Fig. S8C). The decrease in δ*SW observed with decreasing RE suggests that ensembles become more spherical to account for the same RG albeit with smaller RE values. SI Appendix, Fig. S10 shows a comparison of shape characterization in terms of G and δ*. These parameters are weakly coupled although, on average, an increase in〈G〉implies an increase in〈δ*〉. The weak coupling results from the fact that G is highly sensitive to large fluctuations at the ends of chains, whereas δ* is only mildly sensitive to such fluctuations and changes in δ* depend on the sequence separation at which the fluctuations emerge (SI Appendix, Fig. S9 B–D). To extract further insights regarding the distributions of internal distances, we calculated internal scaling profiles that serve as formal order parameters in more nuanced theories of coil-to-globule transitions (48).
Internal scaling profiles quantify the mean spatial separation between all residues i and j that are |j–i| apart along the linear sequence. Fig. 3C shows that all ensembles, irrespective of the target value used for reweighting, show similar scaling in spatial separation for |j–i| < 40. However, the spatial separations start to diverge from one another at larger sequence separations. These internal scaling profiles highlight an important point: based on Lagrange’s theorem (39) we know that the mean-squared RG can be written as the mean-squared sum over all internal distances (definition in SI Appendix, Table S1). Thus, if a majority of internal distances change negligibly, then the value of RG will change minimally. In contrast, the overall shape shows intermediate changes and distances corresponding to larger sequence separations will show large fluctuations (SI Appendix, Fig. S9 C and D).
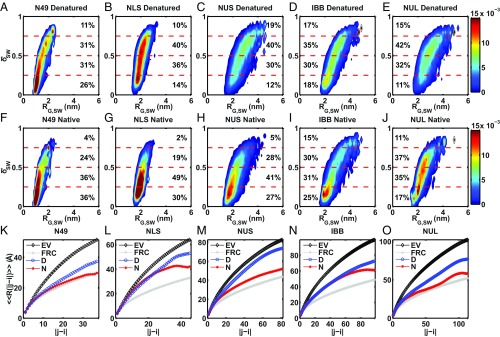
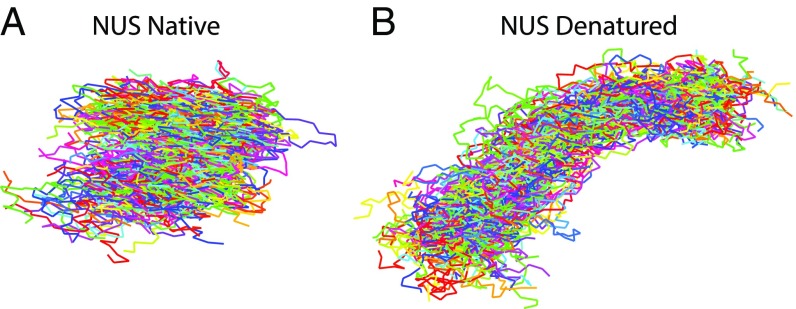
Because we measured RG and for each IDP under native and denatured conditions, we can analyze the ensembles that were reweighted to match both experimental observables. Fig. 4 shows the two-dimensional histograms of RG,SW vs. δ*SW for ensembles reweighted to match both and for each IDP under native (Fig. 4 F–J) and denatured (Fig. 4 A–E) conditions. For all IDPs, δ*SW increases under denaturing conditions, indicating that the ensembles become less spherical. This is consistent with the larger G values extracted from denatured compared with native conditions. Internal scaling plots of the simulated ensembles (Fig. 4 K–O) show that the denatured ensembles diverge from native ensembles to prefer larger spatial separations for larger sequence separations. The sequence separation at which this divergence occurs is specific to each IDP sequence, thus highlighting the contribution of sequence-specific interactions to chain deformations under denaturing conditions. To visualize the change in shape between native and denatured ensembles, we extracted 100 representative conformations with the highest weights for NUS when reweighted to match the experimental observables under either native (Fig. 5A) or denatured (Fig. 5B) conditions. The results show that NUS adopts more elongated and less spherical conformations under denaturing conditions compared with native conditions.
Fig. 4.
Quantification of the shape (δ*SW), size (RG,SW), and scaling of simulated ensembles reweighted to match both <EFRET> and RG,U2 for native and denatured conditions. (A–E) Two-dimensional histograms of RG,SW vs. δ*SW extracted from simulated ensembles reweighted to match both <EFRET> and RG,U2 for denatured conditions. (F–J) Two-dimensional histograms of RG,SW vs. δ*SW extracted from simulated ensembles reweighted to match both <EFRET> and RG,U2 for native conditions. The percentages indicate the percentage of δ*SW that falls between different δ*SW limits (dashed horizontal lines). For all IDPs studied, δ*SW increases under denatured conditions, indicating that the ensembles become less spherical and more elongated. (K–O) Internal scaling plots comparing the native (N) and denatured (D) profiles from simulated ensembles reweighted to match both <EFRET> and RG,U2. FRC and EV denote the internal scaling profiles generated from the Flory random coil and excluded volume coil references, respectively. Error bars denote the SEM over three independent simulations.
Fig. 5.
Representative ensembles of NUS under native and denatured conditions. (A and B) The 100 conformations with the highest weights from the simulated ensembles reweighted to match both <EFRET> and RG,U2 for native (A) and denatured (B) conditions.
We also note that simulations can be used to estimate the error associated with inferences of RE,L from smFRET that are based on the use of the Gaussian chain or other generic polymer models for P(rD,A; RE,L) (49). Fig. 2 G and H shows the distance distributions corresponding to the Gaussian chain model together with the distance distributions obtained from the simulations by restraining the ensembles to match and RG,U2. The results suggest that the Gaussian chain model tends to overestimate RE,L for denatured proteins and underestimate RE,L for IDPs under native conditions (SI Appendix, Table S7). These results are consistent with the findings of O’Brien et al. (49) and Borgia et al. (23). Accordingly, the final α(RE,L) values (SI Appendix, Table S9) are overestimated.
Analysis of the Full SAXS Profiles Beyond RG and ν.
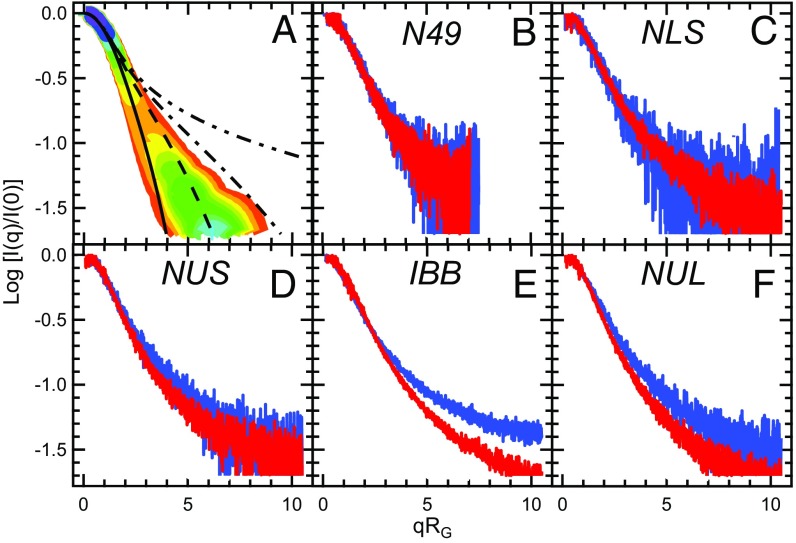
If ensembles of chemically denatured proteins display larger asphericities compared with the native IDPs, then this should be discernible in the SAXS data as well. We tested this by performing a model-independent comparison of the experimental data. Indeed, if one computes a size-independent version of scattering profiles by plotting log[I(q)/I(0)] vs. qRG (Fig. 6A), then the curves corresponding to bodies with changing asphericity display a rather systematic trend, from the right (aspherical polymers) to the left (spherical polymers) of the plot. We plotted the experimental data for unlabeled native and chemically unfolded proteins (Fig. 6 B–F). For the two smallest proteins N49 and NLS, the differences are within the level of statistical noise, whereas the three larger proteins display a systematic shift of the size-independent scattering patterns from the right (higher asphericity for chemically denatured proteins) to the left (more spherical shapes for IDPs under native conditions). The results of this analysis are important because they were obtained solely from the experimental data.
Fig. 6.
The full SAXS profile (but not RG alone) is sensitive to the changes in the ensemble shape that occur upon IDP collapse. To remove the contribution of size to the SAXS profiles and visualize exclusively the influence of shape, size-independent SAXS curves are constructed by plotting the normalized scattering intensities (log[I(q)/I(0)]) as a function of q times RG. (A) Theoretical SAXS profiles predicted for different values of asphericities (δ*): 0.1 (solid line), 0.3 (dashed line), 0.5 (dashed-dotted line), and 0.9 (dashed dotted-dotted line) (see SI Appendix, Fig. S7 for more details on color scale). Note that asphericity increases from left (more spherical) to right (more anisometric) and that a given RG is compatible with many asphericities; i.e., RG is not sensitive to shape. The experimental SAXS profiles of unlabeled native (red lines) and unlabeled denatured (blue lines) IDPs are shown in B (N49), C (NLS), D (NUS), E (IBB), and F (NUL).
We further tested the proposed change in asphericity, using size-independent maps of scattering profiles that were generated using the reweighted ABSINTH ensembles. CRYSOL (50) was used to convert each conformation to a SAXS profile and these were combined to generate the final weighted SAXS profile. The profiles generated from the reweighed ABSINTH ensembles consistently show an increase in asphericity for denatured IDPs compared with native IDPs (SI Appendix, Fig. S11D). This recapitulates the direct calculations of asphericities from reweighted ASBSINTH ensembles, without consideration of the full scattering curves. However, it is noteworthy that ensembles showing divergence in the internal scaling profiles between native and denatured conditions at larger sequence separations show less pronounced differences in the scattering patterns (Fig. 4 and SI Appendix, Fig. S11). This is consistent with the observation that asphericity is mainly sensitive to changes in spatial separation in the intermediate sequence separation regime (SI Appendix, Fig. S9 C and D). Fluctuations at the ends of the chain will have minimal impact on the overall asphericity. It is also noteworthy that the reweighted ABSINTH ensembles, which were reweighted to match RG2 and <EFRET> values, also resemble the experimentally derived SAXS profiles (SI Appendix, Fig. S11A).
In another independent and unbiased approach, the ensemble optimization method (EOM) (51) was used to analyze the SAXS data. The EOM analysis used the unweighted pool of ABSINTH conformations to select subensembles of conformers such that their mixture accurately matches the experimental SAXS data (SI Appendix, Fig. S11A). The EOM-selected ensembles unveiled substantial conformational heterogeneity (displayed as essentially broader, not necessarily monomodal size distributions) compared with the reweighted ABSINTH ensembles. Further, EOM ensembles showed an increase in both conformational heterogeneity (SI Appendix, Fig. S11B) and asphericity (SI Appendix, Fig. S11D) under denaturing conditions compared with native conditions. The increase in asphericity was more pronounced for the longer constructs, in agreement with the results shown in Fig. 6. The EOM ensembles did not always reproduce the experimentally measured <EFRET> values (SI Appendix, Fig. S11C). This highlights the distinctive nature of the information that is gleaned from SAXS vs. smFRET measurements. Specifically, information about the end-to-end distance may be diluted or lost in SAXS profiles. This is not unexpected given that SAXS measurements yield integral information on averaged distance distributions over conformational ensembles as opposed to “differential” averaging of a single end-to-end distance in smFRET. Hence, ensembles generated to match SAXS data are likely to be incompatible with inferences that are based on smFRET measurements, especially away from denaturing conditions. Overall, these results emphasize the importance of gathering SAXS and smFRET data and the joint use of both methodologies for generating mutually compatible ensembles that provide a more complete picture of the overall shapes, sizes, and conformational fluctuations.
Estimating NRES from SAXS and smFRET Data.
Although our work raises caution regarding the use of generic polymer models when analyzing smFRET data for heteropolymers, these models afford the practical convenience required to obtain quick estimates of RE,L from measured values for IDPs as well as denatured states. It is useful to quantify the contribution that dyes (NDYES) make in terms of equivalent residues to the polypeptide chain (NRES). Previous estimates of NDYES have varied from 0- to 20-residue equivalents (3, 12, 52, 53). Given direct access to and estimates of we can quantify NDYES using these data.
Because the scaling behavior of RG,L depends on the actual number of amino acids in both the polypeptide chain (NRES) and NDYES, we rewrite Eq. 3 as follows for RG,L (a similar reasoning can be used for RE,L):
| [7] |
and
| [8] |
Here, the preexponential factors ρE and ρG are related to the size of the repeating unit. Whereas dye labeling does not substantially affect RG as detected by SAXS, we can perform a global fit of the six experimental datasets to extract the contributions that dyes make to RE,L for both denatured proteins and native IDPs. This allowed us to obtain estimates of (SI Appendix, Fig. S6 and Table S11).
Discussion
SAXS and smFRET are two powerful experimental tools that provide useful insights regarding disordered systems such as IDPs and unfolded ensembles of autonomously foldable proteins (7, 54). However, the two measurements yield discrepant inferences when going from denatured to native conditions, with SAXS detecting minimal changes and smFRET suggesting discernible reduction in RE,L as measured by an increase in <EFRET>. We obtained good agreement between inferred RE,L and RG,L values at high denaturant concentrations in terms of the scaling behavior and inferred solvent quality. However, we find a clear “mismatch” in inferences regarding chain sizes in the absence of denaturant: either the inferred values of RE,L appear to be too small or the measured values of RG,L are too large.
Our insights were derived by combining experimentally derived RG values and mean FRET efficiencies with simulations that also include the effects of dyes. A major conclusion from the simulations is that many disordered ensembles with substantially different RE can have similar values of RG (Figs. 3–6). This result was also demonstrated by Song et al. (22) for heteropolymeric systems and it implies that the discrepant expansion factors inferred from SAXS and smFRET measurements are not a consequence of any intrinsic weaknesses of these methods. Instead, they represent a fundamental decoupling between RG, a globally averaged quantity, and RE as well as other distances between dangling ends that are not averaged across the entire sequence. This decoupling is amplified in finite-sized heteropolymeric sequences in the absence of denaturant.
Advanced theories that account for the effects of chain connectivity to describe excluded volume effects demonstrate that chains can undergo nonuniform expansion/compaction (55). The dangling ends of chains (43) experience fewer restrictions on fluctuations. Hence, inferences regarding chain dimensions can be different when quantified in terms of RG vs. RE or distances near the ends of chains. The use of RG and RE as equivalent measures of chain dimensions dates back to Flory-style mean-field theories that reduce polymers to collections of uncorrelated monomers or Kuhn segments (5). This is a powerfully simplifying approach that affords convenient analytical descriptions. In contrast, Lifshitz-style theories recognize the decoupling between RG and RE and rely on the radial density profile (equivalent to the internal scaling profile) as an order parameter for coil-to-globule transitions (56, 57).
Effects of Dyes in smFRET Measurements.
For native and denatured conditions we showed that the behavior of labeled proteins is not different from that of their unlabeled counterparts, at least in terms of the scaling of internal distances manifested by similar values of ν for unlabeled and labeled samples (SI Appendix, Table S6). The parallel axes theorem is a useful theoretical construct to describe the relationship between RG,U, RG,L, and RE,L. A full theoretical treatment of this can be found in SI Appendix, Note S7. The main conclusion from this analysis is that for many values of RE,L, dyes do not cause a measurable change of RG,L relative to RG,U (SI Appendix, Note S7 and Fig. S12A). However, as G increases, the difference between RG,L and RG,U is predicted to increase, with larger changes observed for shorter chain lengths (SI Appendix, Fig. S12B). This prediction is consistent with the experimental trends we observe (SI Appendix, Fig. S12C). Combining the results from SAXS and smFRET with simulations, we estimated the contribution of dyes to RE,L expressed in terms of extra residues as NDYES = 5 ± 3 (SI Appendix, Fig. S6). Such a value is likely to be generally useful for smFRET analysis, irrespective of the particular fluorescent dye pair used, because the actual size of each fluorophore has a limited influence on the inferred distances (SI Appendix, Eq. S29 and Fig. S1C). To further rule out the possibility of artifacts due to the dyes themselves, we discuss potential sources of errors in our experimental design and broader implications.
Case A.
Dyes might experience hindered rotations such that the orientation parameter κ2, and hence the Förster distance R0, deviates from the isotropic averaging condition (58). We tested this via anisotropy measurements. The low values we observe for anisotropies (<0.1, SI Appendix, Table S5) support free dye rotation under all assayed conditions. Therefore, it appears to be reasonable to assume that rotational averaging is allowed, and thus the assumption of κ2 = 2/3 in the FRET equation is valid (SI Appendix, Table S4).
Case B.
The dyes might be drawn toward one another through cohesive forces. The analysis of scaling exponents should make such an effect easy to detect. We do not observe such a trend under either denaturing conditions or native conditions (SI Appendix, Fig. S6).
Case C.
It is known that the dynamics of dyes can affect EFRET measurements (9, 31, 33, 59–63). For unfolded proteins of similar size and in similar solvents to the ones studied here (including NUS), chain reconfiguration times have been shown to be in the range of ∼100 ns (3, 64), which is well above the donor lifetimes of ∼4 ns and well below the transit times through the confocal volume, ∼1 ms. As a result, a major role of dynamics in the measured intensity-based EFRET values seems unlikely. Taken together, we conclude that the dyes alone cannot explain the large changes to RE,L that we observe upon protein denaturation in contrast to the modest changes of RG,L (SI Appendix, Table S9).
Choice of Polymer Models for Analyzing smFRET Data.
Our findings highlight the need for caution in coopting models for distributions of RE or RG that have been designed for infinitely long flexible homopolymers—a point that has been made in previous studies as well (22, 23, 49). Flory’s mean-field theory (5) yields a value of G = 6 for ν = 0.5 in theta solvents and SARWs yield G ∼ 7 for ν ∼ 0.6 (SI Appendix, Note S9 and Table S8). The values of ν (0.57) and the inferred values of G (6.6) for denatured proteins are in accord with the values for SARWs. For the native dataset we obtained GN = 5.2 and νN = 0.5, respectively, when we used smFRET, SAXS, and simulations. This result suggests that according to the inferred value of G, IDPs under native conditions deviate from the Gaussian chain model, whereas the inferred scaling exponent suggests congruence with the statistics of the Gaussian chain model. In SI Appendix, Note S8 and Fig. S13 we show that the same issue persists when using other polymer models, thus highlighting the role of simulations in inferring self-consistent sets of distances and the need for caution in using generic polymer models for estimating RE from measured FRET efficiencies, especially in the absence of denaturants. To overcome difficulties associated with the choice of generic polymer models, O’Brien et al. (49) proposed a self-consistency test that requires the measurement of FRET efficiencies by attaching dyes along different internal positions within a sequence. They showed that the use of multiple, independent measurements provides a rigorous test of the polymer model that is used to extract distance estimates from measured FRET efficiencies.
Connections to Recent Studies.
The discrepant inferences drawn from SAXS and smFRET measurements have stimulated numerous debates and independent investigations. Discrepancies were recently reported for nonbiological homopolymers like polyethylene glycol (PEG) (21). This study compared RG values from SANS experiments to RE values derived from smFRET. Unlike our study, the impact of dyes was not directly investigated as this would have required SANS measurements on PEG molecules with and without dyes. Additionally, the concentrations for SANS measurements correspond to the semidilute regime for PEG in water. In this regime, there are significant nonidealities such as the scaling of osmotic pressure as c9/4, where c is the PEG concentration. The impact of these nonidealities on using PEG as a negative control remains unclear.
Aznauryan et al. (65) performed SAXS and smFRET measurements and combined these with distances extracted from structural ensembles based on data from NMR experiments. Their results point to consistent inferences for average distances and distributions of distances for ubiquitin in high concentrations of denaturant (65). A similar consistency regarding denaturant-mediated expansion was reported by Borgia et al. (23), who used a combination of smFRET, SAXS, dynamic light scattering, and two-focus fluorescence correlation spectroscopy to assess how conformational ensembles change as a function of denaturant concentration. They focused their measurements on the denatured state of the spectrin domain R17 and the IDP ACTR. All of their data support an expansion with increasing denaturant concentration. Borgia et al. (23) also showed that the inferred RE and RG values can be overestimated when using polymer-based models for proteins in denaturant. They argued that the inferred RE and RG appear to have different sensitivities to denaturant. In a third study, Zheng et al. (66) reported results from unbiased simulations to demonstrate consistency between inferences drawn from smFRET and SAXS measurements for proteins in increasing concentrations of denaturant. They noted that the dyes do not materially affect the degree of increase in RG with increases in denaturant concentration. The work of Schuler and colleagues (23, 65, 66) highlights the mutual consistency of inferences from SAXS and smFRET measurements for denatured proteins, the insensitivity of estimates of the changes of RG with denaturant to the presence or absence of dyes, and the possible overestimation of RE and RG values based on the polymer models that are used. Our results for denatured proteins and for IDPs in high concentrations of denaturants are consistent with those of Schuler and coworkers (23, 65, 66).
Working Hypothesis for the Decoupling Between RG and RE.
Flexible polymers can be described using the thermal blob model. RG and RE for a thermal blob will scale as g½, where g is the number of residues per blob (67). By definition, the blob is a length scale where the intrablob interactions and blob-solvent interactions are counterbalanced. The blob size is approximately five to seven residues for most IDPs (41). In mean-field theories for polymers in dilute solutions, there are two interrelated parameters to consider: the surface tension per blob (γB) and the effective pairwise interactions between blobs (67). Depending on solvent quality, γB will be positive (poor solvent), zero (theta solvent), or negative (good solvent) and the pairwise interblob interactions will respectively be, negative, zero, or positive. All blobs are identical in homopolymers, and hence all interactions are uniform and a single parameter suffices to describe the overall chain statistics. Accordingly, in theta and good solvents, RG and RE will provide equivalent descriptions of chain behavior.
For heteropolymers, blobs can be quite different from one another and this depends on the amino acid composition and sequence patterning (68, 69). The chain could have blobs that encode negative, zero, or positive values of γB and these will in turn modulate the pattern of interblob interactions. Attractions can screen repulsions and this can give rise to relatively uniform density profiles that make RG inert to changes in solution conditions but they will be manifest as differences in distances across specific length scales (Fig. 4). The effects of heteropolymericity can be captured as an interaction matrix as opposed to a single interaction parameter, and the key question is whether the variance across the values within the interaction matrix is smaller than, equivalent to, or larger than thermal energy. This variance will encode the extent of convergence or divergence between measures of chain dimensions averaged across the entire sequence (RG) and measures that probe specific length scales, such as RE. The blob-based analysis explains why despite water being a poor solvent for polypeptide backbones (29, 70), we now know that the apparent solvent quality for real IDPs deviates from that for backbones and is actually governed by charge and proline contents as well as the patterning of charged and proline residues (3, 17, 41, 42, 68, 69, 71).
Conclusion and Perspective
Given the high cost required to perform complete SAXS experiments with dye-labeled samples and the small contribution of the commonly used dyes to the total protein size, it is both impractical and unnecessary to measure SAXS profiles for labeled molecules on a routine basis. We have shown that, for many IDPs, RG,U will be a reasonable approximation to RG,L. Given the diversity of IDP sequences (68), it should be stressed that our measured values of GN and δN* are unlikely to be universal. Therefore, RE and RG should be determined for each combination of solution condition and IDP through independent quantification of RE,L by smFRET and RG,U by SAXS or the measurement of multiple internal distances for different sequence separations by smFRET (3, 34) or through the joint use of intramolecular three-color FRET measurements (58). For SAXS measurements, this includes estimates of RG (7) combined with analysis of protein shape preferences from the entire SAXS profile. These measurements can be augmented using methods such as anomalous SAXS (59) that introduce gold labels along the chain for extracting intramolecular distances. Measurements when complemented with computer simulations as performed here and in other efforts (66) can help in converting experimental observables into self-consistent molecular models of the conformational ensembles. The relevance of our work goes beyond IDPs under native conditions. In the protein-folding field there is lingering controversy over the earliest folding events arising from dissimilar FRET and SAXS experiments (15, 34); suggestions have been put forward for chain collapse preceding the folding transition—a view largely supported by FRET measurements—whereas the alternative position is that collapse is intimately coupled with the folding transition—a view supported by SAXS measurements. Based on our data, we propose that the earliest events are likely to be changes in shape (26, 46, 72) within the unfolded ensembles upon dilution from denaturant before folding and the formation of stable local as well as nonlocal contacts; decreased asphericity may be what smFRET measurements pick up as a “collapse” transition. This would be difficult to detect by SAXS using only RG, but the full SAXS profile might be more useful for detecting changes in asphericity and directly estimating the correlation length via the scaling exponent ν. Therefore, we propose that the joint use of smFRET and SAXS, together with other structural biology methods, and the support of computational tools and advanced theories will improve our understanding of heterogeneous conformational ensembles.
Materials and Methods
In total, 10 proteins (abbreviated as N49, BBL, NLS, CSP, NUS, IBB, TRX, NUL, N98, and NSP) bearing a cysteine residue at the second position and the noncanonical amino acid p-acetylphenylalanine at the penultimate position were expressed recombinantly in Escherichia coli BL21 AI cells, purified, and double labeled with Alexa488 hydroxylamine and Alexa594 maleimide. Proteins were measured in two PBS buffer conditions: “denaturing” (in presence of 6 M urea) and “native” (with urea absent). SmFRET was done on a custom-built multiparameter spectrometer, using picomolar concentrations of labeled proteins. FRET efficiencies were analyzed burst-wise. SAXS profiles of labeled and unlabeled proteins at different concentrations (micromolar and beyond) were measured at the BioSAXS P12 beamline of Petra III (DESY). The scattering profiles were analyzed in full to obtain size (mean radius of gyration and its distribution) and shape (asphericity, correlation length) information. Molecular simulations of labeled proteins were performed using the CAMPARI package with the ABSINTH implicit solvation model and force-field paradigm. Experimental observables were used to restrain the conformational space sampled by the simulated ensembles. Comprehensive descriptions of the protein expression, purification, labeling, smFRET and SAXS measurements, atomistic simulations, and theoretical considerations are described in detail in SI Appendix, Notes S1–S9, Tables S1–S11, and Fig. S1–S13.
Supplementary Material
Acknowledgments
We thank Ben Schuler, Robert Best, Andrea Soranno, and Hue-Sun Chan for insightful discussions. G.F. was supported by the EMBL Interdisciplinary Postdocs (EIPOD) Programme and a postdoctoral fellowship “ValI+d” from the Conselleria d’Educació, Formació i Ocupació of the Generalitat Valenciana. M.K. acknowledges support from European Comission (the 7th Framework Programme) Marie Curie Grant IDPbyNMR (Contract 264257). E.A.L. acknowledges funding by the Deutsche Forschungsgemeinschaft, especially the Emmy Noether program. R.V.P. acknowledges support from the US National Institutes of Health through Grants R01NS056114 and R01NS089932. Access to the P12 synchrotron beamline at Petra-3 has received funding from the European Community 7th Framework Programme (FP7/2007-2013) under BioStruct-X (Grant 283570).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704692114/-/DCSupplemental.
References
- 1.Uversky VN. Introduction to intrinsically disordered proteins (IDPs) Chem Rev. 2014;114:6557–6560. doi: 10.1021/cr500288y. [DOI] [PubMed] [Google Scholar]
- 2.Ziv G, Thirumalai D, Haran G. Collapse transition in proteins. Phys Chem Chem Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann H, et al. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman E, Haran G. Coil-globule transition in the denatured state of a small protein. Proc Natl Acad Sci USA. 2006;103:11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flory PJ. Principles of Polymer Chemistry. Cornell Univ Press; Ithaca, NY: 1953. [Google Scholar]
- 6.Sanchez IC. Phase transition behavior of the isolated polymer chain. Macromolecules. 1979;12:980–988. [Google Scholar]
- 7.Bernadó P, Svergun DI. Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol Biosyst. 2012;8:151–167. doi: 10.1039/c1mb05275f. [DOI] [PubMed] [Google Scholar]
- 8.Receveur-Brechot V, Durand D. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr Protein Pept Sci. 2012;13:55–75. doi: 10.2174/138920312799277901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc Natl Acad Sci USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soranno A, et al. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109:17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzmenkina EV, Heyes CD, Nienhaus GU. Single-molecule FRET study of denaturant induced unfolding of RNase H. J Mol Biol. 2006;357:313–324. doi: 10.1016/j.jmb.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 12.Milles S, Lemke EA. Single molecule study of the intrinsically disordered FG-repeat nucleoporin 153. Biophys J. 2011;101:1710–1719. doi: 10.1016/j.bpj.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann A, et al. Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc Natl Acad Sci USA. 2007;104:105–110. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 15.Yoo TY, et al. Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J Mol Biol. 2012;418:226–236. doi: 10.1016/j.jmb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udgaonkar JB. Polypeptide chain collapse and protein folding. Arch Biochem Biophys. 2013;531:24–33. doi: 10.1016/j.abb.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Müller-Späth S, et al. From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob J, Krantz B, Dothager RS, Thiyagarajan P, Sosnick TR. Early collapse is not an obligate step in protein folding. J Mol Biol. 2004;338:369–382. doi: 10.1016/j.jmb.2004.02.065. [DOI] [PubMed] [Google Scholar]
- 19.Walters BT, Mayne L, Hinshaw JR, Sosnick TR, Englander SW. Folding of a large protein at high structural resolution. Proc Natl Acad Sci USA. 2013;110:18898–18903. doi: 10.1073/pnas.1319482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maity H, Reddy G. Folding of protein L with implications for collapse in the denatured state ensemble. J Am Chem Soc. 2016;138:2609–2616. doi: 10.1021/jacs.5b11300. [DOI] [PubMed] [Google Scholar]
- 21.Watkins HM, et al. Random coil negative control reproduces the discrepancy between scattering and FRET measurements of denatured protein dimensions. Proc Natl Acad Sci USA. 2015;112:6631–6636. doi: 10.1073/pnas.1418673112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song J, Gomes GN, Gradinaru CC, Chan HS. An adequate account of excluded volume is necessary to infer compactness and asphericity of disordered proteins by Förster resonance energy transfer. J Phys Chem B. 2015;119:15191–15202. doi: 10.1021/acs.jpcb.5b09133. [DOI] [PubMed] [Google Scholar]
- 23.Borgia A, et al. Consistent view of polypeptide chain expansion in chemical denaturants from multiple experimental methods. J Am Chem Soc. 2016;138:11714–11726. doi: 10.1021/jacs.6b05917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vitalis A, Pappu RV. ABSINTH: A new continuum solvation model for simulations of polypeptides in aqueous solutions. J Comput Chem. 2009;30:673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang GZ, Wu C. Folding and formation of mesoglobules in dilute copolymer solutions. Adv Polym Sci. 2006;195:101–176. [Google Scholar]
- 26.Steinhauser MO. A molecular dynamics study on universal properties of polymer chains in different solvent qualities. Part I. A review of linear chain properties. J Chem Phys. 2005;122:094901. doi: 10.1063/1.1846651. [DOI] [PubMed] [Google Scholar]
- 27.Record MT, Jr, Guinn E, Pegram L, Capp M. Introductory lecture: Interpreting and predicting Hofmeister salt ion and solute effects on biopolymer and model processes using the solute partitioning model. Faraday Discuss. 2013;160:9–44, discussion 103–120. doi: 10.1039/c2fd20128c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brien EP, Ziv G, Haran G, Brooks BR, Thirumalai D. Effects of denaturants and osmolytes on proteins are accurately predicted by the molecular transfer model. Proc Natl Acad Sci USA. 2008;105:13403–13408. doi: 10.1073/pnas.0802113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holehouse AS, Garai K, Lyle N, Vitalis A, Pappu RV. Quantitative assessments of the distinct contributions of polypeptide backbone amides versus side chain groups to chain expansion via chemical denaturation. J Am Chem Soc. 2015;137:2984–2995. doi: 10.1021/ja512062h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo MH, et al. Efficient single-molecule fluorescence resonance energy transfer analysis by site-specific dual-labeling of protein using an unnatural amino acid. Anal Chem. 2011;83:8849–8854. doi: 10.1021/ac202096t. [DOI] [PubMed] [Google Scholar]
- 31.Sisamakis E, Valeri A, Kalinin S, Rothwell PJ, Seidel CAM. Accurate single-molecule FRET studies using multiparameter fluorescence detection. Methods Enzymol. 2010;475:455–514. doi: 10.1016/S0076-6879(10)75018-7. [DOI] [PubMed] [Google Scholar]
- 32.Brustad EM, Lemke EA, Schultz PG, Deniz AA. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J Am Chem Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudryavtsev V, et al. Combining MFD and PIE for accurate single-pair Förster resonance energy transfer measurements. ChemPhysChem. 2012;13:1060–1078. doi: 10.1002/cphc.201100822. [DOI] [PubMed] [Google Scholar]
- 34.Schuler B, Soranno A, Hofmann H, Nettels D. Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Annu Rev Biophys. 2016;45:207–231. doi: 10.1146/annurev-biophys-062215-010915. [DOI] [PubMed] [Google Scholar]
- 35.Flory PJ. Mean-square moments of chain molecules. Proc Natl Acad Sci USA. 1964;51:1060–1067. doi: 10.1073/pnas.51.6.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansen D, Trewhella J, Goldenberg DP. Fractal dimension of an intrinsically disordered protein: Small-angle X-ray scattering and computational study of the bacteriophage λ N protein. Protein Sci. 2011;20:1955–1970. doi: 10.1002/pro.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohn JE, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson MC, Curtis JE. Probing the average local structure of biomolecules using small-angle scattering and scaling laws. Biophys J. 2014;106:2474–2482. doi: 10.1016/j.bpj.2014.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flory PJ. Statistical Mechanics of Chain Molecules. Interscience Publishers; Interscience, NY: 1969. [Google Scholar]
- 40.Debye P. The intrinsic viscosity of polymer solutions. J Chem Phys. 1946;14:636–639. [Google Scholar]
- 41.Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin EW, et al. Sequence determinants of the conformational properties of an intrinsically disordered protein prior to and upon multisite phosphorylation. J Am Chem Soc. 2016;138:15323–15335. doi: 10.1021/jacs.6b10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schafer L. Excluded Volume Effects in Polymer Solutions. Springer; Berlin: 1999. Dilute limit: Details on the internal structure of isolated coils; pp. 305–342. [Google Scholar]
- 44.Holehouse AS, Das RK, Ahad JN, Richardson MO, Pappu RV. CIDER: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys J. 2017;112:16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung HT, et al. A rigorous and efficient method to reweight very large conformational ensembles using average experimental data and to determine their relative information content. J Chem Theory Comput. 2016;12:383–394. doi: 10.1021/acs.jctc.5b00759. [DOI] [PubMed] [Google Scholar]
- 46.Dima RI, Thirumalai D. Probing the instabilities in the dynamics of helical fragments from mouse PrPC. Proc Natl Acad Sci USA. 2004;101:15335–15340. doi: 10.1073/pnas.0404235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theodorou DN, Suter UW. Shape of unperturbed linear polymers: Polypropylene. Macromolecules. 1985;18:1206–1214. [Google Scholar]
- 48.Imbert JB, Lesne A, Victor JM. Distribution of the order parameter of the coil-globule transition. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1997;56:5630–5647. [Google Scholar]
- 49.O’Brien EP, Morrison G, Brooks BR, Thirumalai D. How accurate are polymer models in the analysis of Förster resonance energy transfer experiments on proteins? J Chem Phys. 2009;130:124903. doi: 10.1063/1.3082151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 51.Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 52.McCarney ER, et al. Site-specific dimensions across a highly denatured protein; a single molecule study. J Mol Biol. 2005;352:672–682. doi: 10.1016/j.jmb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 53.Kuzmenkina EV, Heyes CD, Nienhaus GU. Single-molecule Forster resonance energy transfer study of protein dynamics under denaturing conditions. Proc Natl Acad Sci USA. 2005;102:15471–15476. doi: 10.1073/pnas.0507728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferreon AC, Moran CR, Gambin Y, Deniz AA. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 55.Witten TA, Schafer L. 2 critical ratios in polymer-solutions. J Phys A:Math Gen. 1978;11:1843–1854. [Google Scholar]
- 56.Lifshitz IM, Grosberg AY, Khokhlov AR. Some problems of the statistical physics of polymer chains with volume interaction. Rev Mod Phys. 1978;50:683–713. [Google Scholar]
- 57.Grosberg AY, Kuznetsov DV. Quantitative theory of the globule-to-coil transition. 4. Comparison of theoretical results with experimental-data. Macromolecules. 1992;25:1996–2003. [Google Scholar]
- 58.van der Meer BW. Kappa-squared: From nuisance to new sense. J Biotechnol. 2002;82:181–196. doi: 10.1016/s1389-0352(01)00037-x. [DOI] [PubMed] [Google Scholar]
- 59.Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. Proc Natl Acad Sci USA. 2005;102:2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makarov DE, Plaxco KW. Measuring distances within unfolded biopolymers using fluorescence resonance energy transfer: The effect of polymer chain dynamics on the observed fluorescence resonance energy transfer efficiency. J Chem Phys. 2009;131:085105. doi: 10.1063/1.3212602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wozniak AK, Schröder GF, Grubmüller H, Seidel CA, Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc Natl Acad Sci USA. 2008;105:18337–18342. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gopich IV, Szabo A. Theory of the energy transfer efficiency and fluorescence lifetime distribution in single-molecule FRET. Proc Natl Acad Sci USA. 2012;109:7747–7752. doi: 10.1073/pnas.1205120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalinin S, Sisamakis E, Magennis SW, Felekyan S, Seidel CA. On the origin of broadening of single-molecule FRET efficiency distributions beyond shot noise limits. J Phys Chem B. 2010;114:6197–6206. doi: 10.1021/jp100025v. [DOI] [PubMed] [Google Scholar]
- 64.Milles S, et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aznauryan M, et al. Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc Natl Acad Sci USA. 2016;113:E5389–E5398. doi: 10.1073/pnas.1607193113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng W, et al. Probing the action of chemical denaturant on an intrinsically disordered protein by simulation and experiment. J Am Chem Soc. 2016;138:11702–11713. doi: 10.1021/jacs.6b05443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rubinstein M, Colby RH. Polymer Physics. Oxford Univ Press; Oxford: 2003. [Google Scholar]
- 68.Das RK, Ruff KM, Pappu RV. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr Opin Struct Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mao AH, Lyle N, Pappu RV. Describing sequence-ensemble relationships for intrinsically disordered proteins. Biochem J. 2013;449:307–318. doi: 10.1042/BJ20121346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tran HT, Mao A, Pappu RV. Role of backbone-solvent interactions in determining conformational equilibria of intrinsically disordered proteins. J Am Chem Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 71.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tran HT, Pappu RV. Toward an accurate theoretical framework for describing ensembles for proteins under strongly denaturing conditions. Biophys J. 2006;91:1868–1886. doi: 10.1529/biophysj.106.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.