Significance
Invariant natural killer T (iNKT) cells are activated quickly and play a key role in the control of many microbial infections via their ability to rapidly secrete cytokines and chemokines that enhance many immune responses. Microbial glycolipid antigens that activate iNKT cells have been identified as α-anomerically linked glycolipids. However, the nature of the endogenous lipid antigens that are important contributors to the biology of iNKT cells has been unclear. In this study, antigenic lipids from cow’s milk were isolated using a T-cell receptor trap method, and their stereochemical structures were determined to be hexosylceramides with α-linked hexose headgroups, thus identifying dietary lipid antigens for iNKT cells.
Keywords: NKT, iNKT, lipid antigen, CD1d, glycolipid
Abstract
Glycolipid antigens recognized by αβ T-cell receptors (TCRs) drive the activation of invariant natural killer T (iNKT) cells, a specialized subset of innate T lymphocytes. Glycolipids with α-linked anomeric carbohydrates have been identified as potent microbial lipid antigens for iNKT cells, and their unusual α-anomeric linkage has been thought to define a “foreign” lipid antigen motif. However, mammals use endogenous lipids to select iNKT cells, and there is compelling evidence for iNKT cell responses in various types of sterile inflammation. The nature of endogenous or environmental lipid antigens encountered by iNKT cells is not well defined. Here, we sought to identify lipid antigens in cow’s milk, a prominent part of the human diet. We developed a method to directly capture lipid antigens within CD1d–lipid–TCR complexes, while excluding CD1d bound to nonantigenic lipids, followed by direct biochemical analysis of the lipid antigens trapped at the TCR–CD1d interface. The specific antigens captured by this “TCR trap” method were identified as α-linked monohexosylceramides by mass spectrometry fragmentation patterns that distinguished α- from β-anomeric monohexosylceramides. These data provide direct biochemical evidence for α-linked lipid antigens from a common dietary source.
Adaptive mammalian T lymphocytes are well known for their use of combinatorial diversity to generate a broad repertoire of antigen receptors, a process that evolved to recognize newly encountered peptide epitopes from pathogens. However, certain subsets of human T cells, such as invariant natural killer T (iNKT) cells, mucosal-associated invariant T cells, and germline-encoded mycolyl-reactive T cells reliably generate T-cell receptors (TCRs) that are conserved among genetically diverse humans, and in some cases, across species (1). Recent data indicate that T cells expressing such invariant TCRs comprise a significant fraction of all human T cells, estimated in the range of 10–20% (2). Furthermore, these conserved T-cell subsets are not restricted by MHC-encoded antigen-presenting molecules, but instead can recognize different chemical classes of antigens presented by nonpolymorphic antigen-presenting molecules, including CD1, MR1, or HLA-E (3). One of these conserved T-cell populations, iNKT cells, recognizes specific glycolipids presented by CD1d molecules. Following activation, iNKT cells respond rapidly with vigorous cytokine production that can profoundly shape immune responses (4).
Although a glycolipid antigen for iNKT cells was identified from a marine sponge more than 20 y ago as part of a chemical library screen (5), the origins and chemical identities of physiologically relevant antigens that drive selection and activation of iNKT cells remain controversial. Natural antigenic glycolipids have been detected from various foreign sources, including nonpathogenic α-proteobacteria (6–8), spirochetes (9), and other microbes, such as Streptococcus pneumoniae (10) and Bacteroides fragilis (11, 12). These known microbial lipid antigens for iNKT cells share a key biochemical feature, namely an α-anomeric linkage of one hexose sugar to the lipid backbone. Mammals abundantly produce β-linked hexosylceramides, glucosylceramide and galactosylceramide, but were not previously thought to produce α-linked hexosylceramides. Therefore, a major principle of antigen recognition by iNKT cells has been that self-glycolipids are β-linked and not strongly recognized, whereas pathogens produce lipids that are recognized as foreign based on their α-anomeric linkage (5). However, pathogen-derived lipids presumably do not explain the TCR- and CD1d-dependent activation of iNKT cells during positive selection in the thymus, or in settings such as viral infection, cancer, or sterile inflammation (4). Endogenous lipids have long been proposed to activate iNKT cells in these settings.
It is possible that the longstanding view that mammalian monohexosylceramides are always β-linked is incorrect. Kain et al. showed that antibodies raised against α-linked glycosphingolipids could stain mammalian tissues (13). We recently fractionated mammalian milk-derived lipids, finding that antigenic substances were enriched in the hexosylceramide-containing fraction, and that antigenicity was not abrogated by β-specific glycosidases or chromatographic depletion of β-glucosylceramides (14, 15). Both studies point to the possibility that α-linked lipids are produced in mammalian tissues. However, if they exist, they are likely scarce compounds and they have not been isolated in sufficient yield to confirm α-anomeric structures through direct biochemical analysis. Given the importance of the “α-linked self-lipid hypothesis” to iNKT cell biology, we sought to identify mammal-derived antigenic lipids.
Here, we used soluble iNKT TCR and soluble CD1d to capture the subset of lipids from cow’s milk that form a stable TCR–lipid–CD1d complex through direct analysis of TCR–lipid–CD1d complexes. Combining this TCR “trapping” of CD1d–lipid antigen complexes with nanoelectrospray ionization (ESI) mass spectrometry (MS) and liquid chromatography-mass spectrometry (LC-MS), we could detect monohexosylceramides from cow’s milk as lipid antigens. Using MS-based methods, we determined that the captured lipid antigens were α-linked monohexosylceramides. This study identifies the biochemical structures of natural cow’s milk-derived lipid antigens as α-anomeric hexosylceramides, supporting the hypothesis that α-linked lipids are iNKT cell antigens even in the absence of pathogens.
Results
Prepurification of Milk Lipid Antigens for iNKT Cells.
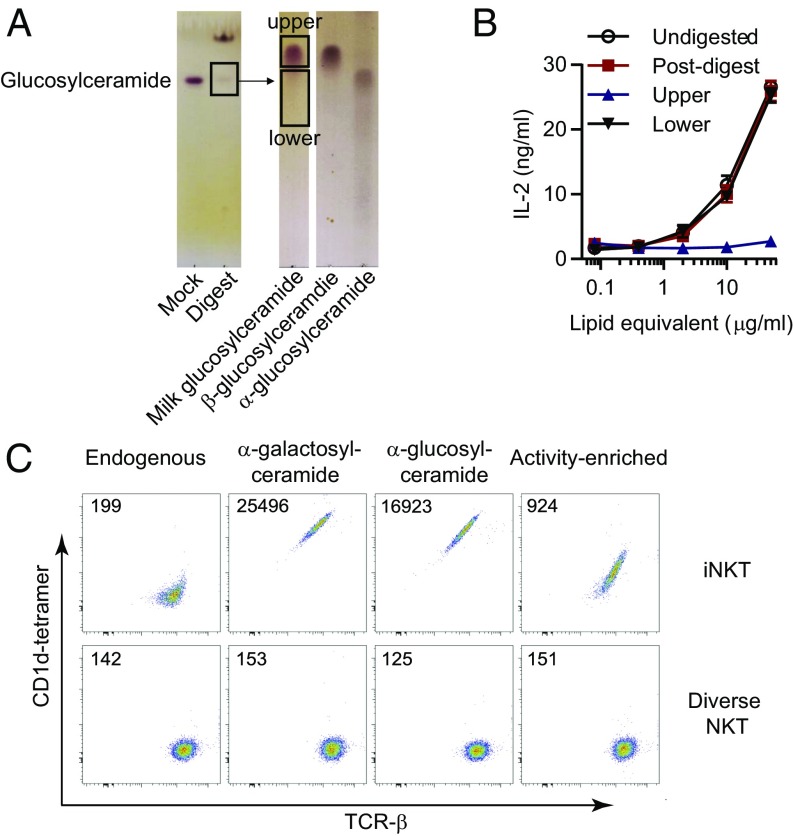
We previously showed that the glucosylceramide-containing fraction of cow’s milk contains antigenic lipids for iNKT cells (14, 15). To further purify and identify the milk-derived lipid antigens, we digested a milk hexosylceramide lipid fraction with β-glucosidase to destroy the β-linked glucosylceramides. Thin-layer chromatography (TLC) showed that this treatment markedly reduced the total mass of glycolipids comigrating with a glucosylceramide standard, yet did not reduce the potency of iNKT cell activation (Fig. 1A). This result could be explained if the starting material was predominantly composed of nonantigenic β-linked glucosylceramides, and antigenic α-linked hexosylceramides were present in milk that survived the enzyme treatment. To address this possibility, we used a TLC separation method that resolved monohexosylceramide α- and β-anomers (15), yielding two bands. Only the lower band, which comigrated with an α-glucosylceramide standard, stimulated an iNKT cell response (Fig. 1B), suggesting that α-linked hexosylceramides could be present. However, this interpretation is contrary to the prevailing view that mammalian products contain only β-linked hexosylceramides. The lower band that activated iNKT cells (Fig. 1 A and B), which we refer to as the “activity-enriched” fraction, represented a very small fraction of total milk hexosylceramides, which provided an obstacle to formal biochemical identification of the antigen. Thus, we considered new approaches to antigen discovery.
Fig. 1.
iNKT cell activation by milk hexosylceramides. (A) A cow’s milk glucosylceramide-fraction was digested with recombinant lysosomal glucosidase to reduce β-glucosylceramide. The remaining material detected at the monohexosylceramide retention time was purified by preparative TLC and further resolved to upper and lower fractions. (B) TLC fractions were tested for activity by coculture with the DN32 iNKT cell hybridoma and CD1d-transfected RAW-264.7 cells. (C) CD1d monomers loaded with the indicated lipids were assembled into tetramers that were used to stain BW58 cells transfected with an iNKT cell TCR or with a diverse NKT cell TCR that does not recognize α-galactosylceramide. Numbers indicate geometric mean fluorescence values. Data are representative of two independent experiments performed with separate starting material.
Milk Lipids Mediate CD1d–TCR Binding.
Nearly all prior approaches to antigen discovery, including that described above (Fig. 1A), have relied on a correlation between two assays that separately measure T-cell activation and biochemical content. The final antigen identification relies on the premise that the most readily detectable molecules are also the T-cell stimulants. By definition, antigenic lipids ligate TCRs to antigen-presenting molecules with higher affinity than nonantigenic lipids, so we reasoned that a recombinant iNKT TCR with relatively high affinity for CD1d–α-galactosylceramide complexes, known as 2C12 (16), might selectively trap those CD1d complexes carrying milk-derived antigens, while leaving behind CD1d molecules that were bound to nonantigenic lipids. Such a TCR trap approach was recently used to identify CD1a autoantigens (17). We hypothesized that this approach might be feasible for CD1d and milk-derived antigens, because partially enriched milk fractions could mediate the physical binding of CD1d tetramers to cells expressing an iNKT cell Vα14Jα18 Vβ8.2Jβ2.1 TCR, but not to cells expressing a sulfatide-reactive diverse NKT cell Vα1Jα26 Vβ16Jβ2.1 TCR (18, 19). Staining of iNKT TCR-expressing cells with CD1d tetramers loaded with activity-enriched milk lipids was stronger than staining with unloaded CD1d tetramers, but weaker than tetramers generated with known potent lipid antigens α-galactosylceramide and α-glucosylceramide (Fig. 1C). Thus, cow’s milk contained compounds capable of mediating CD1d–TCR binding.
TCR Trap Assay.
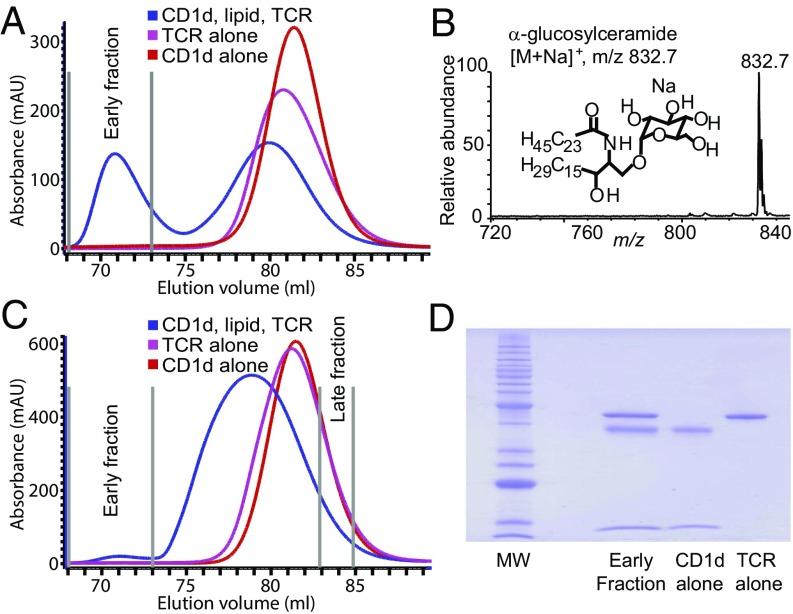
To develop a cell-free assay to detect lipid antigens, we tested the TCR trap system by loading recombinant soluble CD1d with synthetic α-glucosylceramide, and then mixing this antigen-loaded CD1d with recombinant soluble iNKT cell TCR monomers. The subset of CD1d proteins carrying α-glucosylceramide was expected to bind TCR heterodimers, leading to the formation of a higher molecular weight ternary complex, which could be purified by size-exclusion gel chromatography. A mixture of α-glucosylceramide-treated CD1d and TCRs showed a large early fraction that eluted before separate CD1d and TCR monomers (Fig. 2A), consistent with a larger protein complex. After eluting lipids from this fraction using a mixture of chloroform and methanol, positive-mode nano ESI-MS identified a prominent ion at m/z 832.7, corresponding to α-glucosylceramide [(M+Na), M = C48H91NO8], as well as its isotopes. The lack of other ions present above background levels indicated that lipids from culture media or those potentially carried by the outer surface of CD1d or TCRs did not substantially confound the analysis (Fig. 2B). Thus, α-glucosylceramide performed its expected role in ligating CD1d to TCR, and direct analysis of CD1d–lipid–TCR complexes was sufficient to identify the ligating molecule without interference by other substances adherent to the lipid–protein complex.
Fig. 2.
TCR trap assay. (A) Synthetic α-glucosylceramide was loaded in recombinant CD1d molecules, mixed with recombinant iNKT cell TCRs (2C12), and run over a size-exclusion chromatography column. (B) Lipids were eluted from the early fraction excluded from the column and analyzed by linear ion trap MS. A dominant ion corresponding to the α-glucosylceramide standard was recovered and few other ions were detected. (C) The activity-enriched milk monohexosylceramide fraction was loaded in CD1d and treated as in A. (D) The early fraction, CD1d alone, or TCR alone, was resolved on an SDS/PAGE gel and total proteins stained, yielding bands corresponding to TCR, CD1d, and β2M. Data are representative of two independent experiments performed with separate starting material. mAU, milliabsorbance unit.
Next, to identify unknown lipids, we used this TCR trap assay with CD1d molecules that had been loaded with activity-enriched cow’s milk monohexosylceramides (Fig. 1 A and B). A small but detectable peak was observed in the early size-exclusion fractions and this showed no overlap with CD1d or TCR monomer fractions (Fig. 2C). Coomassie staining of the early fraction revealed three proteins matching the apparent molecular weights of CD1d, β2m, and TCR with band-similar intensity, consistent with 1:1:1 stoichiometry (Fig. 2D). The peak height of the protein trace of the early eluting fraction made with milk lipids (Fig. 2C) was substantially lower than that of the early fraction made with CD1d carrying synthetic α-glucosylceramide (Fig. 2A), suggesting that TCRs bound to a small fraction of the total pool of CD1d–lipid complexes present. However, because this peak was observed at the same elution time as the synthetic α-glucosylceramide standard-loaded CD1d, it suggested that the milk lipid-derived ternary complex carried a high-affinity antigen.
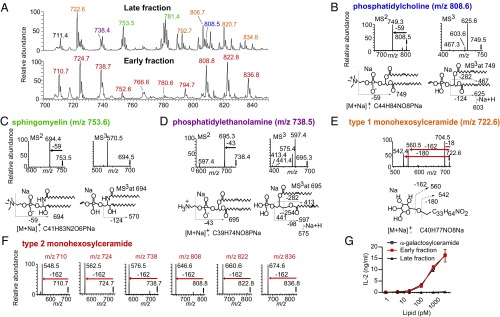
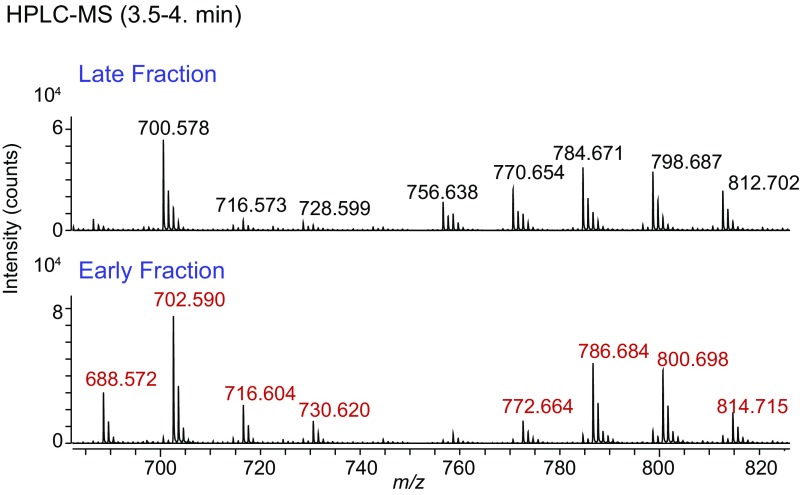
Nanoelectrospray analysis of all eluted milk lipids in the late fraction containing CD1d and TCR monomers found numerous ions, and the mass intervals among the most abundant ions were diverse, suggesting the presence of structurally diverse lipids from many classes (Fig. 3A, Upper). In contrast, the early fraction, which contained CD1d–TCR complexes, showed fewer eluted ions (Fig. 3A, Lower). The lower peak height from the size-exclusion tracing and the less-complex MS profiles suggested that the iNKT cell TCR selected small subset of CD1d–milk lipid complexes from among all CD1d–lipid complexes present in the starting material. In fact, nearly all nano ESI-MS signals derived from the early-eluting CD1d–TCR complexes could be accounted for by a single alkane series comprised of lipids of m/z 710.7, apparent chain-length analogs (m/z 724.7, 738.7, 752.6, 794.7, 808.8, 822.8, 836.8), and the isotopes of these lipids.
Fig. 3.
Linear ion-trap MS analysis of CD1–lipid–TCR complexes. (A) Lipids extracted from the TCR trap assay fractions indicated in Fig. 2C were analyzed by linear ion-trap MS. Prominent ions from the late fraction were identified as (B) phosphatidylcholine, (C) sphingomyelin, (D) phosphatidylethanolamine, and (E) type 1 monohexosylceramide. (F) All major ions in the early fraction were an alkane series, and collision of six of the most abundant ions generated type 2 monohexosylceramide patterns. (G) Lipids were extracted from the early fraction, quantified, and cocultured with the DN32 iNKT cell hybridoma and CD1d-transfected RAW-264.7 cells. Experiments were performed on lipids extracted two independent TCR trap assays.
TCR Ligands Identified Using Nano ESI-MS.
Next we identified the lipid ligands trapped-in or excluded-from CD1d–TCR complexes using shotgun nanoelectrospray with multistage ion-trap MS. This method provides high sensitivity, broad detection, and structural information from collision-induced dissociation (CID), but offers relatively low mass accuracy. Considering the lipids eluted from the late fraction containing CD1d and TCR monomers, CID-MS of selected ions at m/z 738.4, 753.5, and 808.5 identified phosphatidylethanolamine, sphingomyelin, and phosphatidylcholine (Fig. 3 B–D). In addition, fragments from the ion at m/z 722.6 corresponded to a hexosylceramide whose two fatty-acid chains had a combined lipid tail length of C34 with one unsaturation (Fig. 3E). This “type 1” monohexosylceramide collision pattern was characterized by a dehydration [loss of 18 u (atomic mass unit), m/z 704.5], loss of a hexosyl unit (162 u, m/z 560.5), and loss of both fragments (loss of 180 u, m/z 542.4). In contrast, ions from early fraction-containing CD1d–TCR complexes did not show detectable signals for phosphatidylethanolamine, sphingomyelin, or phosphatidylcholine. Instead, complexes in the early fraction released matched expected masses of a “type 2” hexosylceramide, which unlike type 1 patterns, lacked a dehydration ion but did show a loss of 162 u. In fact, the abundant ions in the early fraction were all accounted for by single alkane series corresponding to an monohexosylceramide, with an overall lipid length ranging between C33 (m/z 710.7) and C42 (m/z 836.8). Separate analysis of six members of this alkane series demonstrated highly reproducible type 2 collision pattern (Fig. 3F).
The detailed profiles of these released lipids suggested several general conclusions. First, monomeric CD1d proteins were bound to phospholipids, sphingomyelin, type 1 hexosylceramide, as well as other lipids that were not specifically identified (Fig. 3A). Second, TCR–CD1d complex formation largely or completely excluded CD1d bound to phosphatidylcholine (m/z 808.6 with the m/z 749.3 product), sphingomyelin (m/z 753.6 with m/z 694.4 product), phosphatidylethanolamine (m/z 738.5 with m/z 695.3 product), and the type 1 hexosylceramide with a monounsaturated lipid tail (m/z 722.6 with m/z 542.4 fragment). Third, lipids eluted from the TCR–CD1d complexes contained type 2 monohexosylceramides ranging from C33–C42 in chain length (Fig. 3A). Furthermore, the characteristic patterns of loss in CID analyses by type 1 (18, 162, and 180 u) and type 2 (162 u) hexosylceramides implicated a differing conformation of these ceramide-based lipids. This interpretation is consistent with the TCR discriminating structural classes of lipids including glycoslyceramides, and possibly α- vs. β-linked hexosylceramides.
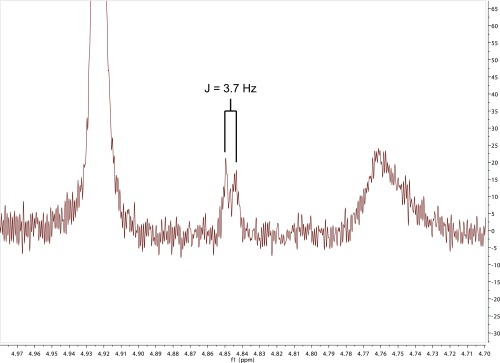
The molar concentrations of hexosylceramides eluted from the early and late complexes were determined using HPLC-MS, in comparison with standards, and this material was tested for the ability to elicit cytokine production from an iNKT cell hybridoma, DN32. The late fraction did not have detectable activity, whereas the molar activity observed in the early fraction was similar to that of a synthetic antigen for iNKT cells known as α-galactosylceramide, KRN7000 (Fig. 3G). Thus, our approach was successful as a TCR-based affinity trap for selecting antigenic glycolipids that ligate TCR to CD1d out of a complex mixture of natural lipids. The equipotency of type 2 hexosylceramides and α-galactosylceramide was consistent with the type 2 hexosylceramides being an α-linked lipid. Although only trace amounts of the type 2 monohexosylceramide could be recovered from ternary complexes of CD1d–lipid–TCR, 1D-proton NMR spectroscopy carried out on a 900-MHz instrument identified a doublet with a chemical shift of 4.85 ppm, consistent with the anomeric proton resonance of α-monohexosylceramide (Fig. S1) (15).
Fig. S1.
One-dimensional proton NMR. Lipids eluted from the TCR trap early fraction were analyzed by 1D proton NMR. A resonance with a chemical shift of 4.85 ppm was observed, characteristic of α-monohexosylceramide. This analysis was performed on one of two independent early-fraction lipid eluates.
Identification of Ligands at High Mass Resolution.
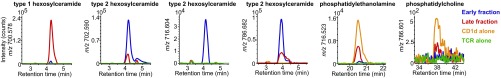
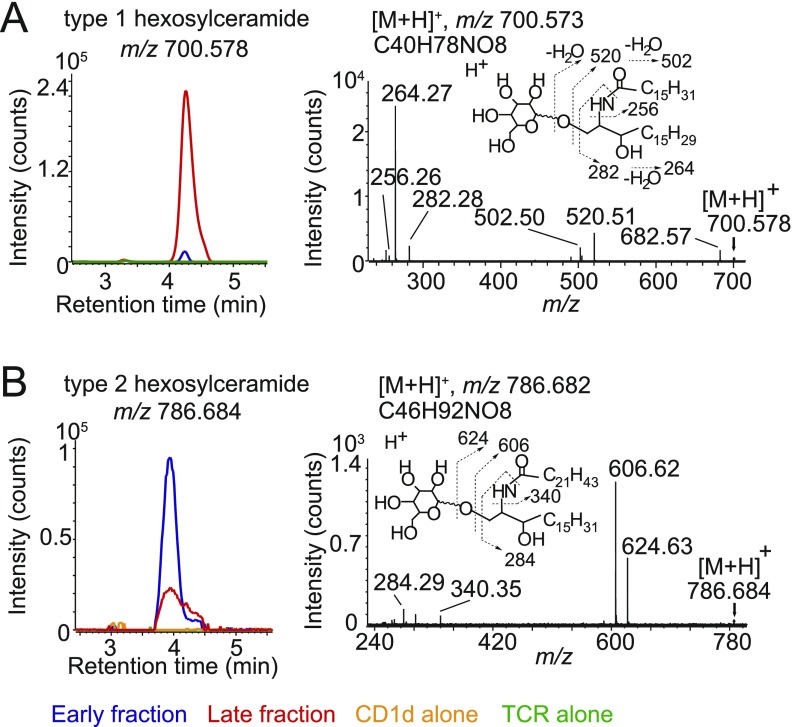
To further biochemically characterize the lipids that promoted the formation of TCR–lipid–CD1d complexes, we separately analyzed eluents from milk lipid-treated protein monomers present in the late fraction, untreated CD1d, and untreated TCR (Fig. 4). Here, we took advantage of the higher mass accuracy of a quadrupole time-of-flight (Q-ToF) mass detector coupled with chromatographic separation in normal-phase HPLC-MS to confirm the identity of the key ions. Using this method, the major ions previously identified by nano ESI-MS were confirmed and assigned based on high mass accuracy and HPLC retention time, compared with standards as phosphatidylcholine (38.1 min, m/z 786.601), phosphatidylethanolamine (20.5 min, m/z 716.523), type 1 hexosylceramide (4.1 min, m/z 700.578), and type 2 hexosylceramide (4.1 min, m/z 702.590, 716.604, and 786.682) (Fig. 4). No substantial false-positive signals were derived from lipids released from the TCR, indicating that the lipids detected were carried by CD1d. As seen with the nanospray method (Fig. 3), CD1d alone and mixed CD1d and TCR monomers bound phosphatidylcholine, phosphatidylethanolamine, and type 1 monohexosylceramides (Fig. 4 and Fig. S2), but these were excluded from CD1d–TCR complexes. Conversely, type 2 hexosylceramides eluted with low or undetectable signals from milk lipid-treated monomers, but were highly enriched in CD1d–TCR complexes. Thus, CD1d bound many lipids, but the iNKT TCRs selectively captured CD1d bound to type 2 hexosylceramides (Fig. S3).
Fig. 4.
HPLC-MS analysis with Q-ToF MS detection. Lipids extracted from TCR trap assay fractions were analyzed by Q-ToF HPLC-MS experiments to provide high mass accuracy confirmation of the structures deduced in Fig. 3, to measure lipid binding to TCR and to measure retention time in comparison with standards. HPLC tracings are shown for selected targeted ions based on predicted masses for the structures identified in Fig. 3. Experiments were performed on lipids extracted two independent TCR trap assays.
Fig. S2.
HPLC Q-ToF MS collision spectra for type 1 and -2 hexosylceramides. TCR trap assay (A) late-fraction type 1 monohexosylceramides and (B) early-fraction type 2 monohexosylceramides were analyzed by HPLC coupled to Q-ToF MS shown as extracted ion chromatograms (Left) and CID-MS (Right). Experiments were performed on lipids extracted two independent TCR trap assays and representative HPLC tracings are shown.
Fig. S3.
Type 1 and 2 hexosylceramide ions determined by Q-ToF MS. TCR trap assay late-fraction type 1 (Upper) monohexosylceramides and early-fraction type 2 (Lower) monohexosylceramides were analyzed by high mass accuracy Q-ToF MS. HPLC elution times of 3.4–5 min are shown, corresponding to the elution time of monohexosylceramide standards. Experiments were performed on lipids extracted two independent TCR trap assays, and representative HPLC tracings are shown.
Structural Identification of Type 1 and 2 Hexosylceramides.
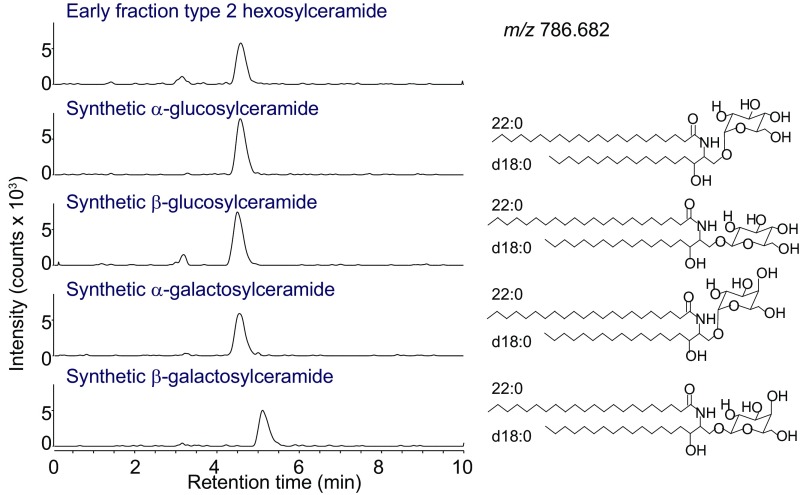
Despite their strikingly different patterns of binding within—or being excluded from—CD1d–TCR complexes, normal-phase HPLC-MS showed that type 1 and 2 hexosylceramides had equivalent retention time and the same underlying molecular composition, except for variations in lipid tail length and unsaturation. CID-MS experiments using the ion-trap architecture, however, showed different fragmentation patterns for type 1 and type 2 hexosylceramides (Fig. 3). We next asked whether anomeric variants could be distinguished by CID-MS. First, we produced synthetic standards differing only in the stereochemistry of their hexose attachment. Both glucosyl and galactosyl variants were synthesized, with dihydrosphingosine (d18:0) as their lipid backbones based on the molecular composition of type 2 hexosylceramides identified in the milk-derived early fraction. These synthetic standards had a similar retention time by normal-phase HPLC to the early-eluting milk lipids (Fig. S4). The synthetic standards were tested for their ability to activate the DN32 iNKT cell hybridoma after loading in CD1d-expressing RAW264.7 cells. For both glucosyl and galactosyl synthetic standards, activity was observed for the α-linked variants, but not for the β-linked variants (Fig. 5A).
Fig. S4.
TCR trap early fraction coelutes with a panel of synthetic monohexosylceramides. HPLC Q-ToF MS tracings of the synthetic monohexosylceramide standards with [M+H]+, m/z 786.682 (Fig. 5A) and TCR trap assay early fraction are shown. Experiments with synthetic lipid standards were performed at least three times. Experiments were performed on lipids extracted two independent TCR trap assays and representative HPLC tracings are shown.
Fig. 5.
Linear ion-trap MS distinguishes α- from β-hexosylceramides. (A) Lipid standards were synthesized based on the structures of the type 2 monohexosylceramides and tested in coculture with the DN32 iNKT cell hybridoma and CD1d-transfected RAW-264.7 cells. (B) MS-MS of sodiated adducts demonstrated characteristic fragmentation patterns for α- and β-glucosylceramide synthetic standards when studied over a range of collision energy settings. (C) Linear ion-trap MS-MS was performed on the cow’s milk unenriched, activity-enriched, and TCR trap early fraction lipids. Three experiments were performed for A and B. Experiments in C were performed on lipids extracted two independent TCR trap assays.
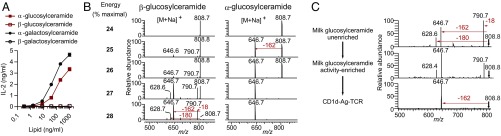
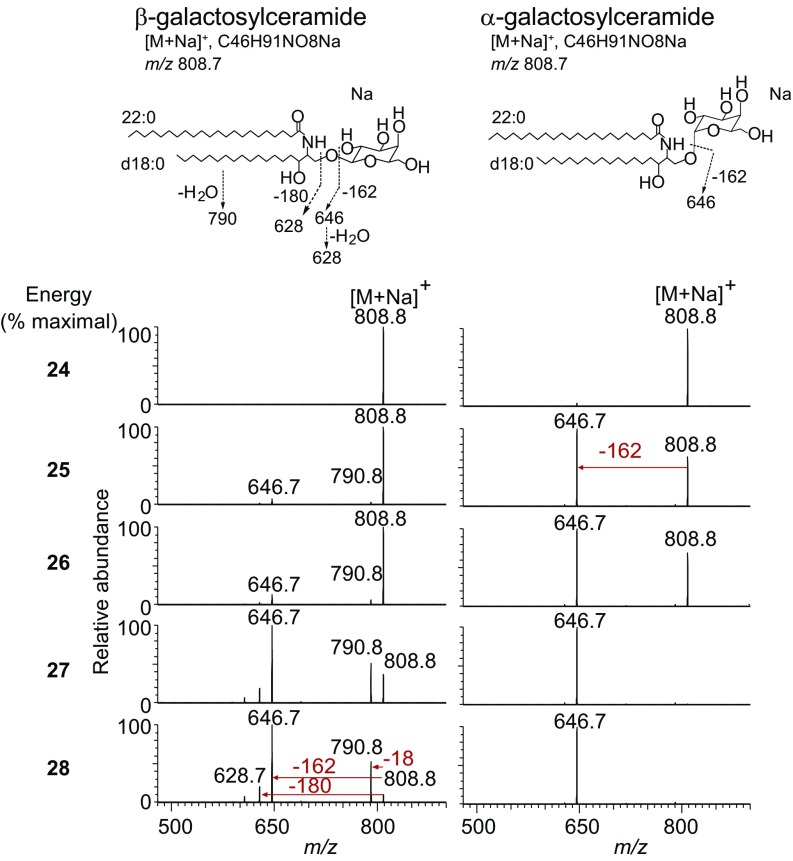
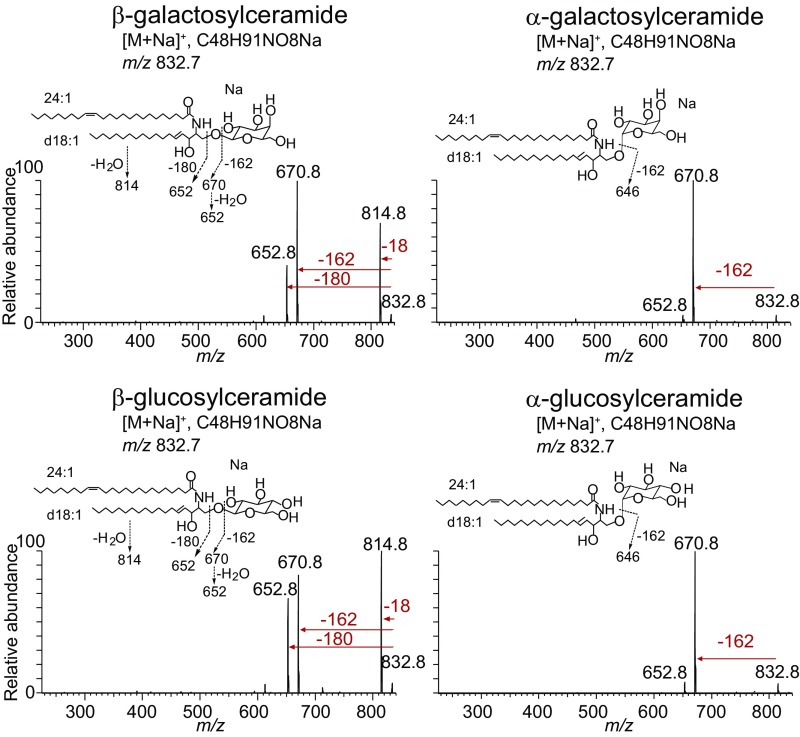
Positive-mode ion-trap CID-MS of sodiated adducts of these synthetic hexosylceramides was carried out under conditions that matched those used for milk-derived compounds (Fig. 3), and over a range of collision energy settings (Fig. 5B). Collision of the α-glucosylceramide synthetic standard demonstrated almost exclusively a primary loss of 162 u, matching the pattern for type 2 milk-derived hexosylceramides. In contrast, β-glucosyl standards matched the fragmentation pattern seen for type 1 hexosylceramides from milk, showing a loss of 18 u at a similar frequency as loss of 162 u. Although both patterns involved the loss of 162 u as a hexosyl unit, analysis over a range of collision energy demonstrated that β-glucosylceramide also lost 18 u and 180 u, whereas α-glucosylceramide showed neither of these fragmentations at a substantial frequency. Parallel analyses of α- and β-galactosylceramide standards showed similar characteristic α- and β-collision patterns (Fig. S5). Matched synthetic α- and β-glucosylceramide and galactosylceramide standards based on the type 1 hexosylceramide lipid backbone structure showed similar characteristic α- and β- collision patterns, demonstrating that the anomeric linkage discrimination by this technique was not limited to dihydrosphingosine-based glycosphingolipids (Fig. S6). Ion-trap CID-MS of α-galactosylceramide with a phytosphingosine base also fragmented with the pattern matching type 2 hexosylceramides. We concluded from these experiments that linear ion-trap MS could sensitively distinguish α- from β-hexosylceramides under the conditions measured for a range of lipids with differing tail lengths and unsaturations.
Fig. S5.
Linear ion-trap MS distinguishes α- from β-galactosylceramides. As in Fig. 5B, α- and β-galactosylceramide synthetic standards were performed over a range of collision energies. Chemical structures of α- and β-galactosylceramide with deduced fragmentations are shown. MS-MS of sodiated adducts demonstrated characteristic fragmentation patterns, like those observed for α- glucosylceramide and β-glucosylceramide synthetic standards. Three independent experiments were performed.
Fig. S6.
Linear ion-trap MS distinguishes α- from β-glycosylceramides with a sphingosine base. Using linear ion-trap CID-MS, the synthetic standards with a d18:1 sphingosine base (structures shown) were analyzed. MS-MS tracings are shown. Three independent experiments were performed.
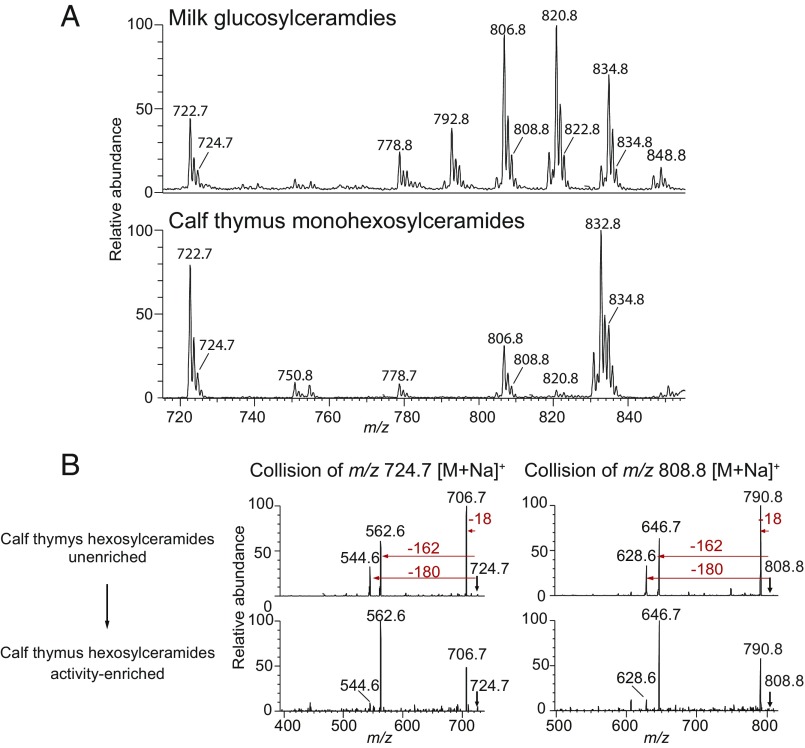
We compared the collision patterns of the major hexosylceramide ions from cow’s milk lipids. Unfractionated cow’s milk glucosylceramides demonstrated a fragmentation pattern characteristic of β-hexosylceramide standards (Fig. 5C), consistent with the main lipid in the starting material being β-glucosylceramide. The activity-enriched cow’s milk glucosylceramide showed a β-hexosylceramide collision pattern, but with an increased frequency of a type 2 pattern characterized by the loss of the sugar headgroup (162 u). The antigenic lipids captured in the TCR trap assay showed the fragmentation pattern typical of α-hexosylceramides. We concluded from these experiments that the TCR trap captured type 2 α-linked hexosylceramides from cow’s milk. To address the possibility that these type 2 hexosylceramides were derived from microbes present in cow’s milk, we purified the monohexosylceramide fraction from calf thymus, a mammalian lipid source previously shown activate iNKT cells (15). Several ions with the m/z seen in cow’s milk were also present in calf thymus, including ions corresponding to type 2 hexosylceramides m/z 724.7 and 808.8 (Fig. S7). Linear ion-trap CID-MS of untreated calf thymus hexosylceramides showed a characteristic β-hexosylceramide pattern. After activity enrichment of thymus hexosylceramides by treatment with β-glucosidase and TLC fractionation, as in Fig. 1, we observed a collision pattern consistent with the presence of α-hexosylceramides, based on a substantial increase in the abundance of the ion representing a primary loss of the sugar headgroup.
Fig. S7.
Monohexosylceramides from calf thymus. (A) The monohexosylceramide fraction from calf thymus was purified by normal-phase TLC and analyzed by linear ion-trap MS. Monohexosylceramides were detected in the thymus with the same m/z as those identified in cow’s milk, including the type 2 monohexosylceramides m/z 724.7 and 808.8. (B) Calf thymus hexosylceramides were subjected to activity enrichment as in Fig. 1 A and B. Collision of the m/z 724.7 and 808.8 before (Upper) and after activity enrichment (Lower) demonstrated an increase in loss of 162 u following activity enrichment, as observed for cow’s milk (Fig. 5C). MS experiments were performed twice.
Discussion
Uncovering the specific antigens recognized by iNKT cells is central to understanding their roles in health and disease. Although lipid antigens for iNKT cells have been identified in the microbial world (6–10), experimental evidence has demonstrated important roles for endogenous and environmental lipid antigens. Several candidate endogenous lipid antigens have been proposed (13, 14, 20–22), although technical limitations in the ability to detect these antigens has hampered our ability to determine their contextual roles. Here, we provide direct biochemical evidence for α-hexosylceramide derived from a mammalian source, cow’s milk.
There is increasing evidence that iNKT cells encounter lipid antigens at mucosal surfaces. CD1-dependent immunomodulatory lipids have been identified in gut microbes (11, 12), pollen (23), house dust (24), and in our work, cow’s milk. We have previously demonstrated that cow’s milk contains substantially more lipid antigen activity than human or mouse milk. Although native triglycerides are removed from infant formula during processing, bovine polar lipids, including lipid antigens for iNKT cells, are retained in the final product (15). Studies in germ-free mice suggest that the iNKT cell compartment can be shaped by CD1d–lipid signals in the enteral tract (11, 25), and thus early-life exposure to cow’s milk-based infant formula might similarly shape human mucosal iNKT cells in early life. In adult mice, orally delivered antigen activates iNKT cells in the gut (26), suggesting that exposure to dietary lipid antigens in adult life may also have consequences. Interestingly, it is unclear whether cows have iNKT cells (27), as their CD1d does not present the long-chain α-galactosylceramide commonly used as a tool in mice and humans (28). Bovine CD1d can present short-chain α-galactosylceramides (28, 29), and it is possible that an increased presence of α-hexosylceramdies in cow's milk and tissues could lead to thymic deletion of iNKT cells, as has been observed in mice in the presence of exogenous α-galactosylceramide (30).
Although the bulk solids in milk derive overwhelmingly from mammary glands, we cannot formally rule out that colonizing microbes synthesized, or played a role in the synthesis of the α-hexosylceramides present in cow’s milk. Several features of our data argue against possible contamination of milk by microbial products. We have previously observed iNKT cell-stimulating activity in the monohexosylceramide fractions of the milks, sera, and internal organs of multiple animal species (14, 15). The N-acyl chain use of the α-hexosylceramides that we identified in this report mirrors that of the bulk β-glucosylceramides in cow’s milk, suggesting that these lipids are of mammalian origin. We also identified type 2 hexosylceramides from calf thymus with the same m/z as those in cow’s milk. These thymic hexosylceramides, like those found in cow’s milk, shifted from a β-hexosylceramide pattern toward a characteristic α-hexosylceramide collision pattern after activity enrichment. Currently, the level of α-hexosylceramide identified from cow’s milk, a relatively abundant and potent natural source of lipid antigen, is near the limit of detection for the TCR trap assay system. However, we are continuing to improve the system so that lipids can be captured and identified from smaller amounts of source material.
The stereochemistry of organic molecules can have significant biological consequences. Determination of stereochemistry has largely been accomplished by NMR. However, for rare compounds, such as endogenous iNKT cell lipid antigens, purification of sufficient material for complete, multidimensional NMR analysis is currently not possible. As shown in this report and others (31, 32), CID-MS can also be used to determine stereochemical features. We tested α- and β-linked synthetic standards with both sphingosine and dihydrosphingosine bases, and found consistent characteristic α- and β-fragmentation patterns, showing that this detection method was not limited to certain lipid backbones. Interestingly, these collision patterns were only observed with selected positive-mode adducts, including sodium and rubidium. We hypothesize that for the β-hexosylceramide structure, positive adducts with a sufficiently large valence, such as sodium and rubidium, can coordinate elements on both sides of the glycoside bond, stabilizing the glycoside bond during collision, thus reducing the propensity for cleavage of the sugar headgroup. For α-hexosylceramide, this headgroup stabilization may be sterically hindered, thus allowing for fragmentation across the glycoside bond at a lower energy.
Identification of CD1d-presented lipid antigens has largely relied on indirect evidence, such as extrapolation from cells deficient in lipid synthesis genes (22, 33), or fractionation of crude lipid sources with activity tracking within these fractions (14, 20). These approaches have been useful, but do not directly demonstrate recognition by the TCR, and rely on correlations between two or more assays. Here, we used a direct method for antigen discovery, the TCR trap assay. This approach uses ternary complex formation both as the preparative method to recover lipids of interest and as the assay demonstrating the lipid's role in ligating CD1d and TCR. The TCR trap assay has also been used successfully to identify CD1a ligands (17), and could be used to identify ligands for other CD1 isoforms, or even MR1. This approach will allow tracking of rare lipids from virtually any source, including from complex mixtures, or limited starting materials, which represent common challenges when starting with mammalian tissues and cells.
Materials and Methods
Initial lipid activity enrichment was performed as previously described (15). MS analyses were performed on a Thermo-Fisher LXQ ion-trap mass spectrometer or an Agilent 6520 Accurate-Mass Q-ToF with HPLC. For details of experimental conditions and analysis, see SI Materials and Methods.
SI Materials and Methods
Lipids.
Starting with 50 mg cow’s milk glucocerebrosides (Matreya), two independent experiments were done to enrich the lipids in the starting material that activated iNKT cells (referred to as the “activity-enriched” fraction) (15). Briefly, lipids were first digested with recombinant human lysosomal glucocerebrosidase (Velaglucerase Alfa, Shire). The monohexosylceramides remaining after digestion were recovered by preparative TLC. The active lipid fraction was further isolated using the HICMW system (15). Lipids were resolved with normal-phase TLC and visualized by staining with an α-naphthol sulfuric acid (Sigma) solution and charring. Calf thymus (Research 87) monohexosylceramides were purified as previously described (15), and activity enrichment was carried out as for cow’s milk lipids. β-Glucosylceramide (d18:1/24:1) was purchased from Avanti polar lipids. Synthetic lipids, including α-galactosylceramide KRN7000 (d18:0-C26:0, containing a phytosphingosine base with a 26 carbon unsaturated N-acyl chain) were synthesized, α-glucosylceramide (d18:0-C22:0, containing a dihydrosphingosine base), β-glucosylceramide (d18:0-C22:0, containing a dihydrosphingosine base), α-galactosylceramide (d18:0-C22:0, containing a dihydrosphingosine base), and α-galactosylceramide (d18:0-C22:0, containing a dihydrosphingosine base) were synthesized as previously described (34).
CD1d Tetramer Staining of Invariant and Type 2 NKT Cell Lines.
iNKT (Vα14Jα18 Vβ8.2Jβ2.1, CDR3α DRGSALGRLHFGAG, CDR3β GDAGGNYAEQFFGPG) and diverse NKT (Vα1Jα26 Vβ16Jβ2.1, CDR3α SEQNNYAQGLTFGLG, CDR3β SFWGAYAEQFFGPG) BW58 cell lines were generated using retroviral transduction (35). Recombinant mouse CD1d tetramers (36) were loaded with lipids dissolved in 0.5% Tyloxapol (Sigma) with Tris-buffered saline pH 8.0 at a 3:1 lipid:protein molar ratio. Cell lines with invariant and diverse NKT cell TCRs costained with TCRβ (clone H57-597) and CD1d tetramers loaded with C24:1 α-galactosylceramide (PBS44) (kindly provided by Paul Savage, Brigham Young University, Provo, UT), α- glucosylceramide, activity-enriched milk antigen, or unloaded CD1d tetramer.
TCR Trap Assay.
Recombinant Vα14Jα18 Vβ8.2Jβ2.5 TCR (2C12), CDR3β GDEGYTQYFGPG was prepared as previously described for the production of recombinant Vα14Jα18 Vβ7 TCR (37). Following lipid loading, excess Tyloxapol was removed from recombinant mouse CD1d using ion exchange chromatography (Mono Q 10/100GL; GE Healthcare). Mouse CD1d and soluble 2C12 TCR were passaged separately over a Superdex-200 16/60 gel-filtration column (GE Healthcare), just before the generation of ternary complexes, to remove any protein aggregates. CD1d-milk Ag-TCR and CD1d–α-glucosylceramide–TCR ternary complexes were generated by mixing equal amounts of CD1d and by passage over Superdex-200 column at a flow rate of 0.5 mL/min. Gel-filtration fractions were analyzed on a 12% SDS/PAGE gels (Bio-Rad) using Coomassie blue stain.
iNKT Cell Activation Assay.
Lipid activity assays were performed with the DN32 iNKT cell hybridoma (38) and CD1d-transfected RAW-264.7 cells (39). Briefly, lipids were dried under a nitrogen stream, then sonicated in serum-containing assay media. Next, 5 × 104 DN32 cells and 5 × 104 CD1d-transfected RAW-264.7 cells were cocultured with lipids for 16 h at 37 °C. Assays were performed in RPMI-1640 media supplemented with 10% FCS (Gemini Bio), penicillin/streptomycin, l-glutamine, and β-mercaptoethanol (Life Technologies) at 37 °C in 5% CO2. For x-y graphs, SEM is shown.
Nanospray ESI-MS.
All organic solvents were HPLC grade. Lipids were extracted from CD1d proteins were with chloroform, methanol, and water according to the method of Bligh and Dyer (40). Briefly, protein samples in 200 µL 10 mM Tris, pH 8.0, 150 mM NaCl were transferred to 15-mL glass tubes, treated with 0.75 mL of chloroform:methanol [1:2 (vol:vol)], and vortexed for 1 min to denature the protein and extract lipids. The mixture was treated with 0.25 mL of chloroform and vortexed for 30 s, and then treated with 0.25 mL of water and vortexed for another 30 s. The mixture was centrifuged at 850 × g for 10 min to separate the layers. The upper aqueous layer was removed, and the lower organic lipid containing layer was transferred to another clean glass tube. The organic solvents were dried under a liquid nitrogen stream and the lipid residue was redissolved in chloroform/methanol (1:2) and stored at −20 οC. For a shotgun lipid analysis, the extracted lipids were loaded onto a nanospray tip for ESI-MS and multistage CID-MS using linear ion-trap MS (LXQ, Thermo Scientific). Collision energy was 20–30% of maximum and product ions were trapped with a q value of 0.25.
HPLC-MS Analysis.
To obtain the accurate mass and retention time information, the extracted lipids from 25–400 μg of CD1d protein and normalized to 25 µM based on the concentrations of input protein. The normalized samples (25 µL) were dried and redissolved in 25 µL of starting mobile phase B (see below), and 20 µL were injected for HPLC-ESI-MS analysis (Agilent 6520 Accurate-Mass Q-ToF and 1200 series HPLC system using a normal-phase Varian MonoChrom Diol column, 3 µm × 150 mm × 2 mm, and running at 0.15 mL/min) according to the published methods (41). The solvent mobile phases were (A) isopropanol/methanol [70:30, (vol:vol)] and (B) hexane/isopropanol [70:30, (vol:vol)]. Both mobile phases were supplemented with 0.1% formic acid and 0.05% ammonium hydroxide. The gradient system was following: 0–10 min, 100% B; 10–17 min, 100–50% B; 17–22 min, 50% B; 22–30 min, 50–0% B; 30–35 min 0% B; 35–40 min, 0–100% B; 40–44 min, 100% B; followed by additional 6 min 100% B postrun. The glycolipid concentrations in the extracted fractions were estimated by comparing the peak areas of the extracted ion chromatograms to those of the external standard curves (β-glucosylceramide d18:1/24:1). All monohexosylceramide variants were assumed to have a similar response factor for ionization. For the synthetic glucosyl- or galactosylceramides, the normal-phase HPLC elution system was optimized for resolution of individual compounds using an isocratic condition with mobile-phase solvents of hexane/isopropyl alcohol [70:30 (vol:vol)], containing formic acid 0.1% (vol:vol) and ammonium hydroxide 0.05% (vol:vol). The run was set at 0.15 mL/min for 10 min. The synthetic compounds were eluted from 4 to 6 min. CID-MS experiments were performed with collision energy of 30 V and 1.3 m/z isolation width.
NMR Analysis.
NMR was performed at the University of Georgia Complex Carbohydrate Research Center, as previously described (15). Samples were dissolved in 2:1 CDCl3-CD3OD followed by 1D proton NMR acquired on a Varian VNMRS 900 MHz spectrometer at 27 °C.
Acknowledgments
We thank C. Costello, C. Heiss, and P. Azadi for helpful discussions and J. Buter for assistance with figure production. This study was supported in part by NIH Grant AI102945 (to P.J.B.); the Violin and Karol families (P.J.B.); NIH Grants AR048632 and AI111224 (to D.B.M.); the Bill and Melinda Gates Foundation (D.B.M.); NIH Grants AI113046 and AI063428 (to M.B.B.); J. Badrick, the Royal Society, and the Medical Research Council (G.S.B.); National Health and Medical Research Council of Australia Grants 1013667, 1113293, 1020770, 1117766, and 1054431 (to D.I.G. and D.G.P.); Australian Research Council Grant CE140100011 (to D.I.G. and D.G.P.); and Australian Research Council Grants FL160100049 and CE140100011 (to J.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705882114/-/DCSupplemental.
References
- 1.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 2.Young MH, Gapin L. Group 1 CD1-restricted T cells take center stage. Eur J Immunol. 2011;41:592–594. doi: 10.1002/eji.201141408. [DOI] [PubMed] [Google Scholar]
- 3.Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15:643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 5.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 6.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 7.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 8.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 9.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 10.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12:966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieland Brown LC, et al. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian α-linked glycosylceramides. Immunity. 2014;41:543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan PJ, et al. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011;12:1202–1211. doi: 10.1038/ni.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan PJ, et al. Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci USA. 2014;111:13433–13438. doi: 10.1073/pnas.1415357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burdin N, et al. Selective ability of mouse CD1 to present glycolipids: Alpha-galactosylceramide specifically stimulates V alpha 14+ NK T lymphocytes. J Immunol. 1998;161:3271–3281. [PubMed] [Google Scholar]
- 17.Birkinshaw RW, et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol. 2015;16:258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardell S, et al. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel O, et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol. 2012;13:857–863. doi: 10.1038/ni.2372. [DOI] [PubMed] [Google Scholar]
- 20.Facciotti F, et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol. 2012;13:474–480. doi: 10.1038/ni.2245. [DOI] [PubMed] [Google Scholar]
- 21.Fox LM, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 23.Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wingender G, et al. Invariant NKT cells are required for airway inflammation induced by environmental antigens. J Exp Med. 2011;208:1151–1162. doi: 10.1084/jem.20102229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YJ, et al. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity. 2015;43:566–578. doi: 10.1016/j.immuni.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Rhijn I, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, et al. Crystal structures of bovine CD1d reveal altered αGalCer presentation and a restricted A′ pocket unable to bind long-chain glycolipids. PLoS One. 2012;7:e47989. doi: 10.1371/journal.pone.0047989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen TK, et al. The bovine CD1D gene has an unusual gene structure and is expressed but cannot present α-galactosylceramide with a C26 fatty acid. Int Immunol. 2013;25:91–98. doi: 10.1093/intimm/dxs092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellicci DG, et al. Intrathymic NKT cell development is blocked by the presence of alpha-galactosylceramide. Eur J Immunol. 2003;33:1816–1823. doi: 10.1002/eji.200323894. [DOI] [PubMed] [Google Scholar]
- 31.Kanie O, et al. Analysis of behavior of sodiated sugar hemiacetals under low-energy collision-induced dissociation conditions and application to investigating mutarotation and mechanism of a glycosidase. Proc Jpn Acad, Ser B, Phys Biol Sci. 2009;85:204–215. doi: 10.2183/pjab.85.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moody DB, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 33.Darmoise A, et al. Lysosomal alpha-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity. 2010;33:216–228. doi: 10.1016/j.immuni.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jervis PJ, et al. Synthesis and biological activity of alpha-glucosyl C24:0 and C20:2 ceramides. Bioorg Med Chem Lett. 2010;20:3475–3478. doi: 10.1016/j.bmcl.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uldrich AP, et al. CD1d-lipid antigen recognition by the γδ TCR. Nat Immunol. 2013;14:1137–1145. doi: 10.1038/ni.2713. [DOI] [PubMed] [Google Scholar]
- 36.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muindi K, et al. 2010. Activation state and intracellular trafficking contribute to the repertoire of endogenous glycosphingolipids presented by CD1d [corrected]. Proc Natl Acad Sci USA 107:3052–3057; Erratum in Proc Natl Acad Sci USA 107:6118.
- 40.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 41.Layre E, et al. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol. 2011;18:1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]