Significance
Complex I is a redox-driven proton pump, central to aerobic energy conversion in most living organisms. To elucidate the mechanism of its pumping machinery, we need a detailed molecular picture of how access across the membrane is established and regulated. In this work we find that proton pathways in complex I form at symmetry-related locations near broken transmembrane helices. The channel opening allows influx of water molecules, catalyzing rapid Grotthuss-type proton transfer reactions. The hydration of these channels is sensitive to the protonation state of conserved buried lysine residues, which are in turn coupled to conformational changes in conserved ion pairs within each subunit. Our results provide mechanistic insight into the function of the long-range proton-pumping machinery in complex I.
Keywords: NADH:ubiquinone oxidoreductase, proton pumping, Grotthuss mechanism, multiscale simulation, bioenergetics
Abstract
Complex I functions as the initial electron acceptor in aerobic respiratory chains of most organisms. This gigantic redox-driven enzyme employs the energy from quinone reduction to pump protons across its complete approximately 200-Å membrane domain, thermodynamically driving synthesis of ATP. Despite recently resolved structures from several species, the molecular mechanism by which complex I catalyzes this long-range proton-coupled electron transfer process, however, still remains unclear. We perform here large-scale classical and quantum molecular simulations to study the function of the proton pump in complex I from Thermus thermophilus. The simulations suggest that proton channels are established at symmetry-related locations in four subunits of the membrane domain. The channels open up by formation of quasi one-dimensional water chains that are sensitive to the protonation states of buried residues at structurally conserved broken helix elements. Our combined data provide mechanistic insight into long-range coupling effects and predictions for site-directed mutagenesis experiments.
Complex I (NADH:ubiquinone reductase) is the largest enzyme of the respiratory chain, generating a proton motive force (pmf) that is used for synthesis of adenosine triphosphate (ATP) and active transport (1, 2). Complex I catalyzes electron transfer (eT) between nicotine adenine dinucleotide (NADH) and quinone (Q), and couples the energy released to pumping of four protons across the membrane (3–9). The distance between the electron and proton transferring modules extends up to approximately 200 Å. It currently remains unclear, however, how complex I catalyzes this remarkable long-range proton-coupled electron transfer (PCET) process. In addition to its central role in biological energy conversion, elucidating the molecular mechanism of complex I is of great biomedical relevance due to dysfunction of this enzyme in about half of all mitochondrial disorders (4, 8, 10).
In recent years, several structures of complex I have been resolved from both bacteria (11–13) and eukaryotes (14–19). During the completion of this study, two new structures of the mammalian enzyme were released (18, 19), resolving at near atomic resolution the structure of the supernumerary subunits in active and deactive states, complementing information from previously resolved structures (17). Similar to other respiratory enzymes (20, 21), the inner part of complex I, comprising 14 central core subunits, is well conserved, suggesting that the overall proton-pumping process might be similar in the bacterial and eukaryotic enzymes (but also see refs. 17, 22–24).
Complex I consists of a hydrophilic domain, responsible for the eT activity, and a membrane domain, which catalyzes proton transfer (pT) across the membrane (Fig. 1A). In the hydrophilic domain, a chain of 8–9 iron–sulfur (FeS) centers connects the NADH/flavin mononucleotide (FMN) site with the Q site (11, 25, 26), which locates approximately 30 Å above the membrane surface (14). The FeS centers form an electron conduction wire, in which the eT takes place on a microsecond–millisecond timescale between the two ends of the hydrophilic domain (27, 28). The Q head group is located approximately 10 Å from the terminal N2 FeS center and interacts with the conserved His-38 and Tyr-87 residues of subunit Nqo4 (T. thermophilus nomenclature) (13, 17), as originally identified in biochemical studies (29). Recent computational work (30) suggests that reduction of Q is coupled to deprotonation of the His-38 and Tyr-87, leading to formation of QH2 that further triggers coupled protonation and conformational changes in the Nqo8 subunit, supporting previous suggestions from biochemical and structural data (6, 31, 32).
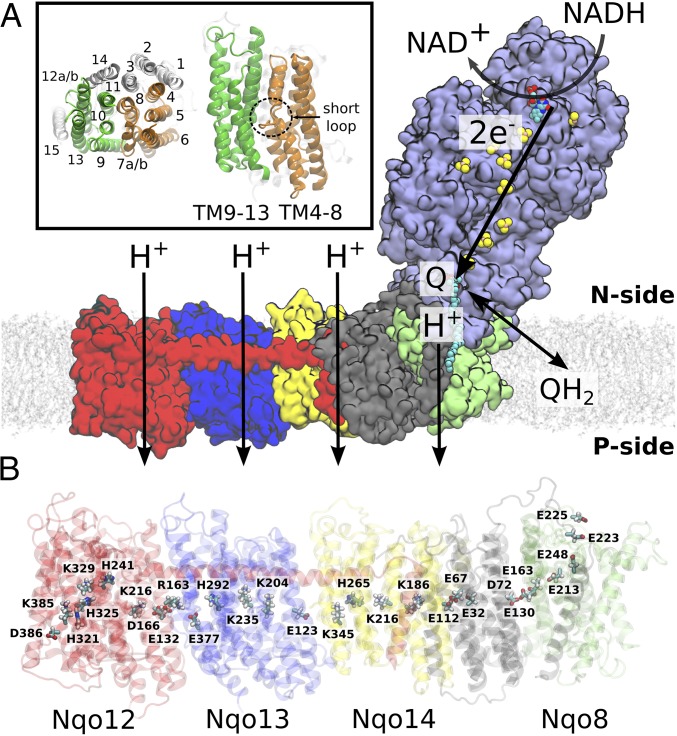
Fig. 1.
Overall structure and function of bacterial complex I from T. thermophilus (PDB ID: 4HEA). (A) Electron transfer from NADH to Q in the hydrophilic domain (in purple) couples to proton transfer across the membrane domain, shown in red (Nqo12), blue (Nqo13), yellow (Nqo14), gray (Nqo11, Nqo10, and Nqo7), and green (Nqo8). (Inset) Transmembrane (TM) helix numbering of the antiporter-like subunits. Symmetry-related transmembrane helices TM4–8 (in orange) and TM9–13 (in green) in Nqo13, and TM15 (in transparent white) from Nqo12. (B) Array of polar and charged residues on the central axis in the membrane domain.
The membrane domain is composed of seven subunits, Nqo7–8 and Nqo10–14 (NuoA/H/J/K/L/M/N in Escherichia coli), arranged in an approximately 180-Å array linked to the hydrophilic domain by subunit Nqo8 (Fig. 1B). This domain comprises three antiporter-like subunits, Nqo12 (NuoN), Nqo13 (NuoM), and Nqo14 (NuoL) that are homologous to each other and also to multiresistance and pH adaptation (Mrp) Na+/H+ antiporters (12). These subunits are most likely responsible for pumping three protons, whereas the fourth proton channel might reside in the Nqo8 subunit (13, 17).
Each of the antiporter-like subunits have a unique internal pseudosymmetry (12) involving a twofold screw axis and comprising five transmembrane (TM) helical bundle segments, TM4–8 and TM9–13 (Fig. 1A, Inset). TM7a/b and TM12a/b are broken by short loops (Fig. 1A, Inset), a motif that is also used in ion translocation in carrier-type transporters (33). Nqo8 contains the same five-helical TM segment, but in contrast to the Nqo12/13/14 subunits, it is not symmetrical with respect to the other helices of the subunit. Structural studies (12, 13, 17) also identified a chain of highly conserved buried charged/polar residues, spanning the entire membrane domain, and creating a connecting element between the Q site and Nqo12. Interestingly, this chain contains motifs that are common in each of the Nqo12–14 subunits: a lysine/glutamate pair, a central lysine, one or two histidine residues, and a terminal lysine (Nqo12 and Nqo14) or glutamate (Nqo13) (Fig. 1B). These residues may provide important functional elements in the proton-pumping machinery, because their mutation reduces the pumping activity (12, 13). Molecular simulations performed on the membrane domain of E. coli complex I (34) suggested that water molecules are involved in the formation of the proton pathways along these residues, but complete connections within all subunits were not resolved. The membrane domain of complex I also contains a long, approximately 110-Å, transverse horizontal helix (HL) from Nqo12 that spans along the domain and interacts with all three antiporter-like subunits. The helix is likely to have a clamping function and forms an element important for structure stability (35–37).
Here we explore the proton-pumping pathways in complex I from T. thermophilus (13) by large-scale classical molecular dynamics (MD) simulations on a microsecond timescale. To probe the proton transfer dynamics in the pumping process, we use a combination of MD simulations, hybrid quantum mechanics/classical mechanics (QM/MM) MD simulations, and electrostatic Poisson–Boltzmann/Monte Carlo calculations for sampling changes in protonation states. Our combined approach provides important suggestions on the pumping elements and long-range coupling effects in complex I that can further stimulate new experiments.
Results
General Channel Topology.
To probe the dynamics of complex I, we performed seven independent classical atomistic MD simulations, initiated from the X-ray structure of the enzyme from T. thermophilus (13). The protein structure was embedded in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane–water–ion surroundings and comprised in total approximately 1 million atoms. The buried conserved residues were modeled in different protonation states (SI Appendix, Tables S1 and S2), and the simulations extended in total 3.8 μs. In the MD simulations, we observe spontaneous hydration of the Nqo8 and Nqo12–14 subunits on a 0.2–0.4 μs timescale from the negatively charged (N) side of the membrane (Fig. 2 A and B and Movie S1). In Nqo12 and Nqo13, the water chains connect buried charged residues with the positively charged (P) side of the membrane, whereas for subunit Nqo14, we observe also a partial hydration from the P side of the membrane. The simulations suggest that the continuous water connectivity across the membrane is broken near highly conserved histidine residues, His-32512/29213/26514 within the horizontal array of charged/polar residues at the center of the membrane domain (see below and Fig. 2).
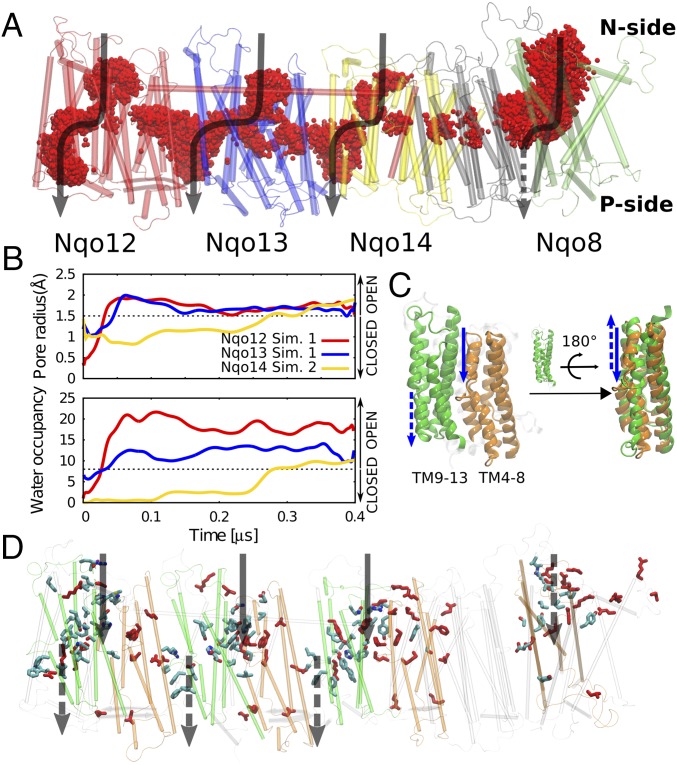
Fig. 2.
Proton channel topology in complex I. (A) The membrane domain of complex I, showing overlapping water molecules (in red) obtained from seven independent simulation setups (3 µs in total). The water channels for each antiporter-like subunit and Nqo8 are shown by black arrows. (B) Water occupancy and channel radius of proton channels from the N side of the membrane to the center buried residues for three antiporter-like subunits calculated from two independent MD simulations (simulations 1 and 2). (C) The internal symmetry of the water input pathway from the N side and output to the P side shown as solid and dotted arrows, respectively, mirrors the internal symmetry of the antiporter-like subunits. Symmetry-related transmembrane helices TM4–8 (in orange) and TM9–13 (in green). (D) Positions of water pathways in the membrane domain, overlapped with conserved residues along the channels. Experimentally characterized residues that affect the proton-pumping activity are shown in red. The input channels from the N side in Nqo8 and output to the P side in Nqo12/13/14 are on the backside of this view.
The water chains form quasi one-dimensional hydrogen-bonded arrays, typical for many proton channels (38–40), and the hydration process increases the channel radius (Fig. 2B). We observe hydrated contacts between the bulk solvent and buried conserved residues Lys-32912/Lys-23513/Tyr-28814 and Lys-38512/Glu-37713/Lys-34514, along the N and P pathways, respectively. These pathways reflect the pseudo twofold internal symmetry of the subunits (Fig. 2C), with water arrays formed at similar locations on each side of the broken helices TM7b/12b (SI Appendix, Fig. S1). We observe that the broken helices undergo conformational changes in the opening/closure of the channels that may be important in controlling the hydration process (SI Appendix, Fig. S2). Due to their dynamic flexibility, broken helix elements also play an important function in carrier-type transporters (41, 42) where they are involved in establishing an alternate access between the two sides of the membrane. In carrier-type transporters, however, the channels often form along linear pathways at a dimeric protein interface, whereas in complex I the channels seem to form at the edge of every half subunit along the domain (Fig. 2D), consistent with their symmetry.
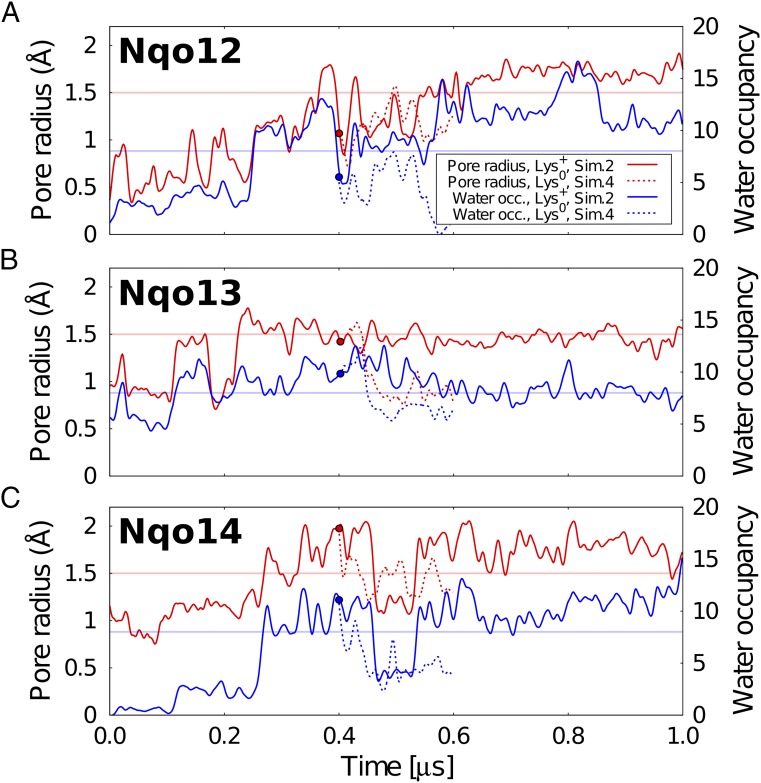
We find that the increase in channel radius in all antiporter-like subunits correlates well with the water occupancy in the channel (SI Appendix, Figs. S3 and S4). The water wires form spontaneously in many independent simulations (simulations 1–3 and 5–7) when the residues located at the end of the channels are in their charged protonation states, suggesting that the overall behavior is statistically significant. Interestingly, upon deprotonation of the central lysine residues Lys-32912/Lys-23513/Lys-21614 located at the end of the N-side pathway (simulation 4), we observe a rapid dehydration of the N-side channels in <100 ns (Fig. 3). This observation suggests that the protonation state of the central buried residues might control the water connectivity across the membrane domain.
Fig. 3.
Pore radius and water occupancy of N-side proton channels in Nqo12–14. The pore radius (in red) and water occupancy (in blue) are shown for (A) Nqo12, (B) Nqo13, and (C) Nqo14 in two different protonation states of the middle lysine (simulations 2 and 4, SI Appendix, Table S2). Circle at 0.4 µs shows simulation 4, started after 0.4 µs of simulation 2 by deprotonating the middle Lys-32912/Lys-23513/Lys-21614. The channel opening threshold was defined based on the pore radius of 1.5 Å, which allows influx of water molecules. Details for simulations 1–7 are shown in SI Appendix.
Based on structural data (12, 13) and site-directed mutagenesis experiments (43–48), the N-side proton channels in Nqo12–14 have previously been suggested to reside either near the Glu/Lys pair within each antiporter-like subunit or at the middle Lys around TM8 (9). The putative channel topology observed in our MD simulations somewhat differs from these sites and resembles more the paths suggested for Yarrowia lipolytica (17), with an entry site near TM7b in the middle of the two bundle segments TM4–8 and TM9–13 and output site near TM12b. Moreover, the Glu/Lys pairs, which were previously suggested to take part in the pumping process (12), are not located along the N-side input channels, and therefore the direct involvement of these residues in the proton uptake seems unlikely, based on the current simulation data. The dissociation of these residue pairs might, nevertheless, be important for propagating electrostatic forces along the central axis of conserved residues (see below).
In Nqo8, we observe hydration along the broken helix TM5 within the TM2–6 bundle, although this subunit has a somewhat different structure and sequence compared with the other antiporter-like subunits Nqo12/13/14 (13). These findings support that a fourth proton pathway may indeed reside within Nqo8, as previously suggested by Baradaran et al. (13).
Our putative channel locations thus coincide with the position of several conserved and functionally important residues, identified in previous biochemical studies (Fig. 2D and SI Appendix, Table S3). Importantly, our putative channels also comprise several residues that have not yet been experimentally probed. To this end, we have summarized central residues along the N- and P-side pathways in SI Appendix, Table S3. In the following sections, we present structural details about the putative proton pathway in each subunit.
Proton Pathways in Nqo12 (NuoL).
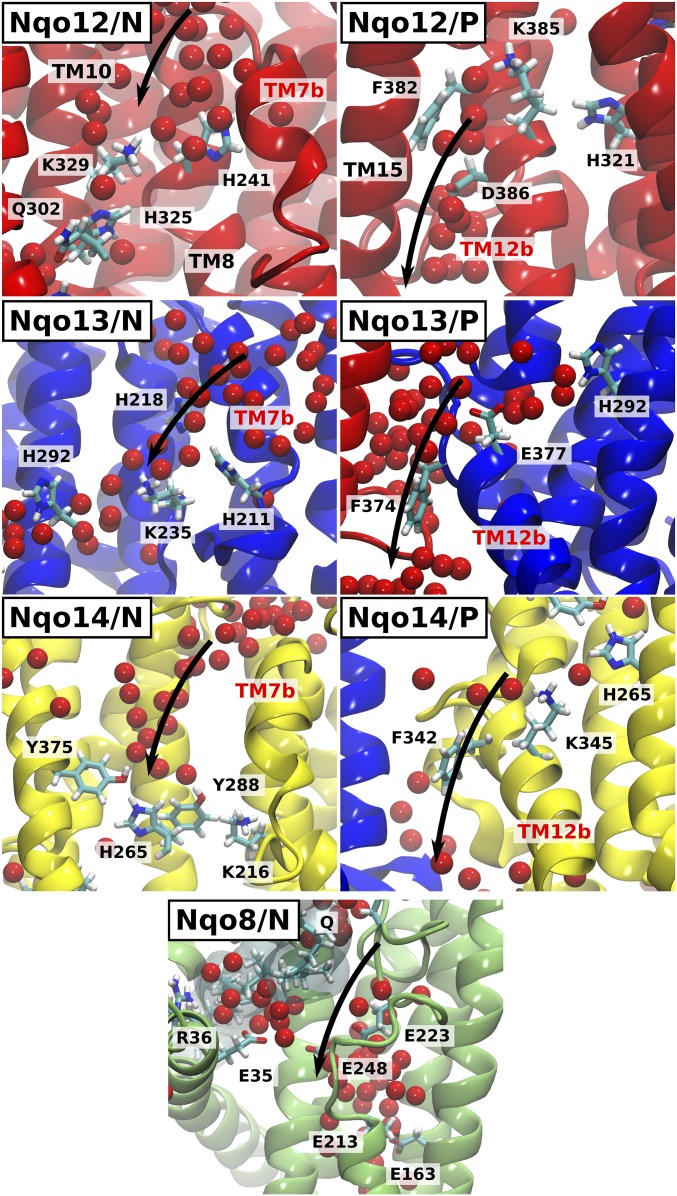
Water molecules enter the Nqo12 subunit (simulations 1, 2, and 5–7) by a crevice between the helices TM7b, TM8, and TM10. This conduit provides a hydrogen-bonded connectivity between the bulk and the conserved buried residue Lys-329, which further connects via His-325 and His-321 to Lys-385/Asp-386 (Fig. 4, Nqo12/N). The channel from the P side of the membrane becomes hydrated in most simulations (SI Appendix, Fig. S4), and forms around the tip of the broken helix TM12b, near Lys-385 and Asp-386 (Fig. 4, Nqo12/P). This pathway lines up with TM15, close to the edge of the membrane subunit.
Fig. 4.
Proton pathways from the N and P sides of the membrane in complex I. Water molecules enter between TM helices 7b, 8, and 10 (Nqo12–14) and TM5 and TM6 (Nqo8) from the N-side bulk and establish a conduction pathway to the buried residues Lys-32912/Lys-23513/Tyr-28814/Glu-1638. Nqo12/P, Nqo13/P, Nqo14/P: Water pathways next to TM12b lead from Lys-38512/Glu-37713/Lys-34514 to the P side of the membrane. Some helices are not shown for visualization purpose.
Although the two water pathways from the N and P sides come in close contact near Lys-329, Phe-328 and Leu-373 tightly seal the channel, and do not allow water exchange between the two sides of the membrane (SI Appendix, Fig. S5). A dry area is also observed in the proximity of Asp-386, suggesting that this residue might control the access to the P side of the membrane. The simulations show that the conserved Gln-302 of TM10 dynamically switches its hydrogen-bonding contacts between Lys-329 and His-321 (SI Appendix, Fig. S6). Although the glutamine cannot undergo protonation changes itself, its side chain can act as a hydrogen bond donor/acceptor and thus stabilize protonated intermediates in the transfer process.
Proton Pathways in Nqo13 (NuoM).
The MD simulations suggest that movement of the broken helix TM7b on an approximately 0.1-μs timescale (simulations 1, 2, 6, and 7) opens a pathway for water molecules to enter Nqo13 from the N side of the membrane (Fig. 4, Nqo13/N). This channel leads to the buried conserved Lys-235 (TM8), as also previously suggested as a potential input channel in the E. coli structure (34). Connection to the P side forms at the interface between Nqo12 (TM5) and Nqo13 (TM12b) (simulation 6) from Glu-377 (Fig. 4, Nqo13/P). Formation of this hydrogen-bonded network to the P side is triggered by a conformational change in Phe-374, a residue that is conserved in all antiporter-like subunits of complex I.
During the opening of the N-side channel, we observe a dissociation of a backbone hydrogen bond between His-211 and Leu-214 (SI Appendix, Fig. S7). To further probe the involvement of these residues, we performed steered molecular dynamics (SMD) simulations in which this hydrogen bond was broken. These simulations suggest that the channel opening/closure can indeed be induced by perturbation of this reaction coordinate (SI Appendix, Fig. S7). Our unbiased MD simulations also indicate that when the N-side channel is closed (simulation 5), water molecules establish a bridging pathway between Lys-235 and Glu-377 via His-292, providing a conduction pathway toward the P side of the membrane (see below).
The channel from the N side passes close to Asp-557 (Nqo12), which is a conserved residue on the transverse HL helix. Although site-directed mutagenesis data (35) suggest that the HL helix might provide a clamping function in complex I rather than the earlier suggested piston function (12), the mutation of this aspartate to a glutamate strongly decreases the proton-pumping activity by approximately 50% (49).
Proton Pathways in Nqo14 (NuoN).
The Nqo14 subunit undergoes the slowest hydration process in our MD simulations on an approximately 0.3-μs timescale (Fig. 2B and SI Appendix, Fig. S3). We find that water molecules enter this subunit through a crevice between TM7b, TM8, and TM10 (Fig. 4, Nqo14/N) leading to the conserved Tyr-288 and Lys-216 residues. This pathway thus forms at locations that closely resemble the channels observed in Nqo12. Similar to the P-side channel formed in Nqo13, we also observe a partial hydration at the interface between Nqo13 and Nqo14 (Fig. 4, Nqo14/P), but Phe-342 seems to partially block the pathway between the Lys-345 and the P side. The channel from the N side in Nqo14 is closed in the crystal structure by the bulky side chain of Leu-258, but its side-chain movement correlates with formation/disruption of the water wire (SI Appendix, Fig. S8), suggesting that this residue might have a potential gating function. In previous simulations of the E. coli structure (34), a channel leading to the middle Tyr-288/Lys-216 pair was also observed.
We do not observe substantial hydration in Nqo14 at the symmetry-related position where the channel forms in Nqo13. In contrast, this analogous site in Nqo14 is covered by the C-terminal loop of Nqo11 (SI Appendix, Fig. S9) that was recently suggested to form an element important for structural stability based on mutagenesis and cross-linking studies in E. coli (50). Moreover, in contrast to the water channel in Nqo13, where the HL helix inserts Asp-55712 near His-211, His-193 in Nqo14 lacks such an interaction partner with the transverse HL helix.
Proton Pathways in Nqo8 (NuoH).
We observe in all independent MD simulations that the Nqo8 subunit undergoes large hydration changes on approximately 0.1-μs timescales (SI Appendix, Fig. S3). The Nqo8 channel contains many charged and polar residues, and it is also wider compared with the hydration sites in the other subunits. The hydration extends from the N side to Glu-213/163 within the so-called E channel (13), located approximately 15 Å below the membrane surface. This region was previously suggested to undergo proton uptake induced by Q reduction (30). Our simulations suggest this hydrated channel coincides with the lower part of the quinone cavity (Fig. 4, Nqo8/N), supporting that proton uptake/release might couple to movement of within its tunnel. Despite a significant N-side entry channel, we observe no clear exit pathways to the P side of the membrane within Nqo8.
Proton Transfer Dynamics in Nqo13.
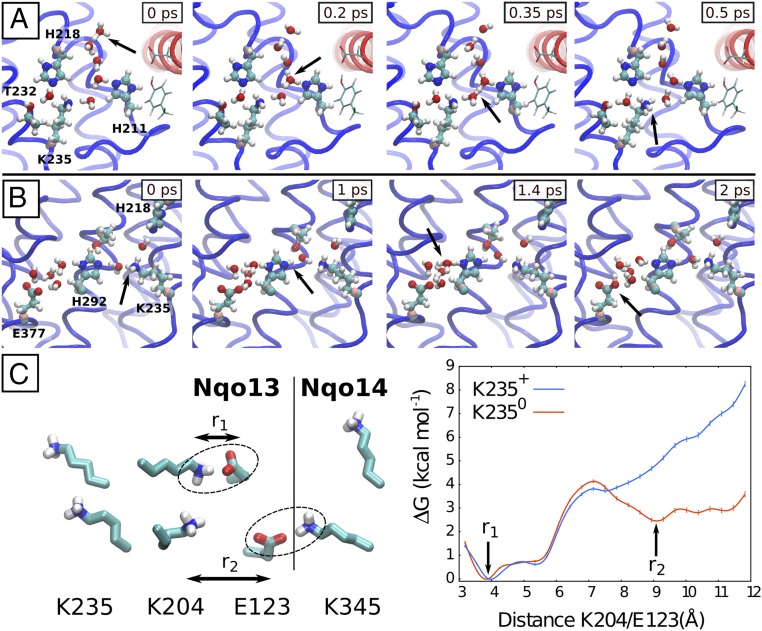
To probe the mechanism of proton transfer along the formed water channels, we performed QM/MM MD simulations of two putative steps of the proton-pumping cycle in the Nqo13 subunit. To this end, we studied the proton uptake from the bulk to the buried Lys-235 residue, and then further along the lateral water pathway from Lys-235 to Glu-377 (Fig. 5 A and B and SI Appendix, Table S4). To study the proton transfer from the solvent to Lys-235, we used a starting geometry obtained after a 0.1-μs classical MD simulation in which the channel from the N side was well hydrated (simulation 6) and Lys-235 was neutralized. We observe a rapid transfer of the excess proton placed at the entrance of the channel (N side) to the deprotonated Lys-235 (Fig. 5A). The transfer process takes place by a Grotthuss-type proton exchange along Zundel and hydronium species, typical for proton transfer in quasi one-dimensional water wires (40, 51) and subsequent rearrangement of the hydrogen bonded network (52). His-211 does not seem to directly take part in the transfer process, but stabilizes the water network and the proton in the cavity (Movie S2). To further probe the dependence of this proton transfer process on the channel hydration state, we performed several independent QM/MM simulations (320 ps in total), starting from different starting structures along the hydration trajectory. We find that, whereas the proton transfer takes place on the picosecond timescale in the simulations that are initiated from well-hydrated structures, the simulations initiated from dry or broken hydrogen-bonded networks have a lower probability for productive proton transfer reactions to the lysine on the sampled simulation timescale (SI Appendix, Fig. S10 and Table S5). This behavior suggests that, whereas there is a large thermodynamic driving force for the proton to transfer from the hydronium to the deprotonated lysine, the slow (microsecond) channel hydration might be rate limiting in the proton uptake process. Interestingly, for some partially hydrated structures, we also find that the hydronium species does itself participate in establishing a water wire toward the central lysine. Similar proton transfer mechanisms have been previously observed in carbon nanotubes (51).
Fig. 5.
Proton transfer dynamics and conformational switching in Nqo13. (A) A water chain from the N side, formed in the MD simulations (simulation 6), catalyzes Grotthuss-type proton transfer to Lys-235 in Nqo13 in QM/MM MD simulations. (B) Proton transfer from Lys-235 to Glu-377. The simulation was initiated from a state in which the channel from the N side is closed and the residues Lys-235 and Glu-377 are protonated and deprotonated, respectively (simulation 5). His-292 bridges the proton transfer between the K235 and E377. (C) Classical free-energy profiles of Lys-204/Glu-123 (Nqo13) dissociation when Lys-235 is protonated (in blue) and deprotonated (in red). Glu-123 interacts both with Lys-204 and Lys-345 (Nqo14). The second minimum is absent when Lys-235 is protonated, possibly due to electrostatic repulsion with Lys-204.
In another independent QM/MM MD simulation setup, we studied the proton transfer along the Lys-235/His-292/Glu-377 triad to further elucidate the role of Lys-235 in the proton release. Starting from a structure with the N- and P-side channels closed, the proton is transferred rapidly in three independent simulations from Lys-235 via His-292 and water molecules to Glu-377 (Fig. 5B and Movie S3). This result supports the idea that the proton transfer along the central polar axis is indeed possible. Our continuum electrostatic calculations on the crystal structure also suggest that the pKa of Glu-377 is higher than the pKa of Lys-235 (SI Appendix, Table S6), further supporting this hypothesis. The QM/MM MD simulations thus support the idea that the water chains formed in the classical MD simulations provide efficient conduits for proton transfer in the membrane domain of complex I.
The Lysine/Glutamate Pair as an Element for Modulating the Proton Pump.
The MD simulations suggest that the protonation state of the buried residues could regulate the channel hydration (Fig. 3 and SI Appendix, Figs. S3 and S4). More specifically, the N-side access channel in Nqo13 is open when Lys-235 is deprotonated and His-211 is modeled in its protonated state (simulation 1) or when Lys-235 is protonated and His-211 is neutral (δ-tautormer, simulations 6 and 7). In contrast, the channel is significantly less hydrated in simulations where both Lys-235 and His-211 are in their neutral states (simulation 4), suggesting that the protonation states of these residues play a role in the channel opening.
To study how the dissociation of the Lys/Glu pair couples to the protonation state of the middle Lys-235, we performed free-energy calculations using the replica exchange umbrella sampling (REUS) method (53), where we biased the dissociation of Lys-204/Glu-123 in Nqo13 and modeled Lys-235 in both its protonated and neutral forms. The free-energy simulations show a clear preference of the Lys/Glu pair to be associated when Lys-235 is protonated (Fig. 5C). Interestingly, we observe a second minimum in the free-energy profile when Lys-235 is deprotonated (Fig. 5C), in which Glu-123 forms an ion pair with Lys-345 of Nqo14. For the protonated Lys-235, this minimum is absent, possibly due to the repulsion between the positively charged residues. Based on the principle of microscopic reversibility, this difference indicates that dissociation of the Lys-204/Glu-123 ion pair could increase the deprotonation probability of the central Lys-235, and possibly triggering lateral proton transfer toward the P side.
To further explore this coupling effect in all three antiporter-like subunits, we performed SMD simulations where the dissociation of the Lys+/Glu− (Asp− in Nqo12) pair was induced (Methods) in the three antiporter-like subunits, followed by pKa calculations. These calculations suggest that the pKa of the middle lysines (Lys-32912, Lys-23513, and Lys-26514) are affected by the dissociation of the ion pairs (SI Appendix, Fig. S11), further supporting the findings from the REUS simulations. Benchmarking studies suggest that PB electrostatic calculations with MC sampling have a rmsd of approximately 1 pK unit from experimental values (54). These pKa benchmarking studies, however, do not include large membrane proteins such as complex I, which might be subjected to larger errors.
Discussion
Our combined results from classical MD simulations, QM/MM calculations, and PB electrostatic models suggest that proton pumping in complex I takes place by hydration of proton channels that form along conserved broken helices, TM7b and TM12b in Nqo12-14, and TM5 in Nqo8, at symmetry-related locations. The hydration is sensitive to the protonation state of buried residues, with charged and neutral states inducing channel opening and closure, respectively. Our simulations suggest that the electrostatic field induced by these buried residues pull in water molecules from the solvent, and the quasi one-dimensionally oriented chains might further strengthen the electrostatic couplings between the residues. Our free-energy and steered molecular simulations further suggest that the dynamics of the conserved Glu/Lys ion pair within TM5/7a modulate the pKa of the buried central residues. These results indicate that complex I may operate within each antiporter-like subunit by using a unit dedicated for signal transmission (TM4–8), and another unit responsible for the proton-pumping activity (TM9–13). The lateral proton transfer within the TM9–13, as explored in our QM/MM simulations, leads to protonation of the terminal protonatable residue (Lys12/14/Glu13), and could in turn modulate the dynamics of the signal transduction unit in the neighboring subunit, as suggested by the MD data where intersubunit contacts are formed (SI Appendix, Fig. S12).
Although the X-ray structure of complex I from T. thermophilus represents a single state of limited resolution (3.3 Å), it nevertheless provides a good starting point to probe the dynamics of transient states along the pumping cycle. We observe similar hydration dynamics in several independent simulations, supporting the idea that the overall findings are robust and statistically significant. Moreover, although the timescales of the individual trajectories are shorter than the overall millisecond turnover of complex I, computations performed on transient states provide valuable information on the structure and dynamics of the pumping cycle and information that is complementary to that obtained from many experiments. Further work is, nevertheless, needed to elucidate the exact role of the interplay between the protonation states of the central Lys and/or His residues as well as the involvement of the Lys/Glu ion pairs in modulating the hydration and proton transfer energetics.
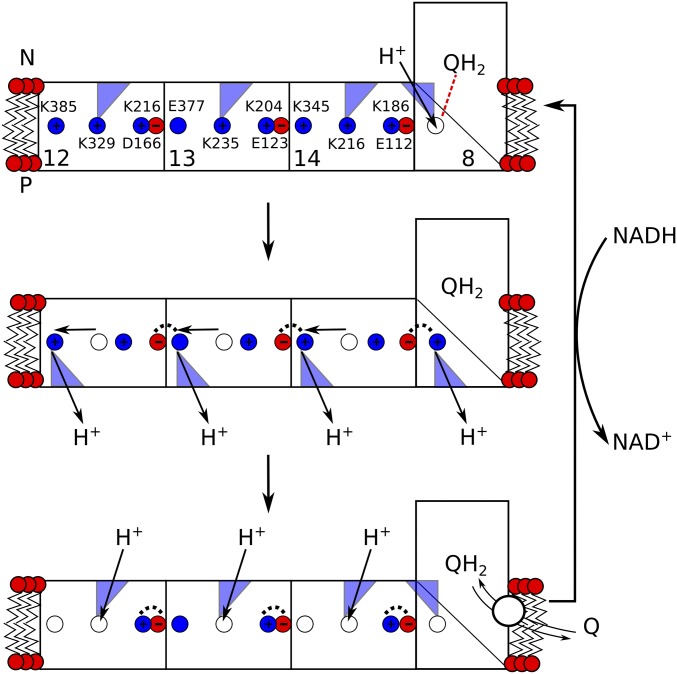
Our findings provide mechanistic ideas into how the force might propagate from the Q site across the antiporter-like subunits, schematically outlined in Fig. 6. In this putative model, the pumping is initiated by reduction of quinone that triggers protonation changes in Nqo8 (30) along water molecules near TM5 (Fig. 4, Nqo8/N). The protonation change further triggers dissociation of the Lys/Glu pair in Nqo14, which in turn could initiate proton transfer and release to the P side along the pathway near TM12b. Deprotonation of the middle lysine in Nqo14 is expected to close the connectivity to the N side (Fig. 3), and could help to prevent the pumped proton from leaking to the N side. Protonation changes in the Nqo14 would then induce dissociation of the Lys/Glu ion pair in Nqo13, which in turn would propagate by similar steps to Nqo12. Relaxation of the Lys/Glu ion pairs reestablishes the original protonation states by protonation from the N side along the TM7b pathways, whereas quinone/quinol exchange and rereduction by NADH initiates a new pumping cycle.
Fig. 6.
Schematic outline of a putative pumping cycle in complex I. (Top) Reduction of Q induces proton uptake in Nqo8 that leads to sequential opening of the K+/E− ion pairs in each subunit. (Middle) The K+/E− opening triggers proton transfer along the later hydrophilic axis and release of the proton to the P side. The open K+/E− ion pairs are stabilized by interactions with neighboring subunits (dashed lines). Deprotonation of the middle buried residues prevents the pumped proton from leaking back to the N side by blocking water connectivity. (Bottom) QH2/Q exchange and relaxation of the K+/E− ion pairs opens up water access from the N side, reforming the initial protonation states. Reduction of Q by NADH reinitiates a new pumping cycle.
This schematic model favors a sequential propagation of the electrostatic force from the Q site along Nqo8 to Nqo14, and further from Nqo14 to Nqo13 and Nqo12. Local electrostatic coupling between neighboring charged residues is strong, which could also allow pumping without large-scale conformational changes within the antiporter-like subunits. Other mechanistic models, such as the wave-spring model (7, 45), have also been presented, in which Nqo8/Nqo13 and Nqo12/Nqo14 pump and release protons during synchronized steps. It is currently unclear, however, whether complex I operates by direct (electrostatic) or indirect (conformational) coupling (6, 55, 56) using one- (22, 27, 57) or two-stroke (31) mechanisms. Our simulations nevertheless support the idea that both electrostatic coupling between titratable residues as well as conformational changes in broken helices might be important for the pumping function in complex I.
Conclusions
We have performed here multiscale classical and quantum molecular simulations based on the experimentally resolved structure of complex I from T. thermophilus to probe how protonation and conformational states of the protein are linked to the proton-pumping process. We observed that hydrated channels form at symmetry-related locations, near TM7b and TM12b in Nqo12–14 and TM5b in subunit Nqo8, mirroring the internal pseudo twofold screw axis of the antiporter-like subunits in complex I. The channels to the N and P sides initiate from buried charged residues, and the protonation states of these residues themselves were found to correlate well with the hydration states of the channels as well as with conformational changes in TM7b/12b. Our simulations suggested that the electrostatic coupling effects are important for the proton-pumping process. We also found that the conformation of the conserved Glu/Lys ion pairs might modulate the pKa of the central buried residues, with important mechanistic implications. Our QM/MM MD simulations further suggested that the proton transfer is rapid, taking place on picosecond timescales, after the water wires have formed on microsecond timescales. The simulations also support a previously postulated lateral proton transfer along the polar buried axis in states where the channel to the N side is closed. These combined results may stimulate new site-directed mutagenesis experiments (SI Appendix, Table S3) and provide insights into the remarkable long-range proton-coupled electron transfer machinery in complex I.
Methods
Classical MD Simulations.
The X-ray structure from T. thermophilus (PDB ID: 4HEA) (13) was embedded in a POPC membrane and solvated with TIP3P water. Sodium and chloride ions were added to give an ionic strength of approximately 100 mM. The Q site was modeled by inserting ubiquinone (Q10) in the cavity, identified using the software HOLE (58), and by placing the Q head group next to His-38 and Tyr-87. The total system size comprised approximately 830,000 atoms. Seven independent simulation systems, each studied for 0.1–1 μs, were created to probe the effect of different protonation states (SI Appendix, Table S2). All classical MD simulations were performed using NAMD2 (59) with the CHARMM27 (60, 61) force field, and density functional theory (DFT)-derived parameters for all cofactors (FeS centers, Q, FMN) (30). The simulations were performed in an NPT ensemble with T = 310 K, P = 1 atm, using a time step of 2 fs, and treating long-range electrostatics by the Particle Mesh Ewald (PME) method. Water occupations were calculated from data averaged over the MD trajectories by selecting water molecules from residues shown in SI Appendix, Table S4. Pore radii were calculated using the software HOLE (58). SMD simulations were performed based on structures obtained after dynamics (SI Appendix, Table S1).
A 20-ns SMD simulation was performed by perturbing the backbone His-211(O)/Leu-214(N) atom distances between 3 and 6 Å using a force constant of 50 kcal mol−1 Å-2 to probe channel hydration in Nqo13 (SI Appendix, Fig. S7). The 3 × 16 ns simulations were performed by perturbing the head groups distances of Lys-216 (Nζ)/Asp-166 (Cγ) in Nqo12, Lys-204 (Nζ)/Glu-123 (Cδ) in Nqo13, and Lys-186 (Nζ)/Glu-112 (Cδ) in Nqo14 between 3 and 12 Å, using force constants of 50 kcal mol−1 Å-2. REUS simulations (53) (simulations 12 and 13, SI Appendix, Table S1) were performed using 25 replicas started from the SMD structures (simulation 10, SI Appendix, Table S1), modeling Lys-235 both in its neutral and charged states. A harmonic biasing potential with a force constant of 10 kcal mol-1 Å-2 was used to restrain the distance of Lys-204 (Nζ) and Glu-123 (Cδ) head groups between 3 and 12 Å. Exchange moves were attempted every 2 ps, and distances were collected every 20 ps. The free-energy profiles converged after 14 × 25 ns REUS simulations and were reconstructed using the weighted-histogram analysis method (WHAM) (62, 63) and estimating errors by bootstrapping analysis. Visual Molecular Dynamics (64) was used for visualization and analysis of all MD simulations. The total MD simulation time was approximately 3.8 μs.
Poisson–Boltzmann Continuum Electrostatics with Monte Carlo Sampling.
pKa values were calculated using PB continuum electrostatic calculations with MC sampling of 2N protonation states of a nine subunit model of complex I, comprising the membrane domain and subunits Nqo4/6. The PB calculations were performed using the Adaptive Poisson–Boltzmann Solver (APBS) (65), and the MC sampling was performed using Karlsberg+ (54, 66). The protein was described using explicit atoms with atomic partial charges, embedded in an inhomogeneous dielectric continuum with a dielectric constant of 4. The bulk water was described by a homogeneous dielectric continuum with a dielectric constant of 80. The boundary interface between the protein and solvent was calculated by the molecular surface routine implemented in APBS, using a solvent probe radius of 1.4 Å, and modeling an implicit ionic strength with 100 mM potassium chloride. Protonation probabilities were probed along all classical simulations every 0.1 ns. The Lys/Glu (Asp) pairs of each antiporter-like subunit were fixed in their standard protonation states in the pKa calculations of the SMD trajectories.
QM/MM MD Simulations.
Proton transfer from the N side to Lys-23513.
Setup 1 was as follows: QM/MM MD simulations were performed based on eight independent structures obtained from simulation 6. A water molecule at the channel entrance was replaced with a hydronium ion and classically relaxed for 10 ps for each of the eight starting structures. A total of 56 independent QM/MM MD simulations were initiated from these relaxed structures with different levels of water chain connectives between the N side and Lys-235. Lys-325, His-211, His-218, and Thr-232 together with the water molecules in the cavity were treated at the DFT level using the BP86 functional (67, 68), the multipole accelerated resolution of identity (MARIJ) approximation (69), and def2-SVP basis sets (70). The QM region comprised approximately 75 atoms and was coupled to the MM region by link atoms, introduced between the Cα and Cβ atoms. Additional eight QM/MM systems comprising 125 atoms were used to model the systems with broken water connectives from the N side (SI Appendix, Table S4). The MM system comprised the Nuo12/13/14 subunits and surrounding lipids and water molecules, with a total system size of approximately 75,000 atoms. The QM/MM MD simulations were simulated for 5 ps each, using a 1-fs integration time step, and a temperature of T = 310 K. The simulations were performed using the CHARMM/TURBOMOLE interface (71–73).
Proton transfer from Lys-23513 to Glu-37713.
Setup 2 was as follows: Structures used for the QM/MM setup 2 were taken from simulation 5, after 0.6 μs of dynamics. Residues His-218, Lys-235, His-292, Thr-322, and Glu-377 together with six bridging water molecules were included in the QM region, comprising 78 atoms, and treated as described above. Three independent QM/MM MD simulations were simulated for 5 ps each, using a 1-fs integration time step at a temperature of T = 310 K.
Supplementary Material
Acknowledgments
We thank Prof. Mårten Wikström and Dr. Gerhard Hummer for insightful discussions. This work was supported by the German Research Foundation. The computing time was provided by the SuperMuc at the Leibniz Rechenzentrum (Computing Grant pr48de).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706278114/-/DCSupplemental.
References
- 1.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida M, Muneyuki E, Hisabori T. ATP synthase: A marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 3.Wikström M. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 1984;169:300–304. doi: 10.1016/0014-5793(84)80338-5. [DOI] [PubMed] [Google Scholar]
- 4.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 5.Dröse S, et al. Functional dissection of the proton pumping modules of mitochondrial complex I. PLoS Biol. 2011;9:e1001128. doi: 10.1371/journal.pbio.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wikström M, Hummer G. Stoichiometry of proton translocation by respiratory complex I and its mechanistic implications. Proc Natl Acad Sci USA. 2012;109:4431–4436. doi: 10.1073/pnas.1120949109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verkhovskaya M, Bloch DA. Energy-converting respiratory Complex I: On the way to the molecular mechanism of the proton pump. Int J Biochem Cell Biol. 2013;45:491–511. doi: 10.1016/j.biocel.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 9.Sazanov LA. A giant molecular proton pump: Structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol. 2015;16:375–388. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 10.Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 12.Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 13.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 15.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirth C, Brandt U, Hunte C, Zickermann V. Structure and function of mitochondrial complex I. Biochim Biophys Acta. 2016;1857:902–914. doi: 10.1016/j.bbabio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Zickermann V, et al. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 18.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaila VRI, Verkhovsky MI, Wikström M. Proton-coupled electron transfer in cytochrome oxidase. Chem Rev. 2010;110:7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 21.Wikström M, Sharma V, Kaila VRI, Hosler JP, Hummer G. New perspectives on proton pumping in cellular respiration. Chem Rev. 2015;115:2196–2221. doi: 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- 22.Efremov RG, Sazanov LA. The coupling mechanism of respiratory complex I: A structural and evolutionary perspective. Biochim Biophys Acta. 2012;1817:1785–1795. doi: 10.1016/j.bbabio.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Kotlyar AB, Vinogradov AD. Slow active/inactive transition of the mitochondrial NADH-ubiquinone reductase. Biochim Biophys Acta. 1990;1019:151–158. doi: 10.1016/0005-2728(90)90137-s. [DOI] [PubMed] [Google Scholar]
- 24.Maklashina E, Kotlyar AB, Cecchini G. Active/de-active transition of respiratory complex I in bacteria, fungi, and animals. Biochim Biophys Acta. 2003;1606:95–103. doi: 10.1016/s0005-2728(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi T. Iron-sulfur clusters/semiquinones in complex I. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 26.Hinchliffe P, Sazanov LA. Organization of iron-sulfur clusters in respiratory complex I. Science. 2005;309:771–774. doi: 10.1126/science.1113988. [DOI] [PubMed] [Google Scholar]
- 27.Verkhovskaya ML, Belevich N, Euro L, Wikström M, Verkhovsky MI. Real-time electron transfer in respiratory complex I. Proc Natl Acad Sci USA. 2008;105:3763–3767. doi: 10.1073/pnas.0711249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Vries S, Dörner K, Strampraad MJ, Friedrich T. Electron tunneling rates in respiratory complex I are tuned for efficient energy conversion. Angew Chem Int Ed Engl. 2015;54:2844–2848. doi: 10.1002/anie.201410967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tocilescu MA, et al. The role of a conserved tyrosine in the 49-kDa subunit of complex I for ubiquinone binding and reduction. Biochim Biophys Acta. 2010;1797:625–632. doi: 10.1016/j.bbabio.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 30.Sharma V, et al. Redox-induced activation of the proton pump in the respiratory complex I. Proc Natl Acad Sci USA. 2015;112:11571–11576. doi: 10.1073/pnas.1503761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandt U. A two-state stabilization-change mechanism for proton-pumping complex I. Biochim Biophys Acta. 2011;1807:1364–1369. doi: 10.1016/j.bbabio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Berrisford JM, Sazanov LA. Structural basis for the mechanism of respiratory complex I. J Biol Chem. 2009;284:29773–29783. doi: 10.1074/jbc.M109.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Screpanti E, Hunte C. Discontinuous membrane helices in transport proteins and their correlation with function. J Struct Biol. 2007;159:261–267. doi: 10.1016/j.jsb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Kaila VRI, Wikström M, Hummer G. Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I. Proc Natl Acad Sci USA. 2014;111:6988–6993. doi: 10.1073/pnas.1319156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belevich G, Knuuti J, Verkhovsky MI, Wikström M, Verkhovskaya M. Probing the mechanistic role of the long α-helix in subunit L of respiratory Complex I from Escherichia coli by site-directed mutagenesis. Mol Microbiol. 2011;82:1086–1095. doi: 10.1111/j.1365-2958.2011.07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres-Bacete J, Sinha PK, Matsuno-Yagi A, Yagi T. Structural contribution of C-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of complex I from Escherichia coli. J Biol Chem. 2011;286:34007–34014. doi: 10.1074/jbc.M111.260968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu S, Vik SB. Constraining the lateral helix or respiratory complex I by cross-linking does not impair enzyme activity or proton translocation. J Biol Chem. 2015;290:20761–20773. doi: 10.1074/jbc.M115.660381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pomès R, Roux B. Structure and dynamics of a proton wire: a theoretical study of H+ translocation along the single-file water chain in the gramicidin A channel. Biophys J. 1996;71:19–39. doi: 10.1016/S0006-3495(96)79211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hummer G, Rasaiah JC, Noworyta JP. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 40.Kaila VRI, Hummer G. Energetics and dynamics of proton transfer reactions along short water wires. Phys Chem Chem Phys. 2011;13:13207–13215. doi: 10.1039/c1cp21112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 42.Forrest LR, et al. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci USA. 2008;105:10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamaru-Ogiso E, et al. The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of Escherichia coli complex I. J Biol Chem. 2010;285:39070–39078. doi: 10.1074/jbc.M110.157826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amarneh B, Vik SB. Mutagenesis of subunit N of the Escherichia coli complex I. Identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry. 2003;42:4800–4808. doi: 10.1021/bi0340346. [DOI] [PubMed] [Google Scholar]
- 45.Euro L, Belevich G, Verkhovsky MI, Wikström M, Verkhovskaya M. Conserved lysine residues of the membrane subunit NuoM are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase (Complex I) Biochim Biophys Acta. 2008;1777:1166–1172. doi: 10.1016/j.bbabio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Michel J, DeLeon-Rangel J, Zhu S, Van Ree K, Vik SB. Mutagenesis of the L, M, and N subunits of Complex I from Escherichia coli indicates a common role in function. PLoS One. 2011;6:e17420. doi: 10.1371/journal.pone.0017420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres-Bacete J, Sinha PK, Castro-Guerrero N, Matsuno-Yagi A, Yagi T. Features of subunit NuoM (ND4) in Escherichia coli NDH-1: TOPOLOGY AND IMPLICATION OF CONSERVED GLU144 FOR COUPLING SITE 1. J Biol Chem. 2009;284:33062–33069. doi: 10.1074/jbc.M109.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres-Bacete J, Nakamaru-Ogiso E, Matsuno-Yagi A, Yagi T. Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1: Conserved charged residues essential for energy-coupled activities. J Biol Chem. 2007;282:36914–36922. doi: 10.1074/jbc.M707855200. [DOI] [PubMed] [Google Scholar]
- 49.Steimle S, et al. Asp563 of the horizontal helix of subunit NuoL is involved in proton translocation by the respiratory complex I. FEBS Lett. 2012;586:699–704. doi: 10.1016/j.febslet.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 50.Zhu S, Canales A, Bedair M, Vik SB. Loss of Complex I activity in the Escherichia coli enzyme results from truncating the C-terminus of subunit K, but not from cross-linking it to subunits N or L. J Bioenerg Biomembr. 2016;48:325–333. doi: 10.1007/s10863-016-9655-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao Z, et al. Mechanism of fast proton transport along one-dimensional water chains confined in carbon nanotubes. J Am Chem Soc. 2010;132:11395–11397. doi: 10.1021/ja1046704. [DOI] [PubMed] [Google Scholar]
- 52.Agmon N. The Grotthuss mechanism. Chem Phys Lett. 1995;244:456–462. [Google Scholar]
- 53.Sugita Y, Kitao A, Okamoto Y. Multidimensional replica-exchange method for free energy calculations. J Chem Phys. 2000;113:6042–6051. [Google Scholar]
- 54.Kieseritzky G, Knapp EW. Optimizing pKa computation in proteins with pH adapted conformations. Proteins. 2008;71:1335–1348. doi: 10.1002/prot.21820. [DOI] [PubMed] [Google Scholar]
- 55.Ohnishi T, Nakamaru-Ogiso E, Ohnishi ST. A new hypothesis on the simultaneous direct and indirect proton pump mechanisms in NADH-quinone oxidoreductase (complex I) FEBS Lett. 2010;584:4131–4137. doi: 10.1016/j.febslet.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Treberg JR, Brand MD. A model of the proton translocation mechanism of complex I. J Biol Chem. 2011;286:17579–17584. doi: 10.1074/jbc.M111.227751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verkhovsky M, Bloch DA, Verkhovskaya M. Tightly-bound ubiquinone in the Escherichia coli respiratory complex I. Biochim Biophys Acta. 2012;1817:1550–1556. doi: 10.1016/j.bbabio.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 58.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 59.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 61.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grossfield A (2013) WHAM: the weighted histogram analysis method, version 2.0.9. Available at membrane.urmc.rochester.edu/content/wham. Accessed November 15, 2013.
- 63.Kumar A, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 64.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–8, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 65.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rabenstein B, Knapp EW. Calculated pH-dependent population and protonation of carbon-monoxy-myoglobin conformers. Biophys J. 2001;80:1141–1150. doi: 10.1016/S0006-3495(01)76091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A Gen Phys. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 68.Perdew JP. Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys Rev B Condens Matter. 1986;33:8822–8824. doi: 10.1103/physrevb.33.8822. [DOI] [PubMed] [Google Scholar]
- 69.Sierka M, Hogekamp A, Ahlrichs R. Fast evaluation of the Coulomb potential for electron densities using multipole accelerated resolution of identity approximation. J Chem Phys. 2003;118:9136–9148. [Google Scholar]
- 70.Weigend F, Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys Chem Chem Phys. 2005;7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 71.Ahlrichs R, Bär M, Häser M, Hom H, Kölmel C. Electronic structure calculations on workstation computers. Chem Phys Lett. 1989;162:165–169. [Google Scholar]
- 72.Riahi S, Rowley CN. The CHARMM-TURBOMOLE interface for efficient and accurate QM/MM molecular dynamics, free energies, and excited state properties. J Comput Chem. 2014;35:2076–2086. doi: 10.1002/jcc.23716. [DOI] [PubMed] [Google Scholar]
- 73.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.