Significance
Directional degradation is an important feature of energy-dependent intracellular proteolysis in all cells. Proteolytic machines can unravel protein substrates from either terminus depending on the site of initial recognition. Here, we use single-molecule optical trapping to probe how the bacterial AAA+ proteases ClpXP and ClpAP mechanically degrade a model substrate, titinI27, from the N terminus. N-terminal unfolding of this substrate is often accomplished with one or a few power strokes and is much faster than C-terminal unfolding, although translocation is only mildly affected by the direction. Our results highlight the paramount role of local stability in protein degradation and provide clues as to how the placement of degradation signals on a substrate may evolve to minimize the energetic cost of degradation.
Keywords: protein degradation, AAA+ proteases, directional unfolding, AAA+ motors
Abstract
AAA+ proteases and remodeling machines couple hydrolysis of ATP to mechanical unfolding and translocation of proteins following recognition of sequence tags called degrons. Here, we use single-molecule optical trapping to determine the mechanochemistry of two AAA+ proteases, Escherichia coli ClpXP and ClpAP, as they unfold and translocate substrates containing multiple copies of the titinI27 domain during degradation initiated from the N terminus. Previous studies characterized degradation of related substrates with C-terminal degrons. We find that ClpXP and ClpAP unfold the wild-type titinI27 domain and a destabilized variant far more rapidly when pulling from the N terminus, whereas translocation speed is reduced only modestly in the N-to-C direction. These measurements establish the role of directionality in mechanical protein degradation, show that degron placement can change whether unfolding or translocation is rate limiting, and establish that one or a few power strokes are sufficient to unfold some protein domains.
Molecular machines belonging to the AAA+ superfamily (ATPases Associated with diverse cellular Activities) power the remodeling of proteins, nucleic acids, and nucleoprotein complexes within the cells of all organisms (1). AAA+ proteases carry out the most dramatic example of remodeling by unfolding and degrading protein substrates (2). These destructive molecular machines help to maintain protein homeostasis and often degrade transcriptional factors to facilitate cellular responses to environmental stress. The basic architecture of AAA+ proteases is conserved from bacteria to humans and consists of a ring-shaped ATPase that controls the entry of protein substrates into a self-compartmentalized peptidase. Mechanochemical coupling of the ATPase cycle to conformational changes within the AAA+ ring exerts a pulling force that results in substrate unfolding. Processive translocation of the polypeptide chain into the degradation chamber immediately follows.
The first step in protein unfolding is the recognition by the AAA+ ring of specific sequence tags called degrons, often located at the N or C termini of target proteins. One of the best-studied degrons is the ssrA tag, a short unstructured sequence (AANDENYALAA in Escherichia coli) that becomes appended to the C terminus of incomplete bacterial proteins when ribosomes stall during translation (3, 4). Addition of this sequence to the C terminus of any protein is sufficient to target it to several AAA+ proteases, including ClpXP and ClpAP, which consist of the ClpP peptidase and the ClpX or ClpA ATPases, respectively. Proteomic profiling of ClpXP substrates in E. coli identified a diverse set of degrons (5). Some degrons shared features with the ssrA tag, but others differed dramatically in sequence, especially those found at the N terminus. Because processive degradation proceeds in a unidirectional manner from the site of initial recognition and the direction of pulling may affect unfolding rates (6), the position of the degron on the substrate protein would seem to be of paramount importance.
Here, we use single-molecule optical trapping to directly test the effect of degron placement on the mechanical degradation of titinI27 domains by ClpXP and ClpAP. The titinI27 domain has a simple Ig-like fold consisting of two major β-sheets (7, 8) (Fig. 1A). We selected titinI27 as a model domain for these studies because single-molecule and ensemble experiments have determined its mechanical, thermodynamic, and kinetic stability; less-stable variants have been well characterized; its forced unfolding has been studied by molecular dynamics simulations; and ClpXP and ClpAP unfolding, translocation, and degradation in the C-to-N direction have been analyzed in biochemical and optical-trapping experiments (8–15). We find that the ClpXP and ClpAP AAA+ machines unfold wild-type and destabilized titinI27 domains much more rapidly when pulling from the N terminus than from the C terminus. By contrast, we observe only minor direction-dependent differences in translocation and speed. This work provides direct evidence for the importance of degron placement in determining the speed and ATP cost of substrate unfolding by AAA+ machines.
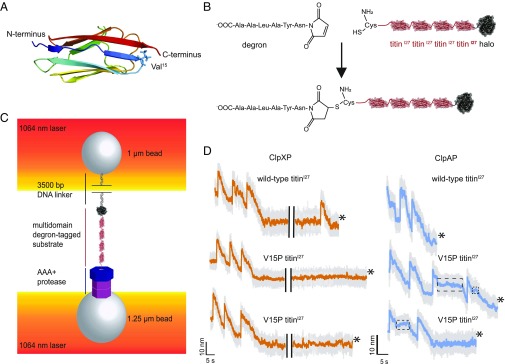
Fig. 1.
Single-molecule N-to-C protein degradation by ClpXP and ClpAP. (A) Structure of the titinI27 domain (Protein Data Bank ID code 1TIT) (cartoon representation), with rainbow coloring from the N terminus (blue) to the C terminus (red). The position of Val15, which is mutated to Pro in the V15P variant, is shown in stick representation. (B) An ssrA-degron peptide was cross-linked to the N-terminal residue of Cys-(titinI27)4-Halo via maleimide-thiol chemistry to generate a substrate for N-to-C degradation. (C) Biotinylated DNA-linked multidomain substrate and ClpAP or ClpXP were attached to separate laser-trapped streptavidin-coated beads in a “dumbbell” configuration for single-molecule optical-trapping experiments. (D) Example traces of single-molecule N-to-C degradation of the multidomain substrate shown in the lower part of B by ClpXP or ClpAP. The bead-to-bead distance changes as the AAA+ protease unfolds (rapid increases) and translocates (gradual decreases) individual substrate domains. The boxed regions of ClpAP traces show pauses during translocation. Traces shown depict raw (gray, 3,000 Hz) and 10 points moving average data (orange for ClpXP and cyan for ClpAP, 300 Hz) under ∼13–16 pN of applied force. In the ClpXP traces, the black bars present a 20-s gap. Asterisks signify tether breaks.
Results
Single-Molecule Degradation of a Substrate Bearing an N-Terminal Degron.
We monitored single-molecule unfolding and translocation by ClpXP and ClpAP using a dual-laser optical trap in passive force-clamp mode as described (14–17). ClpXP or ClpAP complexes containing biotinylated ClpP were immobilized to one streptavidin-coated bead (15). To allow N-to-C degradation, substrates had a unique N-terminal cysteine, to which we cross-linked a synthetic peptide containing the six C-terminal residues of the ssrA tag using maleimide-thiol chemistry, resulting in an N-to-N linkage (Fig. 1B). Following this N-terminal degron, substrates contained four wild-type titinI27 domains or four less-stable V15P titinI27 domains, and a C-terminal HaloTag domain (Halo). The Halo domain was conjugated to a biotinylated 3,500-bp DNA handle to allow attachment of the substrate to a second streptavidin-coated bead (Fig. 1C).
Stable tethers between beads attached to ClpXP or ClpAP and beads attached to substrate formed in the presence of ATP, and enzyme-mediated unfolding of individual domains resulted in very fast increases in bead-to-bead distance (Fig. 1D). This behavior is expected for fully cooperative unfolding and was previously observed for ClpXP unfolding of titin domains initiated at the C terminus (14, 15, 17). Following unfolding by ClpXP or ClpAP, translocation of the denatured domain resulted in a gradual reduction in bead-to-bead distance (Fig. 1D). Completion of translocation of the unfolded domain was typically followed by a short preunfolding dwell with almost no change in bead-to-bead distance before unfolding of the next domain (Fig. 1D).
Analysis of Single-Molecule Unfolding.
Although our substrates contained four titinI27 domains and one Halo domain, no traces had five unfolding events, suggesting that unfolding of some titinI27 domains occurred before data recording could be started and/or that Halo denaturation was too slow to observe. In principle, the last unfolding event in any trace could correspond to titinI27 or Halo unfolding, because covalent linkage of the DNA handle to an internal Halo residue results in similar changes in contour length on unfolding of either domain (18). However, several observations indicate that the vast majority of terminal events correspond to titinI27 or V15P titinI27 unfolding. (i) Distributions of dwells before terminal and nonterminal unfolding events were similar for each domain and enzyme (Fig. 2 A–D). (ii) Ensemble experiments presented below show that ClpXP and ClpAP degrade the titinI27 domains in the multidomain substrates far more rapidly than they degrade the Halo domain. (iii) In most single-molecule traces, we observed long periods between completion of translocation following the last unfolding event and breaking of the enzyme–substrate tether (Fig. 1D). We attribute these terminal dwells to periods in which ClpXP or ClpAP attempt but fail to unfold the Halo domain.
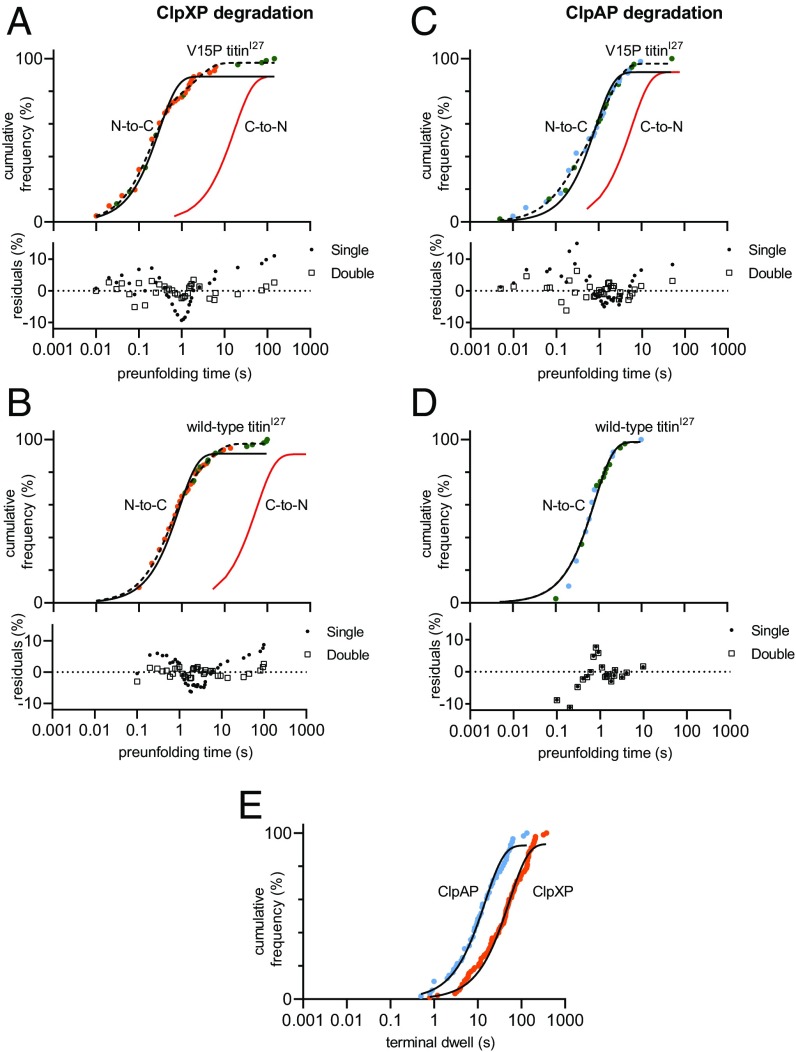
Fig. 2.
N-to-C unfolding of titinI27 domains by ClpXP or ClpAP. (A and B) Distributions of ClpXP preunfolding dwell times for V15P titinI27 (A) or wild-type titinI27 (B). Orange symbols are dwells before nonterminal unfolding events; green symbols are dwells before terminal unfolding events. (C and D) Distributions of ClpAP preunfolding dwell times for V15P titinI27 (C) and wild-type titinI27 (D). Cyan symbols are dwells before nonterminal unfolding events; green symbols are dwells before terminal unfolding events. Fits of cumulative frequency distributions to single exponentials are shown in solid black lines and to double exponentials in dashed lines. The residual plots for individual fits are shown. Solid red lines are fitted distributions for C-to-N degradation taken from Cordova et al. (14) or Olivares et al. (15). Single-molecule C-to-N degradation of wild-type titinI27 by ClpAP has not been characterized. (E) Distributions of terminal dwells for ClpXP (orange; n = 90) and ClpAP (blue; n = 56) traces. The solid lines are single-exponential fits.
The distribution of preunfolding dwells provides information on the kinetics of ClpXP-catalyzed and ClpAP-catalyzed unfolding. For both titinI27 domains and both enzymes the distributions of preunfolding dwells fit reasonably well to single exponentials (R2 ≥ 0.96; Fig. 2 A–D), indicating that substrate unfolding generally follows a single pathway with one rate-limiting step. For ClpXP, fitted unfolding time constants (τ) were ∼0.3 s for V15P and ∼0.9 s for wild-type titinI27 (Table 1). For ClpAP, these time constants were ∼0.8 s for the wild-type domains and ∼0.8 s for the mutant titinI27 domains (Table 1). C-to-N unfolding of these domains is much slower, for example by a factor of ∼50-fold for ClpXP (14, 15). As previously seen for ClpXP and ClpAP unfolding titinI27 domains from the C terminus (14, 15), N-terminal unfolding of the same domains fits better to double exponentials (R2 ≥ 0.99). For ClpXP, time constants for V15P titinI27 unfolding were 0.19 ± 0.01 s (70% amplitude) and 2.1 ± 0.4 s (30% amplitude); and 0.51 ± 0.03 s (67% amplitude) and 3.7 ± 0.4 s (33% amplitude) for wild-type titinI27. For ClpAP, time constants for unfolding of V15P titinI27 were 0.16 ± 0.04 s (28% amplitude) and 1.5 ± 0.1 s (72% amplitude), whereas the wild-type fit yielded parameters similar to those of the single-exponential fit (Fig. 2 A–D). This multiexponential behavior could reflect two unfolding pathways that depend on which part of the titinI27 domain is stochastically destabilized in each population (14).
Table 1.
Preunfolding times and translocation velocities of ClpXP and ClpAP in N-to-C and C-to-N directions
We also quantified the terminal dwells, which were roughly exponentially distributed with time constants of ∼53 s (53 ± 1) for ClpXP and ∼15 s (15.0 ± 0.4) for ClpAP (Fig. 2E). Both values are much longer than observed for N-to-C unfolding of either titinI27 domain, supporting the idea that both AAA+ machines unfold titinI27 much faster than Halo from its N terminus. By contrast, ClpXP or ClpAP unfolded the Halo domain from its C terminus with time constants of less than 10 s, which were similar to or faster than the time constants for C-to-N unfolding of V15P titinI27 (15). In combination, these results suggest that the physical properties of the substrate domain rather than the AAA+ enzyme dictate whether unfolding occurs more rapidly from the N terminus or the C terminus.
Analysis of Single-Molecule Translocation.
Faster N-to-C unfolding of titinI27 domains by ClpXP and ClpAP was not correlated with faster N-to-C translocation. Over a range of applied external forces, the average N-to-C translocation velocities of ClpXP and ClpAP were ∼3.4 nm/s and ∼2.5 nm/s, respectively (Fig. 3 and Table 1). Previously determined C-to-N translocation velocities were ∼30% faster for both enzymes (14, 15).
Fig. 3.
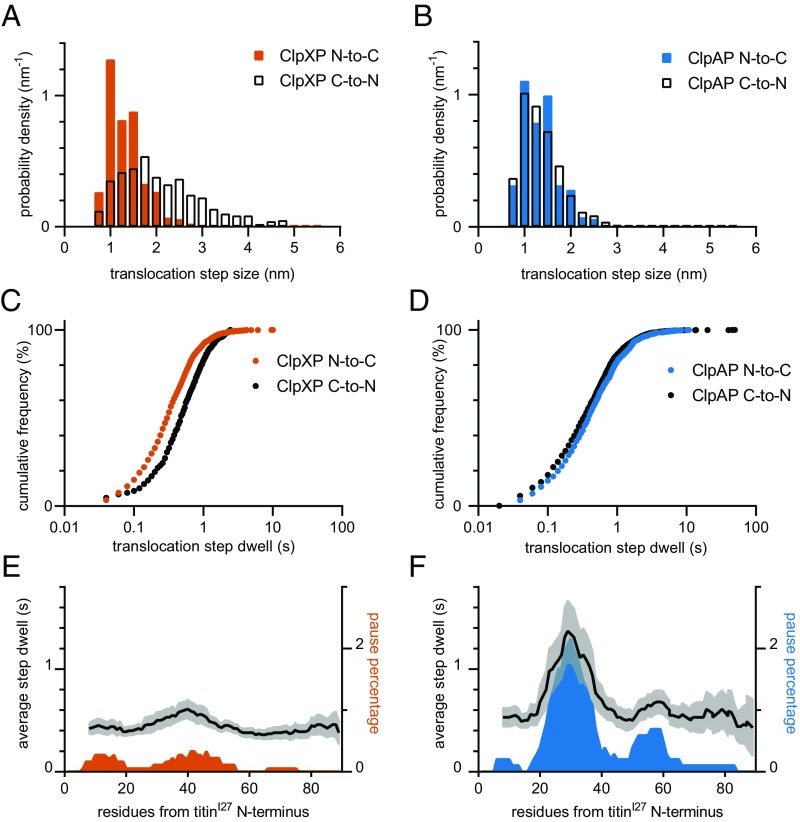
Polypeptide translocation. (A and B) Translocation speeds for ClpXP (A) and ClpAP (B) are averages over a moving window of 30 consecutive values ranked by increasing force. Colored dashed lines represent the average translocation speed over the entire force range. Black dashed lines represent average C-to-N translocation speeds for ClpXP and ClpAP.
To better understand differences in translocation velocities during N-to-C polypeptide translocation, we used a step-finding algorithm to determine the N-to-C stepping mechanics. For both ClpXP and ClpAP, individual steps ranged from ∼1–2 nm (Fig. 4 A and B). For C-to-N translocation, prior studies show that ClpAP steps are also ∼1–2 nm, whereas ClpXP steps are ∼1–4 nm (14, 15). The distributions and time constants for the dwells between individual translocation steps were also similar in both degradation directions (Fig. 4 C and D), with average time constants for individual N-to-C translocation step dwells of ∼0.4 s for ClpXP and ∼0.55 s for ClpAP.
Fig. 4.
Steps during polypeptide translocation. (A and B) Distributions of physical step sizes during directional translocation by ClpXP (orange bars, A) and ClpAP (blue bars, B). Black bars show previously determined C-to-N step sizes for ClpXP (ref. 14, A) and ClpAP (ref. 15, B). (C and D) Distributions of dwell times preceding an individual step during N-to-C translocation are shown for ClpXP (orange symbols, C) and ClpAP (blue symbols, D). Black symbols show previously determined C-to-N step dwells for ClpXP (ref. 14, C) and ClpAP (ref. 15, D). (E and F) Average step dwells and frequency of pauses of 2.5 s or longer along the length of the titinI27 polypeptide during N-to-C translocation by ClpXP (E) or ClpAP (F) were calculated using a 10-aa moving window. The 95% CIs are shown in gray for average step dwells.
We can estimate the ATP-hydrolysis rate during translocation by assuming that each 1-nm step corresponds to hydrolysis of one ATP and an associated power stroke (14, 15). Thus, for ClpXP, the average hydrolysis rate would be ∼3.25 s−1, corresponding to a time constant of ∼0.3 s (average step dwell divided by average step size = 0.4 s/1.3 nm). Notably, this value is the same as the average time required for N-to-C unfolding of V15P titinI27 by ClpXP, suggesting that a single mechanical power stroke results in denaturation. During N-to-C translocation by ClpAP the average ATP-hydrolysis rate would be ∼2.5 s−1, corresponding to a time constant of ∼0.4 s (average step dwell divided by average step size = 0.55 s/1.4 nm). Because the ClpAP unfolding times for wild-type and V15P titinI27 were ∼0.8 s, only a few ClpAP unfolding attempts (i.e., power strokes) are usually sufficient to unfold these domains. Because complete translocation of a single titinI27 domain requires ∼20 individual steps and associated power strokes, the overall process of translocation rather than unfolding seems to be rate-limiting in N-to-C degradation of wild-type and V15P titinI27 by ClpXP and ClpAP.
For both ClpXP and ClpAP, we observed substantial pausing, defined as no translocation for 2.5 s or longer, during N-to-C translocation, which caused an increase in the average translocation step dwell (Fig. 4 E and F). Two pauses in ClpAP translocation are boxed in Fig. 1D. For ClpXP, pausing occurred in ∼8% of all total N-to-C translocation events examined and accounted for ∼8% of the total translocation time, whereas prior studies of C-to-N translocation reported ∼4% pausing (14). Pausing was also higher for ClpAP, occurring in ∼35% of all N-to-C translocation events and accounting for ∼25% of the total translocation time, compared with ∼10% pauses in all C-to-N translocation steps (15). Thus, pausing partially accounts for slower N-to-C than C-to-N translocation. Pauses in the N-to-C direction were not uniformly distributed along the polypeptide (Fig. 4 E and F), suggesting that they result from sequence-specific interactions between the translocating polypeptide and the AAA+ rings of ClpXP or ClpAP. Pausing may occur when ClpX or ClpA transiently switch into an inactive conformation (19). Because ClpA contains two AAA+ rings, pausing could also occur if functional coordination between the top and bottom rings was lost.
Ensemble Degradation of N- and C-Terminally Tagged Substrates.
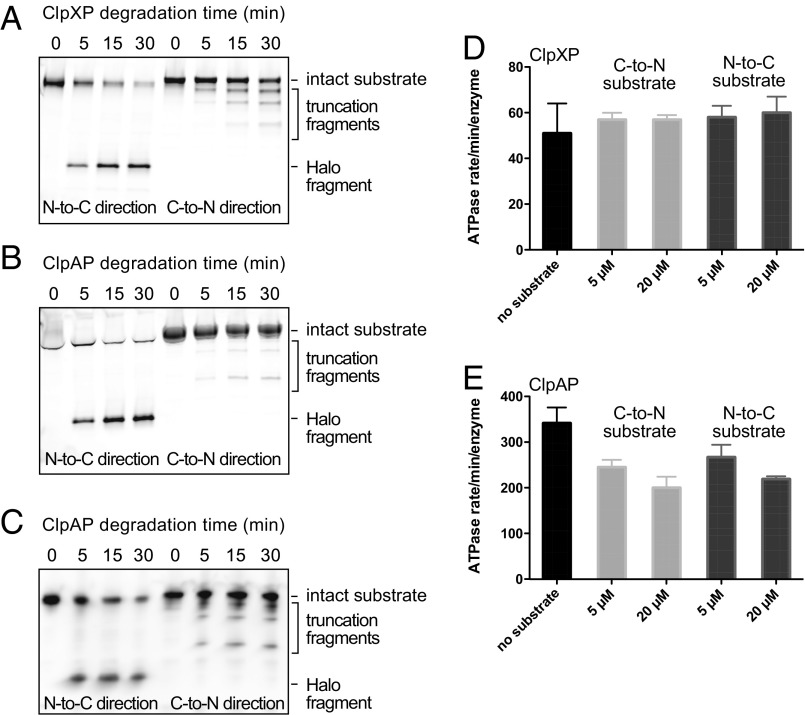
Our results taken with previous studies (14, 15) predict that ClpXP and ClpAP should degrade an N-degron–tagged (titinI27)4-Halo substrate much faster than a C-degron–tagged Halo-(titinI27)4 substrate. In preliminary experiments, however, we found that incomplete cross-linking of the degron peptide to Cys-(titinI27)4-Halo complicated interpretation. To obviate this problem, we cross-linked a peptide containing both an ssrA and H6 sequence to Cys-(titinI27)4-Halo and then used Ni++-NTA affinity to purify the degron-tagged substrate away from the unmodified protein. We then modified the Halo domain of this N-tagged substrate by covalent linkage to a fluorescent HaloTag tetramethylrhodamine (TMR) ligand and assayed the kinetics of ensemble N-to-C degradation by ClpXP or ClpAP by monitoring TMR fluorescence following SDS/PAGE (Fig. 5 A and B, left). In parallel, we assayed ensemble C-to-N degradation of fluorescent Halo-(titinI27)4-ssrA (Fig. 5 A and B, right). As predicted, ClpXP and ClpAP degraded the N-tagged substrate much faster than the C-tagged substrate. For C-tagged substrates, three lower-molecular-weight bands corresponding to fragments with different numbers of titinI27 domains remaining were observed to accumulate during the degradation reaction (20, 21), as expected if C-to-N unfolding/degradation of these domains by ClpXP or ClpAP is slow and enzyme dissociation often occurs, resulting in partially degraded products. By contrast, only one major intermediate or product accumulated during degradation of the N-tagged substrate, which corresponded to a fragment containing the C-terminal Halo domain. This result strongly supports a model in which ClpXP and ClpAP rapidly unfold the titinI27 domains in the N-to-C direction but then stall during attempts to unfold the native Halo domain.
Fig. 5.
Ensemble assays of substrate degradation and ATP hydrolysis. (A–C) SDS/PAGE assays of the kinetics of degradation of N-tagged ssrA-(titinI27)4-Halo or C-tagged Halo-(titinI27)4-ssrA degradation by ClpXP or ClpAP. Initial substrate concentrations were 1 µM, the ClpP14 concentration was 2 µM, the ClpX6 or ClpA6 concentration was 1 µM, and assays contained 5 mM ATP and a regeneration system. The gels shown are representative of three independent experiments. (A and B) The Halo domain was fluorescently labeled, and gels were scanned for fluorescence. (C) The Halo domain was biotinylated, biotinylated proteins were purified by binding to streptavidin beads following degradation, and proteins were detected by Western blotting using an anti-Halo antibody after SDS/PAGE. (D and E) Steady-state rates of hydrolysis of 5 mM ATP by ClpXP (300 nM ClpX6 and 900 nM ClpP14, D) or ClpAP (100 nM ClpA6 and 200 nM ClpP14, E) in the presence of different concentrations of ssrA-(titinI27)4-Halo (N-to-C substrate) or Halo-(titinI27)4-ssrA (C-to-N substrate). Means ± SD, n = 3 independent measurements.
For ClpAP degradation of the N-tagged substrate (Fig. 5B, left), ClpA ran at the same position as the intact, undegraded substrate and partially quenched TMR fluorescence. To monitor degradation in a different way, we labeled the N-tagged substrate with a HaloTag biotin ligand, performed ensemble degradation, isolated Halo-containing proteins using streptavidin beads, separated the mixture by SDS/PAGE, and detected proteins using Western blotting with a polyclonal anti-Halo antibody. The results of this assay (Fig. 5C) closely mimicked those with the fluorescent substrate (Fig. 5B).
Ensemble ATP-Hydrolysis Rates.
We determined the steady-state rate of ATP hydrolysis by ClpXP or ClpAP in the presence of different concentrations of N-degron–tagged (titinI27)4-Halo or C-degron–tagged Halo-(titinI27)4. For ClpXP, the time constant for ATP hydrolysis was ∼1 s, irrespective of the concentration of substrate or direction of degradation (Fig. 5D). This value is approximately threefold slower than the rate estimated during single-molecule translocation (∼0.3 s as previously discussed), but ClpXP has been shown to hydrolyze ATP more slowly during protein unfolding than translocation (13). Thus, for degradation in solution, ClpXP may spend most of the time trying to unfold the Halo domain of the N-degron–tagged (titinI27)4-Halo substrate, consistent with the accumulation of the Halo intermediate in our SDS/PAGE experiments (Fig. 5A). If the ATPase rate of ClpXP also slows approximately threefold when it encounters the wild-type titinI27 domain during N-to-C degradation, then one power stroke may also be sufficient to unfold this domain. If the rate of ATP hydrolysis remains the same as during translocation, by contrast, then an average of approximately three power strokes would be required for unfolding.
For ClpAP, the ATP-hydrolysis rate was suppressed modestly by N-to-C or C-to-N degradation (Fig. 5E). At near-saturating concentrations of N-degron–tagged (titinI27)4-Halo, the time constant for ATP hydrolysis in solution by ClpAP was ∼0.3 s, similar to the ∼0.4 s estimate from mechanical stepping as previously discussed, whereas the time constant for single-molecule unfolding of wild-type titinI27 in the N-to-C direction was ∼0.8 s. These results support a model in which ClpAP uses an average of two or three ATP hydrolysis events and coupled ∼1-nm power strokes to unfold wild-type titinI27 domain from the N terminus.
Discussion
Within cells, AAA+ proteases must degrade a wide variety of substrates that differ in the structure and stability of the native domains proximal to the degron, as well as in the position of the degron, usually at one terminus or the other (22). Ensemble studies using model substrates have shown that the position of the degron can have large effects on the rate of degradation (6, 23–26). However, these studies have not been able to determine unambiguously how directional effects on the mechanical processes of unfolding versus translocation contribute to the observed changes in the rate and the associated energetic cost of degradation. As shown here, single-molecule optical trapping experiments can answer these questions. Based on results reported here and elsewhere (14–17, 27–29), both the ClpXP and ClpAP proteases unfold the titinI27 domain much faster and at far lower energetic costs when pulling from the N terminus as opposed to the C terminus. In principle, many factors could contribute to these differences. For example, ClpXP and ClpAP pull on distinct amino acid sequences in N-to-C versus C-to-N substrate degradation, and their ability to grip these sequences might differ. However, studies of C-to-N ClpXP degradation suggest that sequence-dependent grip effects are relatively modest (30, 31), whereas we observe ∼50-fold faster ClpXP unfolding in the N-to-C direction for the titinI27 domain. It is also highly unlikely that ClpXP and ClpAP are always better at N-to-C unfolding because they can grip the polypeptide chain more tightly in a sequence-independent fashion in this direction. For example, ClpXP degrades Arc repressor tagged with a C degron at almost the same maximal rate as it degrades the same protein tagged with an N degron (24). Moreover, in the C-to-N direction, ClpXP and ClpAP unfold the Halo domain at rates similar to or faster than they unfold V15P titinI27 (15), whereas our results indicate that N-terminal unfolding of Halo by both enzymes is substantially slower than V15P titinI27 unfolding.
The local mechanical stability of a protein near the position of enzymatic pulling has been proposed to be more important than global stability in determining resistance to unfolding by a AAA+ protease (6, 23, 25, 32). Our results strongly support this model for the wild-type and V15P titinI27 domains. Atomic force microscopy (AFM) experiments, molecular-dynamics simulations, mutagenesis, and NMR results suggest that hydrogen bonds between the N-terminal part of the first β-strand and its partner strand break early during unfolding and at lower forces than required to break interstrand hydrogen-bonding interactions at the C terminus (9, 11). Thus, our finding that both ClpXP and ClpAP unfold titinI27 much faster when pulling from the N terminus as opposed to the C terminus supports the idea that N-terminal structure in the titinI27 domain is inherently more fragile than C-terminal structure. We assume that many proteins will be easier to unfold from one end than the other. For example, the rate at which thioredoxin is unfolded and translocated through a nanopore depends on which end is pulled through first (33).
The V15P mutation removes a hydrogen bond that is close in space to the C terminus of titinI27. In the C-to-N direction, ClpXP unfolds wild-type titinI27 (τ ≈ 55 s) about threefold more slowly than the V15P variant (τ ≈ 17 s) (14). In the N-to-C direction, we also find a threefold difference in ClpXP unfolding of wild-type titinI27 (τ≈ 0.9 s) and V15P titinI27 (τ≈ 0.3 s), but at rates ∼50-fold faster than in the C-to-N direction. How does a mutation far from the N terminus alter the rate of N-to-C unfolding? One possibility is that ClpXP unfolding of structure near the N terminus of wild-type titinI27 leaves a folded intermediate that requires additional power strokes to unfold, whereas unfolding of N-terminal structure in the V15P variant results in cooperative unfolding of the rest of the domain. We did not see evidence for multistep N-to-C unfolding of wild-type titinI27 by ClpXP in our single-molecule traces, but an intermediate with a small change in contour length might not have been detected. Another possibility is that the V15P mutation causes minor structural changes that propagate to and reduce the stability of local structure near the N terminus of titinI27. We find that ClpAP unfolds wild-type and V15P titinI27 at essentially the same rate. It is possible that subtle differences in pulling mechanics make ClpXP better able to take advantage of reduced N-terminal stability for this substrate, and related enzyme-specific or substrate-specific features could explain why pulling from the N terminus rather than the C terminus of titinI27 results in ∼50-fold faster unfolding for ClpXP but only approximately eightfold faster unfolding for ClpAP.
For AAA+ proteases and remodeling enzymes translocation of a degron through the narrow axial pore of the ring hexamer provides a mechanism to apply force to attached proteins. ClpXP can degrade model polypeptides with diverse amino acid sequences, d-amino acids, and nonnatural spacings of peptide bonds, suggesting that translocation does not depend on enzyme recognition of precise chemical details of the polypeptide chain (30). Consistently, we find that ClpXP translocation is only a bit slower in the N-to-C than in the C-to-N direction, largely as a result of a smaller average step size and an increased probability of sequence-specific pausing. ClpAP also translocates slightly more slowly in the N-to-C direction along the polypeptide track. It is important to note, however, that these changes in translocation rates and pausing frequencies are minor in comparison with the very large differences in directional unfolding rates.
When ClpXP pulls against the stable C terminus of wild-type titinI27 it cycles through hundreds of power stokes on average before unfolding is successful (13, 14). Nevertheless, a single power stroke eventually causes unfolding, probably because transient thermal fluctuations stochastically destabilize the resisting structure in a minor population of substrate molecules (14, 16). During translocation, each power stroke moves approximately five residues of the substrate through the axial pore, and thus translocation of the ∼100 residues of titinI27 requires hydrolysis of ∼20 ATPs. For C-to-N degradation of titinI27, the predominant ATP cost of degradation is inefficient unfolding. Strikingly, however, ClpXP and ClpAP unfold titinI27 from the N terminus in a far more efficient manner. Based on the average translocation dwells in our optical trapping experiments and the rates of bulk ATP hydrolysis during substrate degradation, we estimate the cost of N-to-C-terminal unfolding to be approximately one to three ATPs, depending on the enzyme and titin variant. Thus, for N-to-C degradation of titinI27, translocation rather than unfolding seems to be rate-limiting and most expensive in terms of ATP hydrolysis.
During a single ClpXP or ClpAP translocation step we estimate the pulling velocity to be ∼3 nm/ms or faster, because data are collected at 3,000 Hz. In AFM studies using comparable pulling speeds, the forces required for unfolding of wild-type titinI27 or the V15P variant are ∼200 pN and ∼150 pN, respectively (10, 12). There are multiple differences between mechanical unfolding in the AFM and by ClpXP or ClpAP. For example, force is applied continuously and increasingly in the AFM, whereas AAA+ enzymes presumably apply an unfolding force only during a power stroke. The direction of pulling may also be different, depending on the attachment points to the surface and AFM tip versus the degron position and how it is engaged during ClpXP or ClpAP unfolding. Nevertheless, our finding that one or a few power strokes are sufficient to enzymatically unfold titinI27 or V15P raises the possibility that ClpXP and ClpAP can generate forces in excess of the 20- to 25-pN forces they work against in optical trapping experiments. Theoretically, hydrolysis of a single ATP could result in a free-energy change of ∼22 kBT, corresponding to a 1-nm step against a force of ∼90 pN.
Why are some cellular substrates degraded via N degrons and others via C degrons? In rare cases, one protein terminus may be involved in an essential catalytic or binding function, dictating degron placement at the opposite end. For the majority of proteins, however, sequence alignments of orthologs show that both termini can be modified by addition or deletion of additional residues sufficient in length to create or remove a degron. For example, placing an accessible Ala–Ala sequence at a protein’s C terminus can make it a substrate for ClpXP degradation (22). If either terminus of a protein can be modified without affecting function, we propose that evolutionary selection will result in placing the degron at the terminus that allows degradation most rapidly and at the lowest ATP cost. One interesting prediction of this model is that natural substrates that have evolved N degrons will have a higher probability of being easier to mechanically unfold from the N terminus, and vice versa.
Methods
Proteins and Peptides.
Full-length E. coli ClpX, a variant of E. coli ClpA lacking its C-terminal tail (∆C9) to prevent autodegradation (34), E. coli ClpP, and a protein substrate consisting of an N-terminal Halo domain, four wild-type titinI27 domains, and a C-terminal ssrA degradation tag were cloned, expressed, and purified as described (35). For optical trapping, we used a tetradecameric ClpP variant (ClpPplatform) consisting of one wild-type, heptameric ClpP ring and one heptameric ring of the M5A ClpP variant containing a C-terminal biotin acceptor peptide sequence that was biotinylated in vitro with the BirA enzyme (15). A precursor of a protein for N-terminal degradation contained an N-terminal His6-SUMO domain, followed by a short linker containing a cysteine, four titinI27 domains (either wild-type or the V15P variant), and a C-terminal Halo domain. Precursor proteins were expressed at 30 °C from a pET23b vector in E. coli strain BL21 (DE3) after induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside at OD600 = 0.6 and 3 h of additional growth. Cell pellets were lysed in lysis buffer (10 mM Tris, pH 8.0, 100 mM NaCl, 10% glycerol, 10 mM imidazole, and 1 mM DTT), and the precursor was purified by Ni++-NTA chromatography (Qiagen), followed by removal of the His6-SUMO domain by cleavage with Ulp1 protease. The Cys-(titinI27)4-Halo protein was separated from His6-SUMO by another step of Ni++-NTA chromatography and then purified further by gel filtration using a HiLoad Superdex 200 16/600 column (GE Healthcare). Proteins were flash-frozen in liquid nitrogen and stored at −80 °C in 10 mM Tris (pH 8), 100 mM NaCl, 10% glycerol, and 1 mM DTT.
A peptide containing six C-terminal residues of the ssrA tag (maleoyl-β-Ala-NYALAA-coo−) was a gift from Karl Schmitz, Massachusetts Institute of Technology, Cambridge, MA. A peptide containing an H6 tag and the full ssrA tag (maleoyl-AH6AANDENYALAA-coo−) was synthesized using standard Fmoc techniques and an Apex 396 solid-phase peptide synthesizer. Peptides were purified by HPLC using a C18 column (Agilent Technologies). Before cross-linking, Cys-(titinI27)4-Halo or Cys-(V15P)4-Halo was incubated in 100 mM Tris (pH 8.0) and 50 mM DTT for 1 h on ice before exchange into labeling buffer (100 mM sodium phosphate, pH 7.0, and 1 mM EDTA) using PD-10 desalting columns (GE Healthcare). Maleoyl-β-Ala-NYALAA-coo− or maleoyl-AH6AANDENYALAA-coo− was then added in 10- to 20-fold molar excess, and the sample was incubated in the dark for 2 h at room temperature. Excess peptide was removed using a PD-10 desalting column. For ensemble experiments, proteins successfully cross-linked to maleoyl-AH6AANDENYALAA-coo− were further purified by Ni++-NTA chromatography.
Single-Molecule Optical Trapping.
Complexes of ClpXP or ClpAP and multidomain substrates were tethered between two beads trapped by 1,064-nm lasers in passive force-clamp mode as described (14–17, 35). Briefly, biotinylated, DNA-linked, multidomain substrates were attached to 1-µm streptavidin-coated polystyrene beads (Spherotech) that were loosely tethered to the surface of a glass coverslip via a DNA-linked glass-binding peptide aptamer. Biotinylated ClpP was attached to a 1.25-µm streptavidin-coated polystyrene bead in the presence of ClpA or ClpX and 5 mM ATP. Free enzymes were removed by centrifugation immediately before use. A weakly laser-trapped ClpXP or ClpAP bead was brought near a surface-tethered substrate bead. On substrate recognition by the enzyme, a stiff laser trap was used to rupture the aptamer–glass attachment of the substrate bead, resulting in a ClpAP-substrate or ClpXP-substrate complex tethered between two laser-trapped beads (Fig. 1C). Experiments were performed at 18–20 °C using 5 mM Mg2+, 5 mM ATP, and ATP-regeneration and oxygen-scavenging systems in PD-T buffer (25 mM Hepes, pH 7.6, 100 mM KCl, 10 mM MgCl2, 10% glycerol, 0.1% Tween-20, and 1 mM Tris-2-carboxyethyl-phosphine) supplemented with 1 mg/mL BSA (Sigma-Aldrich).
Data acquisition was carried out as described (14–17, 35). Custom MATLAB scripts were used to calculate interbead distances, measure the magnitude of unfolding distances, and measure the preunfolding dwell time between the end of one translocation event and the next unfolding event. Translocation events in each trace were fit with a linear equation to determine the average translocation speed, which included pauses. For calculation of preunfolding dwells we excluded dwells before the first unfolding event in a trace unless it was preceded by translocation of 5 nm or more and also separately calculated dwells that included or excluded the final unfolding event in each trace.
Finding Steps in Translocation Traces.
Data were collected at 3-kHz sampling frequency, decimated to 50 Hz, and individual physical steps in translocation traces were determined using a MATLAB implementation of the χ2 minimization method of J. Kerssemakers (Delft University of Technology), which requires input of the number of steps to fit within a given trace (36). We estimated this value by taking the pairwise distribution of decimated data. Steps <0.75 nm and slips or backward steps were combined, and the dwell time preceding a combined step was added to the dwell time of the following step.
To analyze pausing, translocation events were converted to amino acids using the worm-like chain force-extension model (37):
where F is the measured force, kB is the Boltzmann constant, T is temperature, Lp is the persistence length (0.61 nm), Lc is the contour length per amino acid (0.38 nm), and x is the bead-to-bead extension. Associated translocation step dwells were ranked by the calculated residues translocated. Any step dwells above the pause threshold of 2.5 s were mapped to the residue position and pause percentages calculated based on the total number of translocation events analyzed (n = 183 for ClpXP; n = 102 for ClpAP). Translocation step dwell and pause percentage values were averaged with a 10-aa moving window and plotted against average residue translocated. Previous analysis of the robustness and accuracy of the step-finding algorithm with simulated and real data can be found in ref. 14.
Biochemical Assays.
For ensemble ClpXP degradation assayed by fluorescence, 1 µM N-tagged ssrA-(titinI27)4-Halo or C-tagged Halo-(titinI27)4-ssrA was incubated with 0.5 µL of 0.5 mM HaloTag TMR Ligand (Promega) in 25 mM Hepes, pH 7.5, 100 mM KCl, 10 mM MgCl2, 10% glycerol, 0.1% tween-20, and 2 mM DTT at 30 °C for 15 min. Reactions were cooled to 22–24 °C, and ClpX6 (1 µM) and ClpP14 (2 µM) were added. A sample was removed for the 0 time point and 5 mM ATP and a regeneration system (6.25 mM phosphoenolpyruvate and 23.5 U/mL pyruvate kinase) was added to start the degradation reaction. Samples were taken at different times, quenched by addition of SDS-sample buffer, and electrophoresed on a Mini-PROTEAN TGX 4–15% (wt/vol) precast gel (Bio-Rad). TMR fluorescence on the gel was imaged using a Typhoon FLA 9500 scanner (GE Healthcare). Ensemble ClpAP degradation assays were performed by the same procedure, except 1 µM ClpA6 replaced ClpX6.
For ClpAP ensemble degradation assayed by Western blotting, the protein substrates were labeled with HaloTag biotin Ligand (Promega) using the conditions described above. ClpAP degradation was carried out as described for the fluorescent substrates, except that samples were quenched by addition of 6 M guanidinium chloride. Biotinylated substrate and fragments were purified by binding to streptavidin magnetic beads (Pierce) and then eluted by boiling at 95 °C for 5 min in Laemmli SDS/PAGE sample buffer. Samples were electrophoresed on a Mini-PROTEAN TGX 4–15% (wt/vol) precast gel (Bio-Rad), transferred to Immobilon-P membranes (EMD Millipore) in a wet transfer apparatus (Bio-Rad). Membranes were probed with 1:1,000 anti-HaloTag pAb (Promega) overnight at 4 °C, incubated with goat anti-rabbit IgG-AP conjugate (1:10,000 dilution; Bio-Rad) for 1 h at room temperature, and developed with alkaline phosphatase ECF Substrate (GE Healthcare). The blots were exposed with a blue laser using a Typhoon FLA 9500 scanner (GE Healthcare).
Steady-state ATP hydrolysis rates at 24 °C were measured in a coupled assay by monitoring the loss of NADH absorbance at 340 nm (38). Final concentrations were 300 nM ClpX6 and 900 nM ClpP14 for ClpXP assays, 100 nM ClpA6 and 200 nM ClpP14 for ClpAP assays, and varying substrate concentrations. The final reaction mixture contained 5 mM ATP, 0.7 mM NADH, 5 mM phosphoenolpyruvate, 23.5 U/mL pyruvate kinase, and 20 U/mL lactate dehydrogenase in PD-T buffer.
Acknowledgments
We thank Tristan Bell, Matt Lang, Igor Levchenko, Ted Moore, Karl Schmitz, and Ohad Yosefson for materials, advice, and helpful discussions. This work was supported by NIH Grants GM-101988 and AI-15706 and the Howard Hughes Medical Institute. T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: Common structure–diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 2.Olivares AO, Baker TA, Sauer RT. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol. 2016;14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13:285–297. doi: 10.1038/nrmicro3438. [DOI] [PubMed] [Google Scholar]
- 4.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 5.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 7.Improta S, Politou AS, Pastore A. Immunoglobulin-like modules from titin I-band: Extensible components of muscle elasticity. Structure. 1996;4:323–337. doi: 10.1016/s0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- 8.Stacklies W, Vega MC, Wilmanns M, Gräter F. Mechanical network in titin immunoglobulin from force distribution analysis. PLoS Comput Biol. 2009;5:e1000306. doi: 10.1371/journal.pcbi.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marszalek PE, et al. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 10.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler SB, et al. Mechanical unfolding of a titin Ig domain: Structure of unfolding intermediate revealed by combining AFM, molecular dynamics simulations, NMR and protein engineering. J Mol Biol. 2002;322:841–849. doi: 10.1016/s0022-2836(02)00805-7. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 13.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 14.Cordova JC, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivares AO, Nager AR, Iosefson O, Sauer RT, Baker TA. Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat Struct Mol Biol. 2014;21:871–875. doi: 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iosefson O, Olivares AO, Baker TA, Sauer RT. Dissection of axial-pore loop function during unfolding and translocation by a AAA+ proteolytic machine. Cell Rep. 2015;12:1032–1041. doi: 10.1016/j.celrep.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popa I, et al. Nanomechanics of HaloTag tethers. J Am Chem Soc. 2013;135:12762–12771. doi: 10.1021/ja4056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stinson BM, et al. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell. 2013;153:628–639. doi: 10.1016/j.cell.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iosefson O, Nager AR, Baker TA, Sauer RT. Coordinated gripping of substrate by subunits of a AAA+ proteolytic machine. Nat Chem Biol. 2015;11:201–206. doi: 10.1038/nchembio.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 23.Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Farrell CM, Baker TA, Sauer RT. Altered specificity of a AAA+ protease. Mol Cell. 2007;25:161–166. doi: 10.1016/j.molcel.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koodathingal P, et al. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gur E, Vishkautzan M, Sauer RT. Protein unfolding and degradation by the AAA+ Lon protease. Protein Sci. 2012;21:268–278. doi: 10.1002/pro.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Aliaga P, Ramirez L, Kim F, Bustamante C, Martin A. Substrate-translocating loops regulate mechanochemical coupling and power production in AAA+ protease ClpXP. Nat Struct Mol Biol. 2016;23:974–981. doi: 10.1038/nsmb.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen M, et al. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Too PHM, Erales J, Simen JD, Marjanovic A, Coffino P. Slippery substrates impair function of a bacterial protease ATPase by unbalancing translocation versus exit. J Biol Chem. 2013;288:13243–13257. doi: 10.1074/jbc.M113.452524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matouschek A. Protein unfolding–An important process in vivo? Curr Opin Struct Biol. 2003;13:98–109. doi: 10.1016/s0959-440x(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Larrea D, Bayley H. Protein co-translocational unfolding depends on the direction of pulling. Nat Commun. 2014;5:4841. doi: 10.1038/ncomms5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maglica Z, Striebel F, Weber-Ban E. An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity. J Mol Biol. 2008;384:503–511. doi: 10.1016/j.jmb.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 35.Cordova JC, Olivares AO, Lang MJ. Mechanically watching the ClpXP proteolytic machinery. Methods Mol Biol. 2017;1486:317–341. doi: 10.1007/978-1-4939-6421-5_12. [DOI] [PubMed] [Google Scholar]
- 36.Kerssemakers JWJ, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 37.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 38.Kim YI, et al. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]