Significance
Class switch recombination (CSR), a B-cell–specific reaction essential for optimal antibody responses, proceeds through the deliberate generation of DNA breaks. Although these breaks are obligatory intermediates of class switching, their improper resolution not only impairs immune responses but also promotes oncogenic translocations. The pathways and mechanisms that participate in the repair of such lesions are yet to be fully elucidated. In this work, we have identified a BRCT domain containing protein BRIT1 as an effector of the DNA repair phase of CSR. We show that B cells lacking BRIT1 are impaired in undergoing class switching due to an inability to repair DNA breaks efficiently. This work thus has significant implications in both immunity and cancer.
Keywords: class switch recombination, DNA repair, BRCT domains, B cells, MDC1
Abstract
DNA double-strand breaks (DSBs) serve as obligatory intermediates for Ig heavy chain (Igh) class switch recombination (CSR). The mechanisms by which DSBs are resolved to promote long-range DNA end-joining while suppressing genomic instability inherently associated with DSBs are yet to be fully elucidated. Here, we use a targeted short-hairpin RNA screen in a B-cell lymphoma line to identify the BRCT-domain protein BRIT1 as an effector of CSR. We show that conditional genetic deletion of BRIT1 in mice leads to a marked increase in unrepaired Igh breaks and a significant reduction in CSR in ex vivo activated splenic B cells. We find that the C-terminal tandem BRCT domains of BRIT1 facilitate its interaction with phosphorylated H2AX and that BRIT1 is recruited to the Igh locus in an activation-induced cytidine deaminase (AID) and H2AX-dependent fashion. Finally, we demonstrate that depletion of another BRCT-domain protein, MDC1, in BRIT1-deleted B cells increases the severity of CSR defect over what is observed upon loss of either protein alone. Our results identify BRIT1 as a factor in CSR and demonstrate that multiple BRCT-domain proteins contribute to optimal resolution of AID-induced DSBs.
Mature B cells responding to antigenic challenge undergo Ig heavy chain (Igh) class switch recombination (CSR), a deletional-recombination event that exchanges the default Cμ constant region of IgM for one of a set of downstream constant region segments (Cγ, Cε, or Cα). The B cell thereby transitions from expressing IgM to expressing IgG, IgE, or IgA, with each secondary isotype having a distinct effector function during a tailored immune response (1). CSR occurs between repetitive, transcribed switch (S) region DNA elements that precede each of the constant region genes. Prevalent models of CSR posit that activation-induced cytidine deaminase (AID) deaminates cytidines into uridines at transcribed S regions and instigates a cascade of reactions to generate DNA double-strand breaks (DSBs). Ligation of DSBs between donor (generally Sμ) and acceptor S regions by components of the general nonhomologous end-joining (NHEJ) pathway and/or the relatively poorly understood microhomology-dependent “alternative” end-joining processes completes CSR (1).
Although they are obligatory CSR intermediates, DSBs are also one of the most toxic lesions that can occur in a cell; unrepaired DSBs can lead to cell death or potentiate chromosomal translocations that are hallmarks of many kinds of cancer, including B-cell lymphomas (2). It therefore comes as no surprise that multiple mechanisms restrict AID-induced DSB formation specifically at S regions during CSR (3). Once generated, the cellular DNA damage response (DDR) pathways are activated to resolve the DSBs rapidly to promote CSR and maintain genomic integrity (4). Execution of DDR relies on activation of phosphoinositide-3-kinase–related kinases, such as ATM, ATR, and DNA-PKCs, and the subsequent recognition and binding of phosphorylated modules by multiple effectors of DDR proteins, including those proteins with BRCA1 C-terminal (BRCT) domains (5, 6).
The BRCT domains were first identified in the breast and ovarian cancer susceptibility gene product BRCA1 and function as protein–protein interaction modules (5, 6). The architecture of BRCT domains is variable, ranging from a single module to multiple, tandem BRCT repeats. The mouse genome contains at least 27 genes coding for proteins with BRCT domains, with over one-third reported to influence CSR, including 53BP1, PTIP, NBS1, Rev1, MDC1, and DNA ligase IV (7–13). These proteins participate in different phases of CSR. For example, ATM-dependent phosphorylation of 53BP1 facilitates recruitment of factors that protect against extensive end-resection and promote orientation-specific end-joining (14, 15), whereas DNA ligase IV is the essential NHEJ component ligating DSBs at S regions during CSR (12). However, it is unclear whether other BRCT proteins beyond the proteins enumerated above have a role in CSR.
BRIT1 [BRCT-repeat inhibitor of human telomerase (hTert) expression] was isolated in a genetic screen for repressors of the hTert (16). BRIT1 also mapped to one of the 12 loci implicated in primary microcephaly (hence the name microcephalin or MCPH1), an autosomal recessive disease characterized by severely decreased cerebral cortex and varying degrees of mental retardation (17, 18). This ∼110-kDa ubiquitously expressed protein with one N-terminal and two tandem C-terminal BRCT domains has been demonstrated to be a crucial proximal regulator of DDR (19). In cells treated with ionizing radiation (IR), BRIT1 is rapidly recruited to DSBs via its C terminus BRCT repeat-dependent interaction with phosphorylated H2AX (γ-H2AX) (20–22). It is believed that the DSB-bound BRIT1 then serves as a scaffold for Switch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complexes that subsequently render the chromatin accessible to DNA repair factors, including NBS1, 53BP1, MDC1, phosphorylated ATM, BRCA1/Chk1, and BRCA2/RAD51 (19, 23–25). However, the nature of the interactions between these different factors could be cell type-specific. For example, although loss of BRIT1 in the osteosarcoma cell line U2OS abrogates formation of 53BP1 foci (24), 53BP1 foci observed in neurons is independent of BRIT1 (26).
In keeping with its role in DDR, loss of BRIT1 in fibroblasts leads to both spontaneous and IR-induced genomic instability (24, 27). BRIT1-null mice are more susceptible to lymphomagenesis, and BRIT1 expression was found to be significantly decreased in ovarian and breast cancer samples, suggesting BRIT1 acts as a tumor suppressor (19, 24, 27). In spermatocytes undergoing meiotic recombination, loss of BRIT1 leads to a failure to recruit BRAC2/RAD51 to DSBs, leading to infertility in mice (28). Likewise, defects in DDR, in conjunction with uncoupling of mitosis and centrosome cycles, contribute to the attrition of progenitor neurons in BRIT1-depleted mouse models of microcephaly (26, 29, 30). Despite these studies on the requirement of BRIT1 in the repair of induced and spontaneous DSBs in many contexts over the past decade, there is yet to be a comprehensive study on its role in CSR, a process that represents one of the few physiological instances wherein DSBs are deliberately introduced into the mammalian genome. Here, we identify BRIT1 in a targeted short hairpin RNA (shRNA) screen to identify novel effectors of CSR and demonstrate that its loss impairs CSR and increases frequency of B cells with unresolved Igh breaks.
Results
BRIT1 Promotes CSR in CH12 Cells.
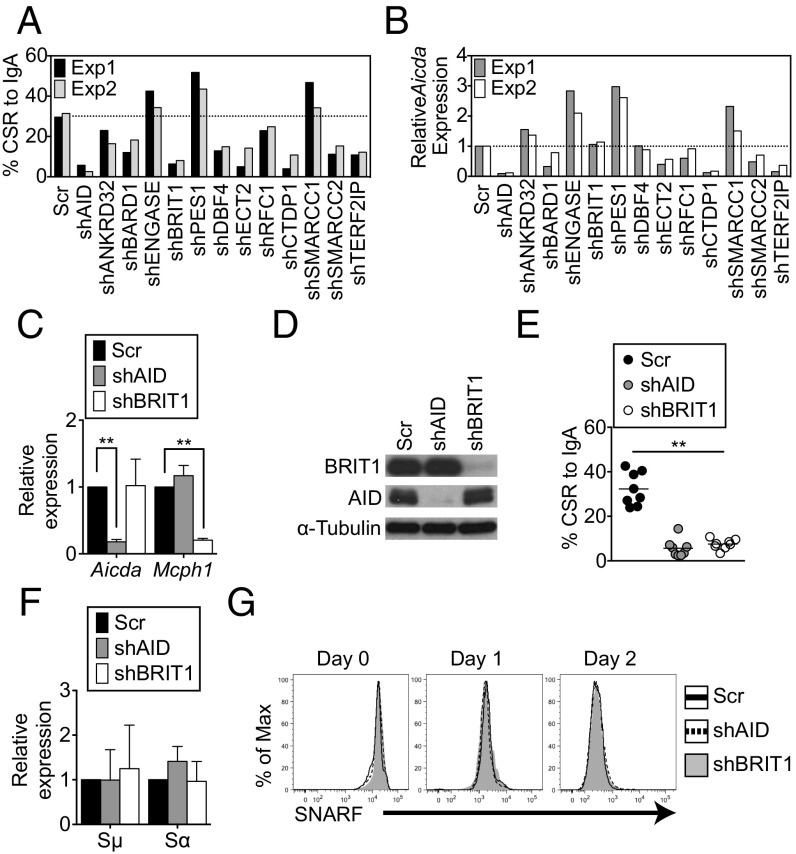
shRNAs against mRNAs encoding 12 known BRCT domain-containing proteins (Table S1) with no reported activity in CSR were individually transduced into CH12 cells, a murine B-lymphoma line that undergoes CSR from IgM to IgA in response to a combination of anti-CD40, interleukin-4 (IL-4), and transforming growth factor β (TGF-β). CSR to IgA was assessed relative to cells transduced with a scrambled shRNA by quantification of cells expressing surface IgA using flow cytometry (Fig. 1A). Consistent with its essential role in instigating DSBs (31, 32), shRNAs against AID led to a marked reduction in CSR (Fig. 1A). Among the 12 candidates examined, shRNAs against nine impaired CSR, and of these shRNAs, only two, shDBF4 and shBRIT1, reduced CSR without markedly altering AID expression (Fig. 1 A and B and Fig. S1). DBF4 participates in initiation of DNA replication (33), and the CSR defect in shDBF4 cells likely reflects impaired proliferation. Because loss of BRIT1 leads to defects in DNA repair during meiotic recombination and increased genomic instability (27), we decided to investigate its role in CSR.
Table S1.
List of qPCR primers and shRNA clones for target genes
| Target | Primer orientation | qPCR primers | shRNA (Sigma) |
| Ankrd32 | F | CACTGAGCCACTCTCTCTTCA | TRCN0000085691 |
| R | GTTTTGCACCATTCAGCATTAGT | ||
| Bard1 | F | AAGGAGCCCGTGTGCTTAG | TRCN0000084437 |
| R | TTGCCCTAGATGTGTTGTCTTTT | ||
| Engase | F | CGGAGGATCAACGAGGATCAA | TRCN0000215757 |
| R | TCGACAGGTAAAAGCTGATAGGT | ||
| BRIT1 | F | TCTGAAAGATGTTGTGGCCTATG | TRCN0000242053 |
| R | GGTCACTTGCTTGTTCAAGGTT | ||
| Pes1 | F | TGCCACCAATTATATCACCCGA | TRCN0000102593 |
| R | GGAGCTTCCGAACAAACACC | ||
| Dbf4 | F | AATAAGATACAGTGTCGGGTCCC | TRCN0000334124 |
| R | GTCCTTCTGGAAATTGGGCTC | ||
| Ect2 | F | AACTTGTGCTTGGCGTCTACT | TRCN0000190489 |
| R | TTCCTCCGATTTTCCAGGACA | ||
| Rfc1 | F | TGCAAGCTGACTCTCCTAAAGA | TRCN0000111302 |
| R | CTACTGCGGTTGCCATGTG | ||
| Ctdp1 | F | AGCACTTCTGGTGAGATTGGA | TRCN0000305855 |
| R | ACCATCAACTCCGGCACAC | ||
| Smarcc1 | F | AGCTAGATTCGGTGCGAGTCT | TRCN0000071389 |
| R | CCACCAGTCCAGCTAGTGTTTT | ||
| Smarcc2 | F | GCTGCCTACAAATTCAAGAGTGA | TRCN0000085542 |
| R | AGGAAAATGTTAGGTCGTGACAG | ||
| Terf2ip | F | CACACCCGAGGAAGACTCAGA | TRCN0000071311 |
| R | TCCGCCCCGACTCTAAGAAG | ||
| AID | F | GCCACCTTCGCAACAAGTCT | TRCN0000112032 |
| R | CCGGGCACAGTCATAGCAC |
Fig. 1.
BRIT1 promotes CSR in CH12 cells. (A) CSR to IgA was assessed in CH12 cells retrovirally transduced with shRNA against indicated target genes. The dotted line represents the average CSR in cells transduced with Scr shRNA (n = 2). Exp, experiment; Scr, scrambled shRNA control. (B) Quantitative real-time PCR analysis of relative levels of AID mRNA (aicda) in each shRNA knockdown experiment. (C) Quantitative real-time PCR analysis of relative mRNA levels of AID and BRIT1 in CH12 cells retrovirally transduced with the indicated shRNAs. (D) Immunoblot analysis of whole-cell extracts of AID or BRIT1 knockdown cells using antibodies against BRIT1, AID, or α-tubulin (loading control). (E) CSR to IgA in shBRIT1 or shAID transduced CH12 cells (n = 8; **P < 0.005). (F) Real-time qPCR analysis of relative levels of Igμ and Igα germline transcripts in CH12 cells depleted for the indicated proteins. (G) Cell proliferation was assessed by staining cells with carboxylic acid acetate, succinimidyl ester (SNARF). One representative flow cytometry plot of SNARF dilution at different time points in live singlet-stimulated CH12 cells is shown (n = 6).
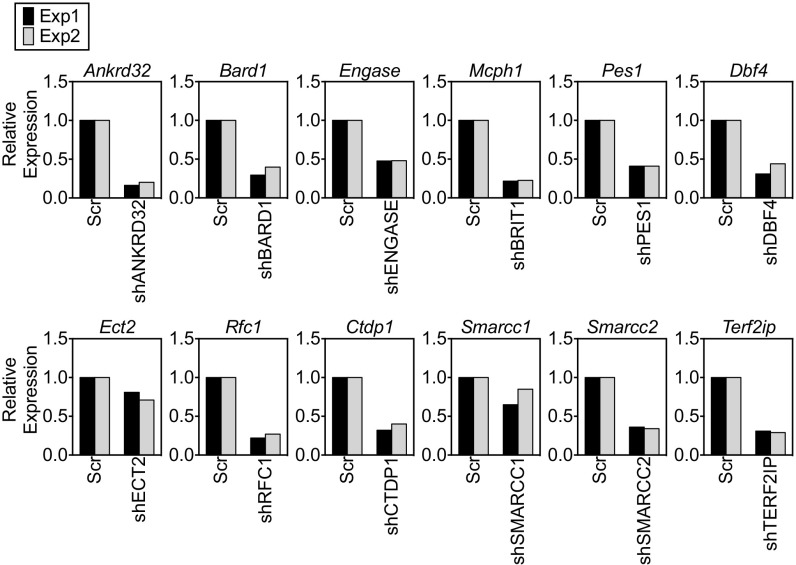
Fig. S1.
Summary of knockdown efficiency with different shRNAs. A real-time qPCR analysis of the relative level of the target gene mRNA for each shRNA used was performed. Scr, scrambled; Exp, experiment.
In multiple independent experiments, shBRIT1 efficiently depleted BRIT1 mRNA (Mcph1) (Fig. 1C) and protein (Fig. 1D) from CH12 cells and led to a severe and significant defect in CSR (∼32% in scrambled versus ∼7% in shBRIT1 cells; P < 0.0001; Fig. 1E). The CSR defect in BRIT1 knockdown CH12 cells approached the CSR defect observed upon AID depletion (Fig. 1E). Impaired CSR was not due to a defect in AID expression (Fig. 1 C and D), germline transcription through Igμ and Igα (Fig. 1F), or cellular proliferation (Fig. 1G). Collectively, these findings suggest that BRIT1 promotes CSR in CH12 cells.
BRIT1 Is Dispensable for B-Cell Development and Maturation.
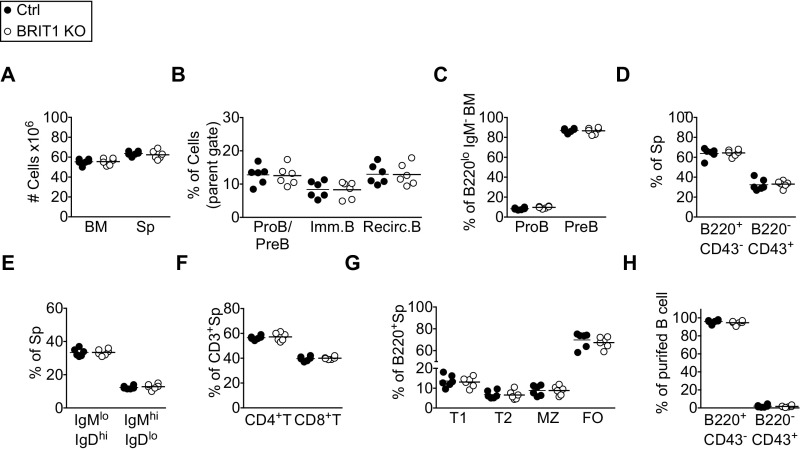
BRIT1-null mice are sterile, growth-retarded, and born at sub-Mendelian ratios (28). Thus, to explore the cell-intrinsic requirement of BRIT1 in B-cell development, BRIT1fl/fl mice (28) were bred to Mb1-Cre (Mb1Cre/+) mice in which Cre recombinase, driven by the Mb-1 promoter, is expressed specifically in B-lineage cells starting from pro-B cells in the bone marrow (34). Mb1Cre/+; BRIT1fl/fl mice (henceforth referred to as BRIT KO) and control mice (Mb1Cre/+; BRIT1+/+ or Mb1+/+; BRIT1fl/fl) had comparable cellularity in the bone marrow and spleen (Fig. S2A). In the bone marrow, the frequency of pro-B cells (B220lo IgM− CD43+), pre-B cells (B220lo IgM− CD43−), immature B cells (B220lo IgM+), and recirculating B cells (B220hi IgM+) was similar to control mice (Fig. S2 B and C). The spleens of BRIT1 KO mice had a normal frequency of B cells (B220+ CD43−) (Fig. S2D), and the frequency of mature (IgDhi IgM+) and immature (IgDlo IgM+) splenic B cells in BRIT1 KO mice was similar to control mice (Fig. S2E). As expected, B-cell–specific expression of the Cre recombinase did not perturb T-cell development in the BRIT1 KO mice (Fig. S2 D and F). Furthermore, the frequencies of splenic B-cell subsets (transitional-1, transitional-2, marginal zone, and follicular) were similar between control and BRIT1 KO mice (Fig. S2G). Thus, BRIT1 deficiency does not impair B-cell development and maturation in bone marrow and spleen.
Fig. S2.
BRIT1 is dispensable for B-cell development. (A) Cell number in bone marrow (BM) and spleen (Sp). (B) Frequency of pro-B (proB)/pre-B (preB), immature B (Imm. B), and recirculating B (Recirc. B) cells in BM from Mb1-Cre control and BRIT1 KO (n = 6) mice. Frequency represents live singlets in the lymphocyte gate. (C) Frequency of proB and preB cells in BM of Mb1-Cre control and BRIT1 KO mice (n = 6). (D) Frequency of B220+ CD43− and B220− CD43+ subsets in spleens of Mb1-Cre control and BRIT1 KO mice (n = 6). (E) Frequency of IgMlo IgDhi and IgMhi IgDlo subsets in spleens of Mb1-Cre control and BRIT1 KO (n = 6) mice. (F) Frequency of CD3+ CD4+ T-cell and CD3+ CD8+ T-cell subsets in spleens from Mb1-Cre control and BRIT1 KO mice (n = 6). (G) Frequency of transitional-1 (T1), transitional-2 (T2), marginal zone (MZ), and follicular (FO) B-cell subsets in spleens from Mb1-Cre control and BRIT1 KO mice (n = 6). (H) Frequency of B220+ CD43− and B220− CD43+ subsets in purified splenic B cells from Mb1-Cre control and BRIT1 KO mice.
Loss of BRIT1 Impairs CSR in Splenic B Cells.
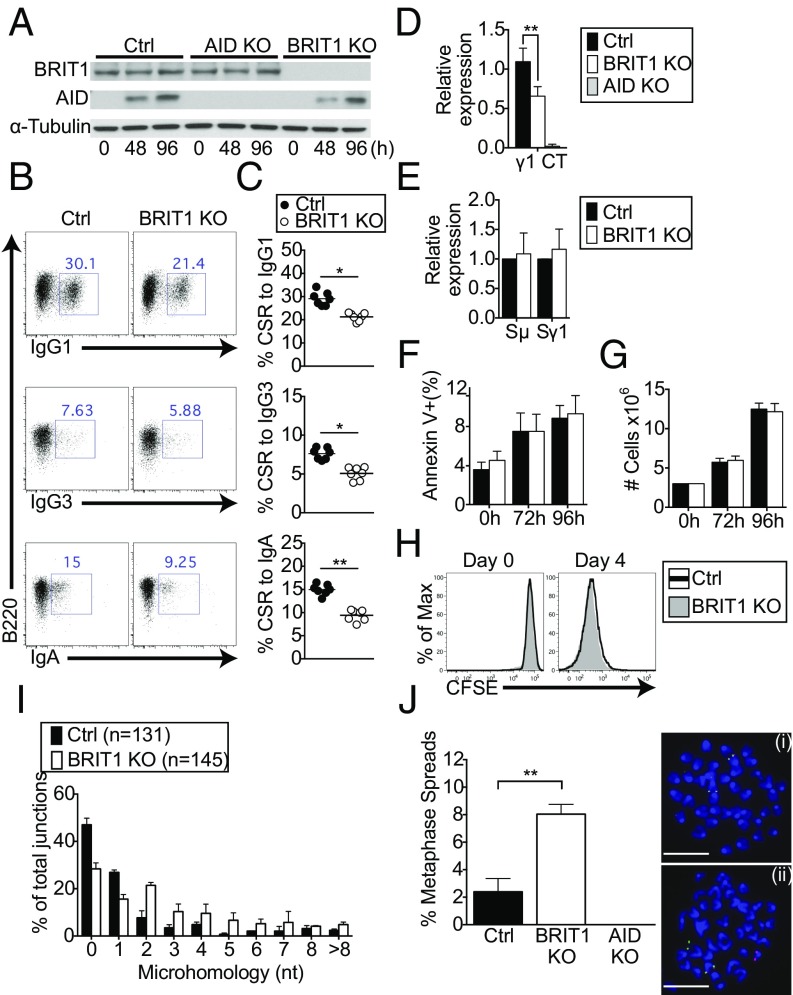
To delineate the role of BRIT1 in CSR, mature naive splenic B cells (B220+ CD43−) from wild-type control and BRIT1 KO mice were purified (Fig. S2H) and stimulated ex vivo with LPS + IL-4 to induce switching to IgG1. The relative abundance of BRIT1 did not change upon stimulation, and, confirming effective deletion of the floxed allele, BRIT1 KO cells had no detectable BRIT1 protein (Fig. 2A). When CSR was assessed by surface expression of IgG1 at 96 h poststimulation, BRIT1 KO B cells were found to be significantly impaired in their ability to undergo CSR to IgG1 relative to control B cells (∼30% for control versus ∼21% for BRIT1 KO; Fig. 2 B and C). Decreased CSR in BRIT1-deficient splenic B cells was also evident in the levels of Iγ1–Cμ circle transcripts derived from the looped-out extrachromosomal DNA generated as a consequence of recombination between Sμ and Sγ1 (35) (Fig. 2D). The defect in CSR was not due to altered AID expression (Fig. 2A), germline transcription (Fig. 2E), apoptosis (Fig. 2F), or B-cell proliferation, as measured by both cell counts (Fig. 2G) and dilution of the cell surface stain carboxyfluorescein succinimidyl ester (Fig. 2H). Finally, to assess CSR to IgG3 and IgA, splenic B cells were cultured ex vivo with LPS or LPS + TGF-β + anti–IgD-dextran, respectively. Similar to what was observed for CSR to IgG1, BRIT1 deficiency significantly impaired class switching to IgG3 and IgA (Fig. 2 B and C). Overall, these findings strongly suggest that BRIT1 positively regulates CSR in ex vivo cultures of stimulated splenic B cells without affecting AID expression, germline transcription, or proliferation.
Fig. 2.
CSR is compromised in BRIT1-deficient splenic B cells. (A) Immunoblot analysis of BRIT1 and AID in whole-cell lysates of splenic B-cells from Mb1-Cre control (Ctrl) and BRIT1 KO mice (0 h, naive; 48 h and 96 h, LPS + IL-4 stimulation). (B) Representative flow cytometry of CSR to IgG1, IgG3, and IgA in splenic B cells from Mb1-Cre Ctrl and BRIT1 KO mice. (C) Quantification of CSR to IgG1, IgG3, and IgA in splenic B cells from Mb1-Cre Ctrl and BRIT1 KO mice. (n = 7; **P < 0.005). (D) Iμ–Cγ1 “circle” transcripts from looped out extrachromosomal DNA in splenic B cells activated with LPS + IL-4 were assessed by real-time qPCR. **P < 0.005. (E) Real-time qPCR analysis of relative levels of Igμ and Igγ1 germline transcripts in splenic B cells activated with LPS + IL-4 at 48 h. (F) Cell viability of LPS + IL-4 activated splenic B cells was assessed by annexin V staining. (G) Cell number at different times following stimulation of splenic B cells with LPS + IL-4 was determined by counting in a hemocytometer. (H) Cell proliferation was assessed following staining with carboxyfluorescein succinimidyl ester (CFSE). Representative flow cytometry plot of CFSE dilution at different time points in live singlet splenic B cells stimulated with LPS + IL-4 (n = 6). Max, maximum. (I) BRIT1 KO and Ctrl B cells were stimulated with LPS + IL-4 for 72 h, genomic DNA was amplified by PCR, and Sμ–Sγ1 junctions were sequenced. The percentage of junctions with the indicated nucleotide overlap is indicated (131 Ctrl and 145 BRIT1 KO sequences were analyzed from three independent experiments). (J, Left) Splenic B cells were treated with colcemid at 72 h after LPS + IL-4 stimulation, and metaphase spreads were hybridized with probes specific for sequences upstream of the Igh variable domain (5′ Igh, labeled for green signal) and sequences immediately downstream of the Igh constant region exons (3′ Igh, labeled for red signal), and then counterstained with DAPI (blue). The percentage of metaphases with split signals was quantified. At least 200 metaphases for each group were analyzed in four independent experiments. **P < 0.005. Error bars represent means ± SD. (J, Right) Representative examples show signal from an intact Igh locus (i) and Igh break (ii). (Scale bar, 1.5 μm.)
BRIT1 Influences Resolution of DSBs During CSR.
To explore the possibility that BRIT1 might influence the repair of DSBs in B cells undergoing CSR, we analyzed the nature of S junctions. The extent of microhomology between recombining S sequences has been used as an effective parameter to assess end-joining pathways in switching B cells (12). In wild-type B cells, the majority of junctions are either blunt or have 1- to 3-bp microhomology, a bias that is largely altered in cells with defects in NHEJ proteins (12). To assess end-joining pathways mediating CSR, we cloned and sequenced Sμ–Sγ1 junctions from B cells stimulated with LPS + IL-4. We observed that BRIT1 KO B cells have fewer S junctions that are either blunt or have single-nucleotide microhomology (Fig. 2I). This alteration suggested an aberration in the end-joining phase of CSR. To substantiate this notion, we performed two-color fluorescence in situ hybridization (FISH) on metaphase spreads from activated splenic B cells with probes specific for sequences upstream and downstream of Igh (36). Although nearly all metaphases from AID KO B cells showed colocalization of the two Igh probes, split signals (separated red and green signals) were significantly higher in BRIT1 KO B cells compared with control B cells (∼8% versus ∼2.4%, n = 4; P < 0.001; Fig. 2J). These results demonstrate that a substantial subset of Igh DSBs was left unrepaired in the absence of BRIT1. Thus, BRIT1 appears to influence CSR by promoting the resolution of AID-induced DSBs.
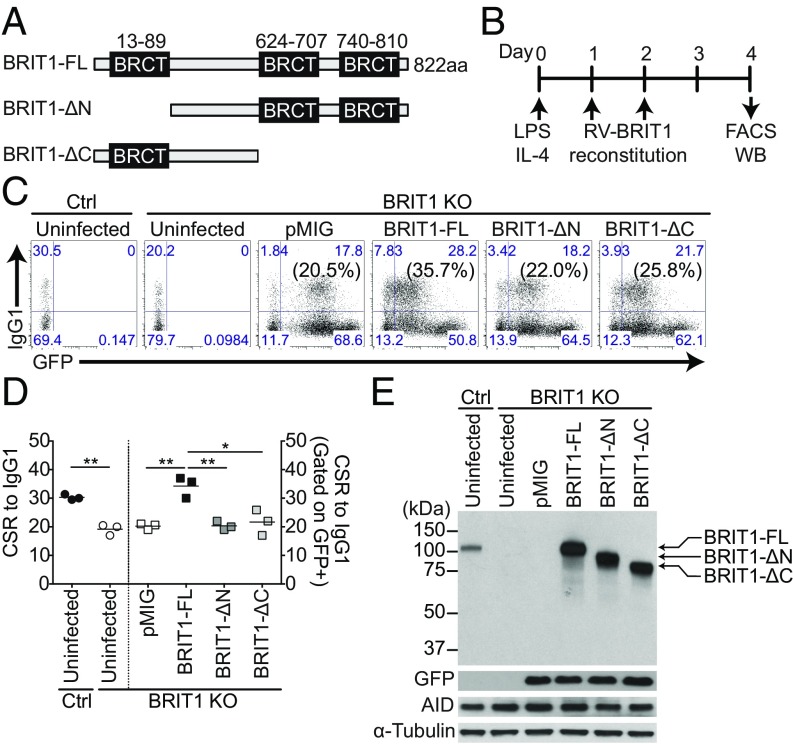
BRCT Domains of BRIT1 Are Required for Efficient CSR.
The influence of BRIT1 on the DNA repair phase of CSR suggested that the BRCT domains might play a crucial role in this process. The correlation between BRCT domains, per se, and molecular functions in CSR is not straightforward. For example, the N terminus BRCT domain of PTIP is required for CSR, whereas the C terminus BRCT repeats of 53BP1 are dispensable (37, 38). BRIT1 contains an N-terminal BRCT domain and two C-terminal BRCT domains (Fig. 3A). To determine the critical BRCT domains that influence CSR, cDNAs encoding full-length BRIT1 (BRIT1-FL), N-terminal BRCT-truncated BRIT1 (BRIT1-ΔN) deletions, or C-terminal BRCT-truncated (BRIT1-ΔC) deletions were cloned into the retroviral vector pMIG (Fig. 3A). These pMIG-BRIT1 constructs were individually transduced into BRIT1 KO splenic B cells activated with LPS + IL-4, and CSR to IgG1 was assessed (Fig. 3B). Although BRIT-FL restored CSR frequency to wild-type levels, neither BRIT1-ΔN nor BRIT1-ΔC could rescue CSR in BRIT1 KO cells to a level higher than observed with pMIG alone (Fig. 3 C and D). Immunoblot analysis showed that the different BRIT1 protein domains, as well as AID, were expressed at similar levels (Fig. 3E). These findings suggest that both the N-terminal and C-terminal BRCT domains of BRIT1 are required for optimum CSR.
Fig. 3.
BRCT domains of BRIT1 influence CSR. (A) Schematic of BRIT1 protein with N-terminal and C-terminal BRCT domains and various deletions. ΔC, C-terminal BRCT domain deletion; ΔN, N-terminal BRCT domain deletion. (B) Schematic of retroviral reconstitution of BRIT1 in ex vivo activated splenic B cells. Splenic B cells were harvested from control and BRIT1 KO mice, activated with LPS + IL-4, and retrovirally reconstituted with BRIT1 variants from pMIG vector, which also expresses GFP as a marker. WB, Western blot. (C) Representative flow cytometry of CSR to IgG1 in activated splenic BRIT1 KO B cells retrovirally transduced with vector alone (pMIG), BRIT1-FL, BRIT1-ΔN, or BRIT1-ΔC. The vectors express GFP (numbers in parentheses indicate IgG1+ cell in the GFP+ gate). Uninfected activated splenic B cells from Mb1-Cre control and BRIT1 KO mice served as negative controls for GFP gating (n = 3). (D) Quantification of the percentage of CSR to IgG1 in splenic B cells from Mb1-Cre control or BRIT1 KO mice retrovirally reconstituted with BRIT1 variants or left uninfected (n = 3; **P < 0.005, *P < 0.05). (E) Representative immunoblot analysis of BRIT1 and AID in transduced cells.
BRIT1 Interacts with γ-H2AX in B Cells Undergoing CSR.
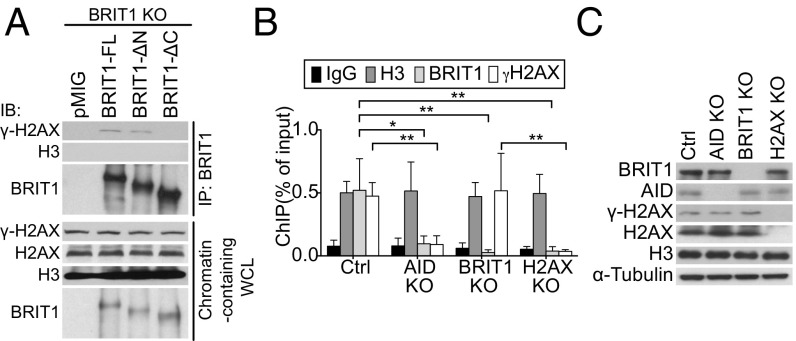
In fibroblasts and in cell lines treated with IR, BRIT1 colocalizes with γ-H2AX foci, and this interaction is dependent on its C terminus tandem BRCT domains (21, 22). To examine if the interaction between BRIT1 and γ-H2AX could be detected in splenic B cells, we immunoprecipitated BRIT1 from BRIT1 KO splenic B cells retrovirally transduced with BRIT1-FL. Phosphorylated H2AX (γ-H2AX), but not histone H3, was readily detected in the immunoprecipitated fraction (Fig. 4A). Additionally, while BRIT1-ΔN could interact with γ-H2AX, deletion of the C terminus BRCT domains abolished the interaction (Fig. 4A). Taken together, these results indicated that in activated B cells, the tandem C terminus BRCT domains of BRIT1 facilitate its interaction with γ-H2AX.
Fig. 4.
BRIT1 interacts with γ-H2AX. (A) BRIT1-FL or BRIT1 deleted for BRCT domains was immunoprecipitated, and the immunoprecipitate was analyzed by immunoblotting with the indicated antibodies. WCL, whole-cell lysates. (B) ChIP-qPCR analysis of H3, BRIT1, and γ-H2AX occupancy at the Sμ region in activated splenic B cells from Mb1-Cre control, AID KO, BRIT1 KO, and H2AX KO mice (n = 4; **P < 0.005, *P < 0.05). (C) Immunoblot analysis of whole-cell extracts from cells that were cross-linked for ChIP analysis.
The interaction between γ-H2AX and BRIT1 in splenic B cells prompted us to explore if BRIT1 associates with S regions in an H2AX-dependent fashion. We performed chromatin immunoprecipitation (ChIP) experiments with BRIT1 antibodies to assess BRIT1 localization at the Igh locus. Antibodies against histone H3 and nonspecific IgG were used as positive and negative controls, respectively. ChIP-quantitative PCR (qPCR) analysis demonstrated that BRIT1 was significantly enriched at Sμ compared with the IgG control and not detected in BRIT1 KO B cells (Fig. 4B). As expected, γ-H2AX was detected at Sμ in control cells but not in AID-deficient B cells (Fig. 4B). Additionally, loss of BRIT1 did not impair occupancy of γ-H2AX at Sμ (Fig. 4B) in keeping with the notion that γ-H2AX foci are not regulated by BRIT1 (24, 30). Significantly, BRIT1 was not bound to S region DNA in either AID-deficient or H2AX-deficient B cells (Fig. 4B), even though BRIT1 protein levels were unaltered in these mutants (Fig. 4C). Taken together, these results strongly suggest that the C terminus BRCT domains of BRIT1 facilitate its recruitment to AID-instigated DSBs at S regions via interaction with γ-H2AX.
Multiple BRCT-Domain Proteins Influence CSR.
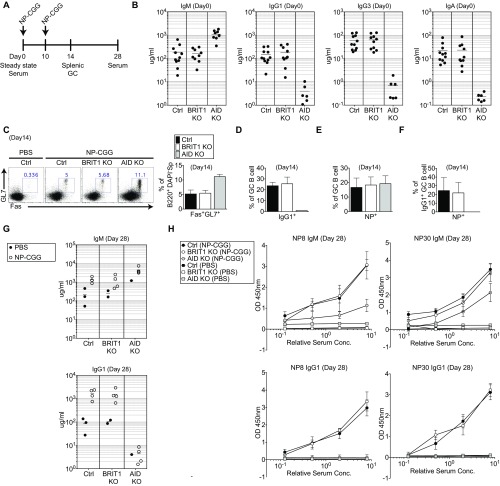
To examine how the defect in DSB resolution in an ex vivo setting is manifested during an immune response in vivo, BRIT1 KO mice were challenged with the model antigen 4-hydroxy-3-nitrophenylacetyl-chicken gamma globulin (NP-CGG) (Fig. S3A). In contrast to the CSR defect observed in splenic B cells activated in culture, no marked difference in serum Ig levels (Fig. S3B), frequency of NP-specific germinal center B cells (Fig. S3 C–E), or NP-specific serum Ig levels (Fig. S3 G and H) were observed. This lack of correlation between the ex vivo and in vivo results suggests the existence of compensatory mechanisms en route to mounting an effective immune response. To explore potential compensatory mechanisms in BRIT1 KO B cells, we used the ex vivo B-cell culture system as a convenient platform for molecular dissection of CSR.
Fig. S3.
BRIT1 is dispensable for immune response in vivo. (A) Schematic of NP-CGG immunization protocol. GC, germinal center. (B) IgM, IgG1, IgG3, and IgA concentrations in serum, determined by ELISA on day 0. Ctrl, control. (C) Flow cytometric analysis of GC B cells (B220+ DAPI− GL7+ Fas+) in control, BRIT1 KO, and AID KO mice following NP-CGG immunization on day 14 and quantification of GC B cells in immunized mice on day 14 (n = 3). (D) Quantification of IgG1+ GC B cells in immunized mice on day 14 (n = 3). (E) Quantification of NP+ GC B cells in immunized mice on day 14 (n = 3). (F) Quantification of IgG1+ NP+ GC B cells in immunized mice on day 14 (n = 3). (G) IgM and IgG1 concentrations in serum from Ctrl, BRIT1 KO, and AID KO mice following NP-CGG immunization on day 28, determined by ELISA. (H) Detection of low-affinity and high-affinity NP-specific IgM and IgG1 serum antibodies in Ctrl (n = 4), BRIT1 KO (n = 4), and AID KO (n = 4) mice following NP-CGG immunization on day 28 by ELISA. Conc., concentration.
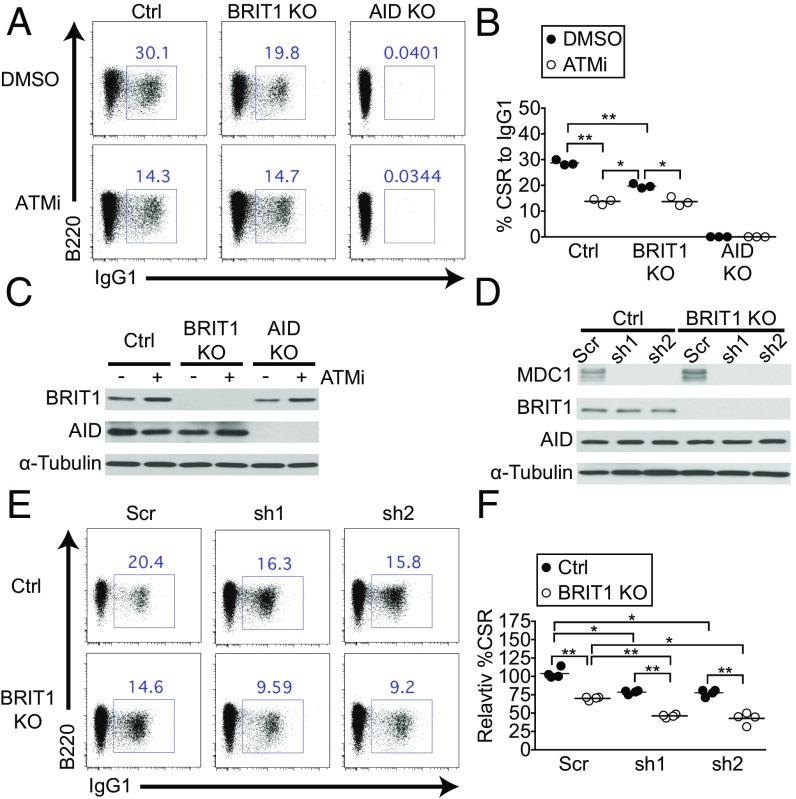
The DDR protein ATM is a major effector of CSR, in part, through its ability to phosphorylate H2AX. To explore the ATM–γ-H2AX–BRIT1 axis in CSR further, we treated splenic B cells with the small-molecule ATM inhibitor ATMi (39). In control cells, ATMi addition led to an ∼50% reduction in CSR to IgG1 over what was observed upon vehicle treatment without altering levels of BRIT1 or AID (Fig. 5 A–C). Inhibition of ATM in BRIT1 KO B cells also led to a significant reduction in CSR. However, loss of BRIT1 did not exacerbate the CSR defect imposed upon ATM inhibition (Fig. 5 A and B), suggesting that ATM and BRIT1 likely participate in overlapping pathways, with ATM acting upstream of BRIT1 by phosphorylating H2AX, and the γ-H2AX subsequently recruiting BRIT1 to DSBs. These results, along with the observation that the CSR defect in ATM-deficient B cells (40–42) is more severe than in BRIT1 KO B cells, lend support to the notion that a putative BRIT1-independent DSB resolution factor would function downstream of the ATM–γ-H2AX axis.
Fig. 5.
BRIT1 and MDC1 could both contribute to the ATM–γ-H2AX axis of DSB resolution during CSR. (A) Representative flow cytometry of CSR to IgG1 in splenic B cells derived from Mb1-Cre control, BRIT1 KO, and AID KO mice treated with ATMi or with DMSO vehicle control. (B) Quantification of CSR to IgG1 (n = 3; **P < 0.005, *P < 0.05). (C) Immunoblot analysis of whole-cell lysates from control, BRIT1 KO, or AID KO cells treated with ATMi or DMSO. (D) Immunoblot analysis of whole-cell lysates from control or BRIT1 KO cells transduced with two different MDC1 shRNAs (sh1 and sh2) or scrambled control. (E) Representative flow cytometry of CSR to IgG1 in splenic B cells of indicated genotypes expressing MDC1 knockdown shRNAs sh1 or sh2. (F) Quantification of CSR to IgG1 (n = 3; **P < 0.005, *P < 0.05).
Besides interacting with BRIT1, γ-H2AX also interacts with the BRCT-domain protein MDC1 to facilitate its recruitment to DSBs (22). In fact, the BRCT domains of both BRIT1 and MDC1 interact with γ-H2AX peptide with similar affinities (22), suggesting that BRIT1 and MDC1 could provide alternate mechanisms to the repair of DSBs downstream of the ATM–γ-H2AX axis (22, 43). We therefore sought to examine if BRIT1 and MDC1 could both contribute to CSR. We used several shRNAs to deplete MDC1 from control or BRIT1 KO splenic B cells (Fig. 5D and Fig. S4). Loss of MDC1 led to a significant yet modest reduction in CSR (Fig. 5 E and F), in keeping with what was observed in MDC1 KO B cells (8). However, loss of MDC1 in BRIT1 KO splenic B cells led to a significantly more profound defect in CSR than observed in B cells lacking either factor (Fig. 5 E and F and Fig. S4 B and C). These results suggest that MDC1 has BRIT1-independent activity in CSR and support the idea that both BRIT1 and MDC1 could process DSBs downstream of the ATM–γ-H2AX axis. The findings, in addition, raise the intriguing possibility that MDC1-mediated mechanisms could be engaged in the absence of BRIT1 in vivo, leading to compensation for DSB-resolving activity and lack of an observable phenotype in BRIT1 KO mice during an in vivo immune response. Further studies using genetic models will be required to evaluate how loss of both BRIT1 and MDC1 impacts an in vivo immune response.
Fig. S4.
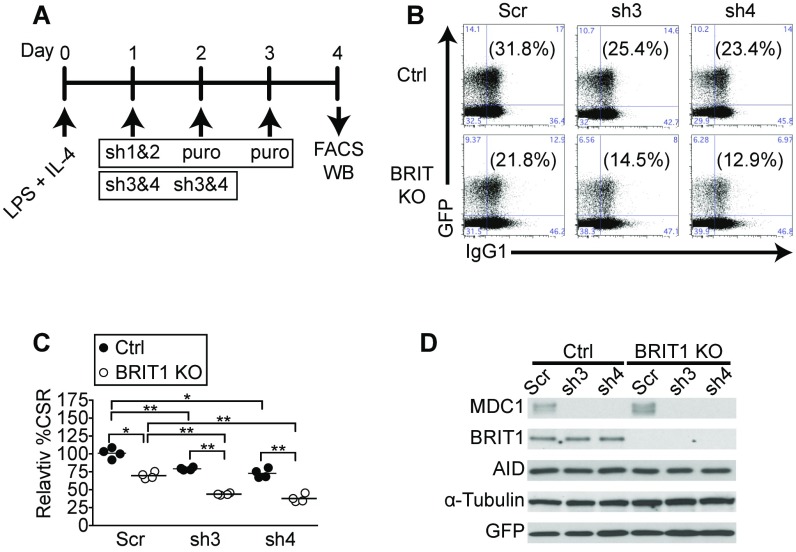
Depletion of MDC1 exacerbates CSR defect in BRIT1 KO B cells. (A) Strategy to deplete MDC1 in ex vivo stimulated splenic B cells. B cells were harvested from control and BRIT1 KO mice, activated with LPS + IL-4, and retrovirally infected with shRNA and a GFP marker (sh3 and sh4). WB, Western blot. (B) Representative flow cytometry of CSR to IgG1 from Mb1-Cre control and BRIT1 KO mice with MDC1 knockdown constructs sh3 and sh4. (C) Quantification of CSR to IgG1 (n = 3; **P < 0.005, *P < 0.05). (D) Immunoblot analysis of BRIT1, MDC1, and AID upon MDC1 depletion.
Discussion
In this work, we identified BRIT1 as an effector of the DDR response during CSR. Our results are consistent with a model wherein AID-instigated DSBs lead to ATM activation, H2AX phosphorylation, and association of BRIT1 at the Igh loci. Bound to Igh DSBs, BRIT1 could serve as a scaffold to recruit factors that resolve S region DSBs to complete CSR; the identity of such factors remains to be elucidated. Our results also demonstrate a previously underappreciated genetic interaction between BRIT1 and MDC1 in CSR. Although loss of either protein alone leads to a significant, albeit modest, reduction in CSR, loss of both proteins together markedly accentuates the defect. These results support the notion that the C-terminal BRCT domains of both BRIT1 and MDC1 could individually interact with γ-H2AX (22) and be recruited to S region DSBs during the resolution phase of CSR. Whether the DSBs processed by MDC1 and BRIT1 use distinct or overlapping sets of proteins to mediate ultimate end-joining is yet to be resolved.
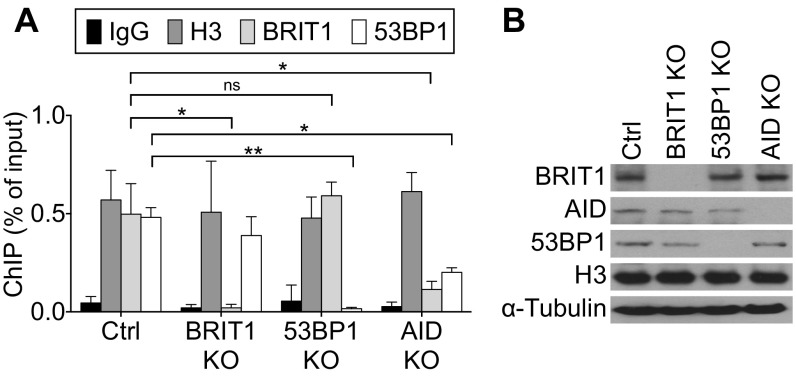
The DDR protein 53BP1 has been implicated in BRIT1-dependent DSB resolution following IR-induced damage; 53BP1 loss also leads to a profound CSR defect (8, 11, 19, 44–46). However, the reduction in levels of 53BP1 bound to S regions in BRIT1 KO B cells is modest and statistically insignificant (Fig. S5), suggesting either that BRIT1-independent mechanisms promote interaction of 53BP1 with DNA or that subtle changes in chromatin–protein interactions could not be evaluated in our ChIP assays. Interestingly, there are significant levels of 53BP1 bound to S regions in AID-deficient B cells (Fig. S5). This observation leaves open the possibility that BRIT1 with associated SWI/SNF might facilitate the recruitment of proteins, such as Rif1 (14), that are required for 53BP1 function in CSR. An additional layer to this complexity is derived from the observation that H2AX-dependent and H2AX-independent pathways exist for the recruitment of factors such as 53BP1 to execute efficient DSB repair (47, 48). This observation could also explain why the CSR defect in 53BP1-deficient B cells is more profound than the CSR defect observed upon loss of BRIT1 (44).
Fig. S5.
Recruitment of BRIT1 and 53BP1 to S region DNA is independent of each other. (A) ChIP-qPCR analysis of H3, BRIT1, and 53BP1 occupancy at the Sμ region in activated splenic B cells from Mb1-Cre control, BRIT1 KO, 53BP1 KO, and AID KO mice (n = 3; **P < 0.005, *P < 0.05). ns, not significant. (B) Immunoblot analysis of whole-cell extracts from cells that were cross-linked for ChIP analysis.
During V(D)J recombination, the RAG proteins remain bound to the DSBs as a postsynaptic complex and shepherd the lesions in such a fashion that end-joining is almost entirely reliant on NHEJ (49). On the other hand, AID, per se, has no reported role in enforcing a specific end-joining pathway during the resolution of DSBs. Although AID can directly promote recruitment of factors such as RPA that protect DNA ends (50, 51), in all likelihood, AID-instigated DSBs can be processed by multiple factors and parallel pathways, including BRIT1 and MDC1. Additional work would be required to delineate the BRIT1-reliant and BRIT1-independent pathways that can resolve Igh breaks to promote CSR and suppress genomic instability.
Materials and Methods
Animals.
Aicda−/− mouse strain (AID KO) was a kind gift of Tasuku Honjo, University of Kyoto, Kyoto, Japan (31). Other mice used in the study, including Mb1Cre/Cre, Mb1 (for breeding), Mb1Cre/+ (for controls), H2AXfl/fl, and Mcph1fl/fl, have been described previously (28, 34, 52). The care and use of mice were performed with the approval of the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee in accordance with institutional guidelines.
Real-Time Quantitative PCR.
To assess germline (switch) transcripts and circle transcripts (10), total RNA was extracted with TriZOL reagent (Invitrogen) from CH12 cells or primary B cells and cDNA was synthesized using SuperScriptIII reverse transcriptase (Invitrogen), followed by real-time PCR analysis (SYBR green; BioRad). Values were normalized to β-actin mRNA. Primers are listed in Table S2. Knockdown efficiency of shRNA in CH12 cells was determined by RT-qPCR analysis, normalized to β-actin mRNA as described previously (54). Primers for target genes are listed in Table S1.
Table S2.
qPCR primer list
| RNA examined | Primer orientation | qPCR primers |
| μ-GLT | F | 5′-TAGTAAGCGAGGCTCTAAAAAGCAT-3′ |
| R | 5′-AGAACAGTCCAGTGTAGGCAGTAGA-3′ | |
| γ1-GLT | F | 5′-GGCCCTTCCAGATCTTTGAG-3′ |
| R | 5′-GGATCCAGAGTTCCAGGTCAC T-3′ | |
| α-GLT | F | 5′-GACATGATCACAGGCACAGG-3′ |
| R | 5′-TTCCC CAGGTCACATTCATCGT-3′ | |
| γ1-CT | F | 5′-TCGAGAAGCCTGA-GGAATGTG-3′ |
| R | 5′-GAAGACATTTGGGAAGGACTGACT-3′ | |
| β-actin | F | 5′-TGCGTGACATCAAAGAGAAG-3′ |
| R | 5′-CGGATGTCAACGTCACACTT-3′ |
F, forward; GLT, germline (switch) transcript; R, reverse.
B-Cell Purification and Retroviral Infection in Splenic B Cells.
Splenic B cells were purified from 8- to 10-wk-old mice by negative selection using anti-CD43 magnetic beads (Miltenyi Biotec) according to the manufacturer’s protocol. A total of 3 × 106 B cells were plated at a density of 1 × 106 cells per milliliter and stimulated with 30 μg/mL LPS (Sigma) plus 25 ng/mL mouse IL-4 (R&D Systems). At 24 h and 48 h poststimulation, cells were layered with retroviral supernatants generated and centrifuged at 2,000 × g for 90 min at 32 °C, after which viral supernatants were aspirated and fresh B-cell media plus LPS and IL-4 were added. B cells were harvested at 96 h for flow cytometry analysis or to prepare lysates for Western blotting. Retrovirus supernatant was prepared by cotransfecting pMIG or pMIG-BRIT1 with packaging vector pCL-Eco into HEK293T cells using calcium phosphate. In MDC1 knockdown experiments, retrovirus supernatant was prepared by pSuperior.retro.puro MDC1 shRNAs (sh1 and sh2) or pGFP-V-RS mouse MDC1 shRNAs (sh3 and sh4) into Phoenix cells. The shRNAs (sh1 and sh2) against mouse MDC1 (53) were obtained from Titia de Lange, Rockefeller University, New York. For MDC1 shRNA (sh1 and sh2) knockdown experiments, puromycin (Sigma) at a concentration of 2.5 μg/mL was added to the culture 24 h after retroviral infection. The shRNAs (sh3 and sh4) against mouse MDC1 were obtained from Origene (TG517490). Target sequences are listed in Table S3.
Table S3.
shRNA sequences against mouse MDC1 and scramble control
| Target | Sequence |
| Mouse MDC1 sh1 | 5′-ACAGCATGCAGTAATTGAA-3′ |
| Mouse MDC1 sh2 | 5′-ACACAGCCGTTCTGTCTAA-3′ |
| Mouse MDC1 sh3 | 5′-GCCGCTTGAGTTGCCAGACGACACCTGCT-3′ |
| Mouse MDC1 sh4 | 5′-GTAGGTCTGCTGTCAAGACTCCTGAAGCG-3′ |
| Scramble for sh1 and sh2 | 5′-GCGAAAGATGATAAGCTAA-3′ (Addgene plasmid no. 30520) |
| Scramble for sh3 and sh4 | 5′-GCACTACCAGAGC TAACTCAGATAGTACT-3′ |
ChIP.
ChIP was performed as described elsewhere (55) with 1 μg of antibodies to H3 (no. 1791; Abcam), γ-H2AX (07-164; Millipore), 53BP1 (Novus Biologicals), and BRIT1 (no. 4120; Cell Signaling Technology and no. 2612; Abcam). Splenic B cells from 8- to 10-wk-old mice were isolated by CD43− selection (Miltenyi Biotec) and stimulated with LPS + IL4 for 48 h. Relative abundance of regions of interest in ChIP DNA was measured by qPCR using Power SYBR-Green PCR master mix (BioRad). Primers for Sµ were as follows: forward, 5′-TAGTAAGCGAGGCTCTAAAAAGCAT-3′, and reverse, 5′-AGAACAGTCCAGTGTAGGCAGTAGA-3′.
Two-Color FISH Analysis.
Two-color FISH was performed as described elsewhere (36). The 3′ end of Igh was detected using BAC199, and the 5′ end was detected using BAC207, a gift from the laboratory of Frederick W. Alt, Howard Hughes Medical Institute, Boston Children’s Hospital, Boston, MA. Chromosome spreads from splenocytes were performed as previously described (36). Briefly, cells were incubated in 50 ng/mL colcemid (KaryoMAX; GIBCO), harvested, hypnotically swollen with 0.075 mol/L KCl for 15 min at 37 °C, fixed, washed in ice-cold 3:1 methanol/acetic acid, and dropped on slides. Slides were denatured at 75 °C for 4 min, hybridized with probes overnight at 37 °C, washed, dehydrated, stained with DAPI, and mounted with VECTASHIELD (Vector Laboratories). Images were captured using a DeltaVision Elite Imaging System with a customized Olympus IX71 microscope stand (General Electric). More than 200 spreads were scored for each genotype.
SI Materials and Methods
Reagents, Cytokines, and Flow Cytometry.
For CSR assays, LPS (30 μg/mL, no. L4130; Sigma), mouse IL-4 (25 ng/mL), TGF-β (2 ng/mL, no. 240-B; R&D Systems), and anti-IgD dextran conjugated (300 ng/mL, no. 0001; Fina Biosolution) in different combinations were used to stimulate B cells ex vivo. For ATM inhibition, 2.5 μM ATMi Ku55933 was added to the cell culture. CH12 cells were stimulated with anti-CD40 (1 μg/mL; eBioscience), mouse IL-4 (12.5 ng/mL), and TGF-β (0.1 ng/mL; R&D Systems). Fluorophore-conjugated antibodies were purchased from BD Biosciences and eBioscience. Stained cells were analyzed using an LSRII flow cytometer (BD Biosciences), and data were analyzed using FlowJo software (TreeStar). Flow cytometric analysis of carboxyfluorescein succinimidyl ester (CFSE) staining was performed according to the manufacturer’s instructions (CellTrace CFSE Cell Proliferation Kit Protocol; Invitrogen). For flow cytometric analysis of carboxylic acid acetate, succinimidyl ester (SNARF) labeling, CH12 cells were stained with 9 μM SNARF in 5% (vol/vol) FBS for 10 min at 37 °C as described previously (54).
Knockdown with Lentiviral shRNA in CH12 Cell Line.
CH12 cells were cultured in RPMI 1640 supplemented with 10% (vol/vol) FBS, 5% (vol/vol) NCTC-109 (Gibco), 10 mM β-mercaptoethanol, 100 U of penicillin per milliliter, and 0.1 mg/mL streptomycin. Lentiviral transduction of CH12 cells with shRNAs was performed as described previously (56). Scrambled shRNA in pLKO.1 was obtained from Addgene (plasmid 1864). All other shRNAs in pLKO.1 were from Sigma. Hairpin sequences are listed in Table S1.
NP-CGG Immunization.
NP-CGG (no. N-5055D-5; Biosearch Technologies) was precipitated with 10% alum. Mice aged 8 to 10 wk were immunized intraperitoneally with 50 μg on day 0 and boosted with 50 μg on day 10. Serum was collected on days 0 and 28. To analyze CSR in vivo, mice were immunized intraperitoneally with 50 μg on day 0 and boosted with 50 μg on day 10. CSR was assessed by flow cytometry at day 14.
ELISA.
Coating antibodies for binding IgM, IgG1, IgG3, and IgA were purchased from Southern Biotechnology Associates (nos. 1020-01, 1070-01, 1100-01, and 1040-01, respectively). Coating antibody for detecting IgE was purchased from BD Pharmingen (no. 553413). The following isotype standards were used to calculate absolute concentration values: IgM (no. 14-4752-81; eBioscience), IgG1 (no. 0102-01; Southern Biotechnology Associates), IgG3 (no. 553486; BD Pharmingen), and IgA (no. 553478; BD Pharmingen). Secondary antibodies for detecting IgM, IgG1, IgG3, and IgA were purchased from Southern Biotechnology Associates (nos. 1020-05, 1070-05, 1100-05, and 1040-05). BSA-conjugated NP (8) (no. N-5050l-10) and NP (30) (no. N-5050H-10) were obtained from Biosearch Technologies. Plates were read at 450 nm on a Biotek Synergy HT detector.
Western Blot Analysis.
Protein extracts from primary or sorted GFP+ splenic B cells were prepared using Nonidet P-40 lysis buffer (20 mM Tris at pH 8, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) plus protease inhibitors. Immunoblotting was done with antibodies for H2AX (Cell Signaling Technologies), γ-H2AX (Bethyl Laboratories), AID (57), α-tubulin (Sigma), BRIT1 (Cell Signaling Technologies), MDC1 (Novus Biologicals), GFP (Thermo Fisher), and 53BP1 (Novus Biologicals).
Preparation of Chromatin-Containing Whole-Cell Lysates and Immunoprecipitation.
Cell pellets were lysed and sonicated to elute whole-cell lysate plus chromatin in NETN buffer [100 mM NaCl, 20 mM Tris⋅Cl at pH 8.0, 0.5 mM EDTA, 0.5% (wt/vol) Nonidet P-40]. The lysates were centrifuged at 15,000 × g for 20 min, and the supernatant was collected as cell-free extracts. Extracts (0.5 mg) were precleared with protein A agarose beads and incubated with anti-BRIT1 antibodies (1.5 μg, #4120 Cell Signaling Technology and 1.5 μg, #2612 Abcam) at 4 °C overnight and then with protein A agarose beads for another 90 min. The immunoprecipitate was washed twice with low-salt buffer (150 mM NaCl), once with high-salt buffer (500 mM NaCl), and then with 1× Tris-buffered saline. The beads were eluted with 2× SDS sample loading buffer, and analyzed by Western blotting.
Microhomology Analysis.
Genomic DNA was prepared from splenic B cells stimulated with IL-4 plus LPS for 72 h. Sµ–Sγ1 junction DNA was amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England Biolabs). Amplification conditions were 10 cycles at 94 °C (10 s), 60 °C (30 s), and 68 °C (1 min), and 20 cycles at 94 °C (10 s), 60 °C (30 s), and 68 °C (1 min and 20 s per cycle). The PCR products were cloned into Zero Blunt TOPO (Invitrogen). DNA from individual clones was sequenced with T7 and Sp6 primer at the Memorial Sloan Kettering Cancer Center DNA Sequencing Core Facility. Sequence analysis was performed using Vector NTI (Invitrogen). Primers for the Sµ–Sγ1 junction were 5μ3 (5′-AATGGATACCTCAGTGGTTTTTAATGGTGGGTTTA-3′) and γ1-R (5′-CAATTAGCTCCTGCTCTTCTGTGG-3′).
Statistical Methods.
Statistical analysis was performed using a two-tailed, unpaired Student’s t test on individual biological replicates in Prism (GraphPad). P values less than 0.05 were considered significant. All error bars represent SD from the mean.
Acknowledgments
We thank members of the J.C. and A.Y.R. laboratories for technical assistance and discussion; A. Bravo and M. Cols for help with the mouse colony; V. Kumar for H2AXfl/fl mice and Igh probes; R. Panchakshari for 53BP1 KO spleens; S. Hemmers for Mb1-Cre mice; C. C. Chen for ATMi; and K. TaKai and T. de Lange for MDC1 shRNAs. This work was supported by NIH Grants 1RO1AI072194, 1RO1AI124186, 5U54CA137788 (to J.C.), and R01CA155151 (to K.L.); NIH/National Cancer Institute Cancer Center Support Grant P30 CA008748 (to J.C. and A.Y.R.); the Hilton-Ludwig Cancer Prevention Initiative (Conrad N. Hilton Foundation and Ludwig Cancer Research) (A.Y.R.); the Howard Hughes Medical Institute (A.Y.R.); and NIH T32 Training Grant 4T32CA009149-40 (to W.T.Y.). J.C. was also supported by grants from the Ludwig Center at Memorial Sloan Kettering Cancer Center (MSKCC), the Functional Genomics Institute at MSKCC, and the Geoffrey Beene Cancer Center at MSKCC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708211114/-/DCSupplemental.
References
- 1.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casellas R, et al. Mutations, kataegis and translocations in B cells: Understanding AID promiscuous activity. Nat Rev Immunol. 2016;16:164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenter AL, Kumar S, Wuerffel R, Grigera F. AID hits the jackpot when missing the target. Curr Opin Immunol. 2016;39:96–102. doi: 10.1016/j.coi.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarrin AA, Goff PH, Senger K, Alt FW. Sgamma3 switch sequences function in place of endogenous Sgamma1 to mediate antibody class switching. J Exp Med. 2008;205:1567–1572. doi: 10.1084/jem.20080451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung CC, Glover JN. BRCT domains: Easy as one, two, three. Cell Cycle. 2011;10:2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerloff DL, Woods NT, Farago AA, Monteiro AN. BRCT domains: A little more than kin, and less than kind. FEBS Lett. 2012;586:2711–2716. doi: 10.1016/j.febslet.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kracker S, et al. Nibrin functions in Ig class-switch recombination. Proc Natl Acad Sci USA. 2005;102:1584–1589. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Matthews AJ, Husain S, Chaudhuri J. Binding of AID to DNA does not correlate with mutator activity. J Immunol. 2014;193:252–257. doi: 10.4049/jimmunol.1400433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc Natl Acad Sci USA. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 13.Zan H, et al. Rev1 recruits ung to switch regions and enhances du glycosylation for immunoglobulin class switch DNA recombination. Cell Rep. 2012;2:1220–1232. doi: 10.1016/j.celrep.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong J, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin SY, Elledge SJ. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113:881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 17.Faheem M, et al. Molecular genetics of human primary microcephaly: An overview. BMC Med Genomics. 2015;8(Suppl 1):S4. doi: 10.1186/1755-8794-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson AP, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin SY, Liang Y, Li K. Multiple roles of BRIT1/MCPH1 in DNA damage response, DNA repair, and cancer suppression. Yonsei Med J. 2010;51:295–301. doi: 10.3349/ymj.2010.51.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffers LJ, Coull BJ, Stack SJ, Morrison CG. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene. 2008;27:139–144. doi: 10.1038/sj.onc.1210595. [DOI] [PubMed] [Google Scholar]
- 21.Lin SY, Rai R, Li K, Xu ZX, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci USA. 2005;102:15105–15109. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282:35416–35423. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Lee J, Stern DF. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem. 2004;279:34091–34094. doi: 10.1074/jbc.C400139200. [DOI] [PubMed] [Google Scholar]
- 24.Rai R, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10:145–157. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng G, Lin SY. BRIT1/MCPH1 is a multifunctional DNA damage responsive protein mediating DNA repair-associated chromatin remodeling. Cell Cycle. 2009;8:3071–3072. doi: 10.4161/cc.8.19.9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou ZW, et al. DNA damage response in microcephaly development of MCPH1 mouse model. DNA Repair (Amst) 2013;12:645–655. doi: 10.1016/j.dnarep.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 27.Liang Y, et al. Mcph1/Brit1 deficiency promotes genomic instability and tumor formation in a mouse model. Oncogene. 2015;34:4368–4378. doi: 10.1038/onc.2014.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Y, et al. BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet. 2010;6:e1000826. doi: 10.1371/journal.pgen.1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber R, et al. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- 30.Brown JA, Bourke E, Liptrot C, Dockery P, Morrison CG. MCPH1/BRIT1 limits ionizing radiation-induced centrosome amplification. Oncogene. 2010;29:5537–5544. doi: 10.1038/onc.2010.302. [DOI] [PubMed] [Google Scholar]
- 31.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 32.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 33.Sheu YJ, Kinney JB, Stillman B. Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Res. 2016;26:315–330. doi: 10.1101/gr.195248.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinoshita K, Harigai M, Fagarasan S, Muramatsu M, Honjo T. A hallmark of active class switch recombination: Transcripts directed by I promoters on looped-out circular DNAs. Proc Natl Acad Sci USA. 2001;98:12620–12623. doi: 10.1073/pnas.221454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franco S, et al. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J Exp Med. 2008;205:557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starnes LM, et al. A PTIP-PA1 subcomplex promotes transcription for IgH class switching independently from the associated MLL3/MLL4 methyltransferase complex. Genes Dev. 2016;30:149–163. doi: 10.1101/gad.268797.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bothmer A, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lumsden JM, et al. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J Exp Med. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zha S, et al. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 44.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 45.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng G, Lin SY. The linkage of chromatin remodeling to genome maintenance: Contribution from a human disease gene BRIT1/MCPH1. Epigenetics. 2009;4:457–461. doi: 10.4161/epi.4.7.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 48.Xie A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 50.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 51.Yamane A, et al. RPA accumulation during class switch recombination represents 5′-3′ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Rep. 2013;3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bassing CH, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci USA. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dimitrova N, de Lange T. MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev. 2006;20:3238–3243. doi: 10.1101/gad.1496606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18:75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng S, et al. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]