Abstract
Background
The analysis of synonymous and nonsynonymous rates of DNA change can help in the choice among competing explanations for rate variation, such as differences in constraint, mutation rate, or the strength of genetic drift. Nonphotosynthetic plants of the Orobanchaceae have increased rates of DNA change. In this study 38 taxa of Orobanchaceae and relatives were used and 3 plastid genes were sequenced for each taxon.
Results
Phylogenetic reconstructions of relative rates of sequence evolution for three plastid genes (rbcL, matK and rps2) show significant rate heterogeneity among lineages and among genes. Many of the non-photosynthetic plants have increases in both synonymous and nonsynonymous rates, indicating that both (1) selection is relaxed, and (2) there has been a change in the rate at which mutations are entering the population in these species. However, rate increases are not always immediate upon loss of photosynthesis. Overall there is a poor correlation of synonymous and nonsynonymous rates. There is, however, a strong correlation of synonymous rates across the 3 genes studied and the lineage-speccific pattern for each gene is strikingly similar. This indicates that the causes of synonymous rate variation are affecting the whole plastid genome in a similar way. There is a weaker correlation across genes for nonsynonymous rates. Here the picture is more complex, as could be expected if there are many causes of variation, differing from taxon to taxon and gene to gene.
Conclusions
The distinctive pattern of rate increases in Orobanchaceae has at least two causes. It is clear that there is a relaxation of constraint in many (though not all) non-photosynthetic lineages. However, there is also some force affecting synonymous sites as well. At this point, it is not possible to tell whether it is generation time, speciation rate, mutation rate, DNA repair efficiency or some combination of these factors.
Background
Rates of DNA sequence evolution vary among taxa and among genes, and the causes of this variation are many. In some cases, generation time has been shown to be correlated with rates in plants. For example, annual plants can sometimes have higher rates of DNA evolution than perennials [1]. In one study, it was shown that long-lived woody grasses exhibit slower rates than short-lived herbaceous ones [2]. However, a more extensive set of 33 phylogenetically independent comparisons failed to find a generation time effect for plants in general [3].
A useful method for distinguishing among the potential causes of rate variation is to separately examine nonsynonymous rates (rN) and synonymous rates (rS). For example, when rN increases relative to rS, relaxation of purifying selection is a possible explanation. However, when rS increases, but the rN/rS ratio is not greatly affected, then an increase in the mutation rate is a possibility. An example of this is Plantago mitochondrial DNA [4]. However, a decrease in DNA repair efficiency could also explain such a change. In addition, population processes, such as genetic drift could play a role. Reduced effective population size (Ne) can increase the fixation rate of neutral and slightly deleterious mutations. Thus, if slightly deleterious mutations are common, both rS and rN are expected to be higher when Ne is low [5,6].
One expectation of this drift-based hypothesis is that rN will vary from protein to protein, as each protein will have different functional constraints and thus a different proportion of slightly deleterious mutations. In contrast, rS is expected to be similar among proteins when the cause is a change in mutation rate or repair efficiency[7].
During founder-effect speciation, genetic drift can be expected to increase the substitution rate, even at silent sites. This has been called the speciation-rate hypothesis [8]. For example, speciation rate has been used to explain the difference in non-coding DNA substitution rates between the sister genera Utricularia and Pinguicula in the plant family Lentibulariaceae [9].
Rates can also vary if the underlying mutation rate varies [10,11] or if DNA repair is impaired [12,13]. Although rates themselves are hard to measure, the number of synonymous (dS) and nonsynonymous (dN) substitutions can be measured and are used to compare rates and calculate rate ratios.
Epifagus virginiana, a nonphotosynthetic plant, has an increased rate of sequence evolution for plastid DNA in general [14-17] and relative rates tests of the plastid rps2 gene indicate a significant increase for both dN and dS. This suggests that purifying selection is at least partially relaxed and that there has been an increase in the rate at which mutations are entering the population in this species, due to increased mutation rate or lax DNA repair. MatK, another plastid gene, is characterized by a partial relaxation of purifying selection in the clade containing Epifagus, Orobanche and Boschniakia [18]. In this paper, we explore rate variation in E. virginiana and 38 of its relatives for three plastid genes: rps2, matK, and rbcL. Each of these genes is present in photosynthetic relatives of Epifagus, is accelerated (or even lost) in Epifagus or related parasitic plants. Although plastid encoded, the three genes encode proteins that participate in different processes in the plastid. rps2 encodes the ribosomal protein S2 in small subunit ribosome, matK is an intron maturase, and rbcL encodes the large subunit in the CO2-fixing enzyme RUBISCO. We ask several questions: When does the rate increase observed in Epifagus begin, relative to the evolutionary loss of photosynthesis? What are the causes? Relaxation of constraint? More mutations entering the population? Are these patterns consistent across multiple plastid genes?
Results
Phylogenies of the Orobanchaceae and relatives were constructed using maximum parsimony (MP) and maximum likelihood (ML). The MP analysis discovered four most parsimonious trees of 3817 steps, with CI = 0.6275, CI (excluding uninformative characters) = 0.5118, and RC = 0.3888. The strict consensus tree was unresolved as to the position of Schwalbea relative to the Alectra-Orobanche clade, the Bartsia-Melampyrum clade and the Castilleja-Pedicularis clade. It was also unresolved concerning the relationships among the outgroups Mimulus, Kigelia, Hemimeris, Verbascum, Antirrhinum and Veronica.
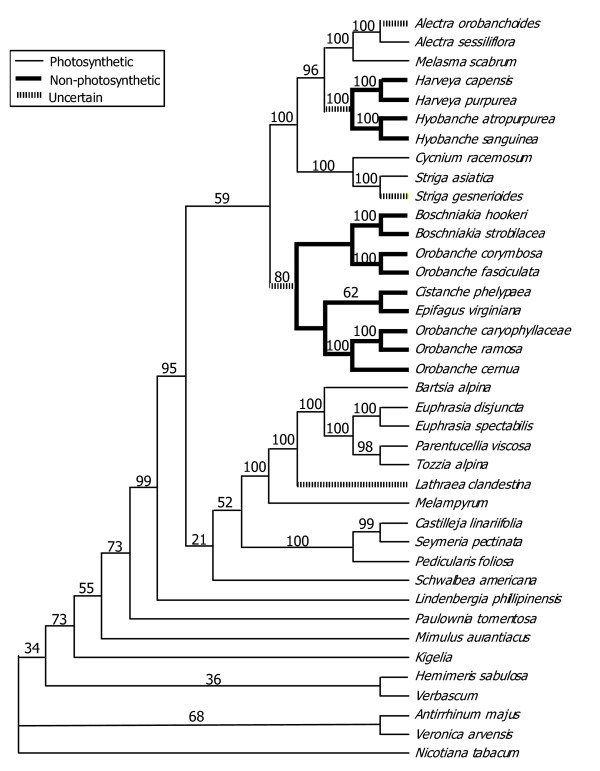
The ML analysis found two trees, with -ln likelihood values of 24525.88663. The strict consensus of these trees was unresolved, but in a different place, regarding the position of the Cistanche-Epifagus clade. When the MP consensus and the ML consensus were combined into a semistrict consensus tree, a completely resolved tree resulted. This tree is shown in Fig. 1.
Figure 1.
MP/ML consensus tree with MP bootstrap values
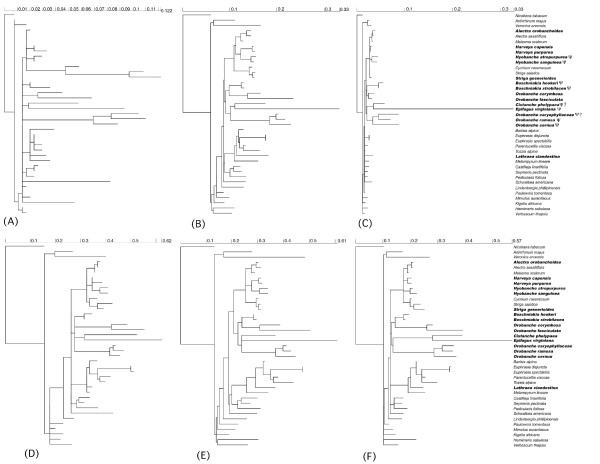
All three gene trees exhibited statistically significant rate heterogeneity (p < 0.0005), as assessed by the Kishino-Hasegawa (K-H) test [19]. Synonymous and nonsynonymous branch lengths for each of the three genes are shown reconstructed in Fig. 2.
Figure 2.
Nonsynonymous and synonymous rates of change in three genes. Nonphotosynthetic taxa are named in bold. (A) rps2 nonsynonymous branch lengths. (B) matK nonsynonymous branch lengths. (C) rbcL nonsynonymous branch lengths. (D) rps2 synonymous branch lengths. (E) matK synonymous branch lengths. (F) rbcL synonymous branch lengths. Taxa with rbcL pseudogenes are identified with Ψ. Uncertain pseudogene status is indicated by "Ψ?" [20, 26].
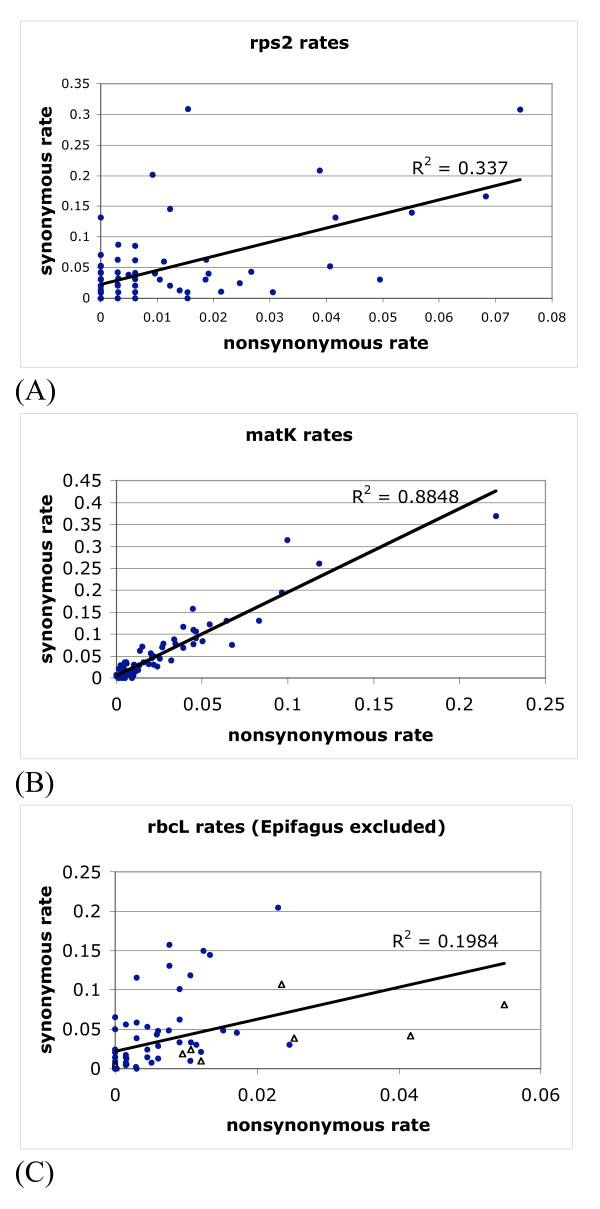
The correlation analyses show that there is a higher correlation of synonymous evolution across genes than nonsynonymous evolution (Fig. 3). They also show that within rps2 and rbcL synonymous and nonsynonymous evolution is poorly correlated, but in matK, the correlation is better (Fig. 4).
Figure 3.
Correlations across each possible pair of genes for synonymous and nonsynonymous rates. (A) rps2 versus matK. (B) rps2 versus rbcL. (C) matK versus rbcL. All nonsynonymous comparisons involving rbcL pseudogenes are indicated with a hollow triangle.
Figure 4.
Correlation plots of synonymous and nonsynonymous rates within each gene. (A) rps2 rates. (B) matK rates. (C) rbcL rates. In 4C, the Epifagus pseudogene has been excluded and the other pseudogenes are indicated with a hollow triangle.
Some of the non-photosynthetic plants, (Epifagus, Cistanche, and the Orobanche species) have increases in both synonymous and nonsynonymous rates. Rates are not, however, increased in Boschniakia, Harveya, Hyobanche, Lathaea, Alectra orobanchoides, and Striga gesnerioides.
Synonymous rates vary markedly among taxa. For example, the branches leading to Epifagus are at least two or three times as long as those for most of the photosynthetic taxa. Despite this among-taxon variation, the pattern for each gene is strikingly similar (Fig. 2D, E, F). For example, sister taxa (e.g. Veronica and Antirrhinum, Euphrasia and Panetucellia/Tozzia, Epifagus and Cistanche, O. fasciculata and O. corymbosa) usually show an identical pattern of who is faster than whom. This indicates that the causes of synonymous rate variation are affecting the whole plastid genome in a similar way.
There is even more extensive variation in nonsynonymous rates, both among taxa and among genes. This is not surprising because these genes have different functions and some of the taxa are photosynthetic while others are not. The scale bars in Figs. 2A, B, and 2C show the overall differences in nonsynonymous rates. MatK is much more rapidly evolving than the other two genes for nearly all taxa, but Epifagus' rbcL pseudogene has a similar rate. When looking at taxa across genes, there is much less consistency than with the synonymous rates. There are some big differences, such as the branch lengths for Striga, Cycnium and Schwalbea, and the Euphrasia species. Overall the picture is more complex, as could be expected if there are many causes of variation, differing from taxon to taxon and gene to gene.
The pattern of matK rate variation is very similar in the synonymous and nonsynonymous figures. This fits well with the fact that it is less constrained overall, as can be seen by comparing the scale bars in Figs. 2A, B, and 2C.
Rates were compared by using two categories at a time and testing for significant differences using likelihood ratio tests. These tests are summarized in Table 2. In general, it was found that the nonphotosynthetic plants have higher synonymous and nonsynonymous rates of change, although when tested separately, it was found that Harveya, Hyobanche and Boschniakia do not have higher synonymous rates.
Table 2.
P-values of the likelihood ratio tests.
| rps2 | matK | rbcL | |||||||
| rN | rS | ratio (ω) | rN | rS | ratio (ω) | rN | rS | ratio (ω) | |
| photo. vs nonphoto. | 1.4 × 10-11 | 0.0014 | 0.0034 | 0 | 0.077 (NS) | 0.001 | 0 | 2.3 × 10-8 | 2.2 × 10-8 |
| photo. vs OEC | 0 | 6.3 × 10-10 | 7.2 × 10-8 | 0 | 7.6 × 10-14 | 0.0038 | 0 | 0 | 2.7 × 10-11 |
| photo. vs HHB | 0.41 (NS) | 0.0066 | 0.24 (NS) | 1.4 × 10-6 | 4.5 × 10-9 | 0.23 (NS) | 0.076 (NS) | 0.0011 | 8.1 × 10-5 |
The two categories at left were compared for nonsynonymous rate differences (rN), synonymous rate differences (rS) or rate ratio (ω) differences. "Photo." refers to photosynthetic branches, "nonphoto." refers to nonphotosynthetic branches, "OEC" refers to Orobanche, Epifagus and Cistanche branches, and "HHB" refers to Harveya, Hyobanche and Boschniakia branches.
Purifying selection is relaxed in the nonphotosynthetic plants for all three genes. The test values are as follows. matK: 2×LR = 10.67, df = 1, p = 0.001; rbcL: 2×LR = 31.3, df = 1, p = 2.2 × 10-8; rps2: 2×LR = 8.56, df = 1, p = 0.00343.
Another way to describe the difference in the pattern of synonymous and non-synonymous rates is to say that the former are more correlated across genes. This can be seen in Fig. 3, which shows plots comparing two genes at a time. In comparisons including rbcL nonsynonymous rates, the data point from the Epifagus pseudogene has been excluded. Its unconstrained evolution is not typical of "nonsynonymous" change and its position on the plot made it an extreme outlier with an enormous influence on the regression line.
For rps2 and rbcL, the synonymous plots are more highly correlated, whereas for matK, which is relatively unconstrained, they are about the same.
Discussion
The dramatic rate increase observed in Epifagus [14], with branches 5–10 times as long as other taxa, can now be seen to have begun earlier in the history of the Orobanchaceae. It is shown to be composed of increases in both synonymous and nonsynomymous rates. The general pattern is that many of the non-photosynthetic plants, such as Epifagus, Cistanche, and the Orobanche species, have increases in both synonymous and nonsynonymous rates, indicating that both (1) selection is relaxed, and (2) there has been a change in the rate at which mutations are entering the population in these species. However, rate increases are not immediate upon loss of photosynthesis, since we do not see increases in Boschniakia, Harveya, Hyobanche, Lathaea, Alectra orobanchoides, and Striga gesnerioides. This pattern is similar to that found using smaller data sets [15,20]. Separate analyses of synonymous and nonsynonymous rates give us some insight into potential mechanisms.
The speciation rate hypothesis predicts that more speciose clades should have a faster rs (and therefore larger ds) than a species-poor sister group. This was suggested as a cause of rate variation in non-coding DNA in the Lentibulareaceae [9]. In this study, not all genera have been sampled, and those that have are often represented by one or two species. In addition, some genera may not be monophyletic (for example, Orobanche is not). Thus, accurate numbers of species cannot be assigned to individual branches or clades. However, a few things can be noted. Euphrasia, with ~170 spp., is clearly more speciose than its sister group, with 3 spp. It has a somewhat faster ds. However, the Schwalbea lineage, with a single species, has a fairly high ds. Its position is not certain, but its sister group is probably the Bartsia – Melampyrum clade (>300 spp.), the Castilleja – Pedicularis clade, (>700 spp.), or the union of the two. These groups have do not have dramatically higher ds values; in fact the Castilleja – Pedicularis clade's value is slightly lower.
Differences in generation time may play some role in the observed rate variation. However, as was found previously [15], the pattern is not clear. Since most of the genera sampled in this study contain both annuals and perennials, it is likely that most branches on the tree actually represent a combination of annual and perennial evolutionary history. However, there are some intriguing details that might merit further study. The clade containing Bartsia, Euphrasia and Melampyrum contains mostly annuals [21] and has some high ds values, as one would expect from a generation time effect. However, both Euphrasia and Melampyrum contain almost exclusively annuals and have very different rates. Likewise, the large clade containing Boschniakia and Epifagus contains mostly annuals and has an overall high ds. The perennials Boschniakia and Cistanche have lower ds than their sister taxa, which also supports the generation time hypothesis, but there is as much variation among categories (perennial, annual) as between categories.
Conclusions
The distinctive pattern of rate increases in Orobanchaceae has at least two causes. It is clear that there is a relaxation of constraint in many (though not all) non-photosynthetic lineages. However, there is also some force affecting synonymous sites as well. At this point, it is not possible to tell whether it is generation time, speciation rate, mutation rate, DNA repair efficiency or some combination of these factors. Clearly, generating additional data from nuclear and mitochondrial genes would help us to more clearly distinguish among these hypotheses. Some of the above-mentioned hypotheses (generation-time, speciation rate) would be expected to affect nuclear and mitochondrial genomes in a similar fashion, whereas factors affecting mutation rate or efficiency of DNA repair would not, as these process involve different, though perhaps overlapping, sets of enzymes in each of the three genomes [22-24].
Methods
Sampling
We sampled 15 photosynthetic and 16 nonphotosynthetic Orobanchaceae, and eight outgroup taxa. The specimens used and their GenBank accession numbers are given in Table 1.
Table 1.
Specimens used for DNA sequencing, with GenBank accession numbers for rps2, matK, and rbcL sequences.
| GenBank accession numbers | |||
| Species | rps2 | matK | rbcL |
| Alectra orobanchoides | U48741 | AF489960 | AF026819 |
| Alectra sessiliflora | U48742 | AF051977 | AF026820 |
| Antirrhinum majus | U48766 | AF051978 | L11688 |
| Bartsia alpina | U48751 | AY849600 | AF190903 |
| Boschniakia hookeri | U48757 | AF051979 | AF026817 |
| Boschniakia strobilacea | U48758 | AF051980 | AF26818 |
| Castilleja lineariifolia | U48739 | AF051981 | AF026823 |
| Cistanche phelypaea | AY849597 | AF056149 | AY849862 |
| Cycnium racemosum | U48745 | AY849601 | AF026826 |
| Epifagus virginiana | EPFCPCG | EPFCPCG, AF051982 | EPFCPCG |
| Euphrasia disjuncta | AY849598 | AY849602 | AY849863 |
| Euphrasia spectabilis | U48752 | AY849603 | AY849864 |
| Harveya capensis | AF055142 | AF489961 | AF026829 |
| Harveya purpurea | U48749 | AF051984 | AF026830 |
| Hemimeris sabulosa | U48765 | AF051985 | AF123668 |
| Hyobanche atropurpurea | AY849599 | AF051986 | AF026831 |
| Hyobanche sanguinea | U48750 | AF051987 | AF026832 |
| Kigelia africana | U48764 | AF051988 | AF102648 |
| Lathraea clandestina | U48755 | AF051989 | AF026833 |
| Lindenbergia phillipinensis | AF055151 | AF051990 | AF123664 |
| Melampyrum sylvaticum | AF055148 | AF051991 | |
| Melampyrum lineare | AF026834 | ||
| Melasma scabrum | U48743 | AY849604 | AF190904 |
| Mimulus aurantiacus | AF055154 | AY849605 | AF026835 |
| Nicotiana tabacum | Z00044 | Z00044 | Z00044 |
| Orobanche caryophyllacea | AF055145 | AF051992 | AY582187 |
| Orobanche cernua | AF055147 | AF056147 | U73968 |
| Orobanche corymbosa | U48760 | AF051993 | U73969 |
| Orobanche fasciculata | AF055143 | AF051994 | U73970 |
| Orobanche hederae | AF055146 | AF051995 | AF078682 |
| Orobanche ramosa | U48761 | AF056148 | U73971 |
| Parentucellia viscosa | U48753 | AY849606 | AY849865 |
| Paulownia tomentosa | AF055255 | AF051997 | L36447 |
| Pedicularis foliosa | U48740 | AF489959 | AF026836 |
| Schwalbea americana | AF055150 | AF051998 | AY849866 |
| Seymeria pectinata | AF055141 | AF051999 | AF026837 |
| Striga asiatica | U48746 | AF052000 | AF026838 |
| Striga gesnerioides | U48747 | AF489963 | AF026839 |
| Tozzia alpina | U48754 | AF052001 | AF026843 |
| Verbascum blattaria | U48763 | ||
| Verbascum thapsus | AF052002 | L36452 | |
| Veronica arvensis | U48768 | AF052003 | |
| Veronica persica | L36453 | ||
Taxonomic authorities, localities and voucher information can be found in [25], with the exception of Euphrasia disjuncta Fernald & Wiegand, Hyobanche atropurpurea Bolus., and Veronica persica Poir.
Amplification and sequencing
We amplified and sequenced rps2 as in [15], matK as in [25], and rbcL as in [26]. A total of 15 new sequences were generated for this study, including 3 rps2, 7 matK, and 5 rbcL sequences.
Alignment
The rps2 alignment was simple, containing only two small indels. For matK and rbcL, a search for the best alignment was conducted using Clustal X and a variety of alignment parameters. Alignments were evaluated according to the following optimality criterion: whichever alignment yields the MP tree(s) with the highest consistency is considered the best alignment[27]. For alignment assessment, MP analyses with and without indel characters were used. When used, indel characters were generated with the program GapCoder [28](available from http://www.tufts.edu/vet/richlab/young/GapCoder), which uses the simple indel coding method of Simmons and Ochoterena [29]. The rescaled consistency (RC) index [30] of the resulting parsimony analyses was used to assess alignment optimality, with one exception: very low gap opening penalties (GOP), such as 3 or less were excluded. These low GOP values lead to inflated RC values, due to the large numbers of gaps, which reduce the treelength and the homoplasy. rps2 had just one small indel and was aligned by eye. For matK, the optimal computer alignment was generated using GOP = 5 and gap extension penalty (GEP) = 1. Transitions were weighed the same as transitions. The RC from the analysis with indel characters included was 0.3693. The RC without indel characters was 0.3759. The alignment was then adjusted by eye. This final alignment yielded RC values of 0.3878 (indel characters included) and 0.3763 (indel characters excluded).
For rbcL, the optimal computer alignment was generated using GOP = 5 GEP = 3. Transitions were weighed the same as transitions. The RC from the analysis with indel characters included was 0.4679. The RC without indel characters was 0.4437. The alignment was then adjusted by eye. This final alignment yielded RC values of 0.4635 (indel characters included) and 0.4499 (indel characters excluded).
Phylogenetic analyses
For rps2, positions homologous to positions 48–660 of the Nicotiana gene were used. For matK, the entire gene was used. For rbcL, positions homologous to Nicotiana gene positions 5–1325 were used. The three genes were then combined into a single data set. PAUP* 4.0b8 [31] was used to conduct a MP heuristic search, including indel characters, and using Nicotiana tabacum as the outgroup taxon, TBR branch swapping and 100 random addition replicates. Bootstrap analyses were conducted with the same settings, except with only 40 random addition orders. 500 bootstrap replicates were performed.
ML analyses excluded indels. Using the hLRT method of the program ModelTest 3.06 [32], the ML model of GTR+G was selected as the best evolutionary model for the combined data set. Base frequencies (A = .298, C = .177, G = .203) and substitution rates (A-C = 1.51, A-G = 2.11, A-T = 0.264, C-G = 0.788, C-T = 2.62, G-T = 1) were obtained from the MP trees. Among-site variation was included in the model, based on a gamma distribution with four categories. A heuristic search was conducted, similar to the MP search, but with only 10 random addition replicates, each limited to the examination of 5000 rearrangements.
Rates of DNA change
Overall rate heterogeneity was assessed using the K-H test as implemented in PAUP, using the same ML analyses, except that the starting tree was a neighbor-joining tree and the analysis was limited to 40 rearrangements. Nonsysnonymous and synonymous changes were reconstructed on branches using the codon-based likelihood model of Muse and Gaut [33], as implemented in HYPHY for MacOS, ver. 0.95 beta [[34] 2004], available at http://www.hyphy.org. At least nine of the rbcL "genes" are probably pseudogenes. These are indicated in Figure 2C. Seven of these have already been discussed elsewhere [20]. The Orobanche caryophyllaceae and Cistanche phelypaea "genes" have internal stop codons and thus are probably also pseudogenes. There may also be other pseudogenes with intact ORFs, making their pseudogene status less obvious [35]. Once a pseudogene is formed, it is no longer constrained for a protein function, so synonymous and nonsynonymous changes can no longer be formally defined. Moreover, changes that would have been synonymous and nonsynonymous are now expected at equal rates. Thus, by including these sequences in the tests, we get additional evidence that constraint is relaxed in nonphotosynthetic plants. Therefore, even for the pseudogenes, we have still calculated the synonymous and nonsynonymous rates separately, assuming a reading frame based on alignment to the other genes in the data set, and have indicated the pseudogenes in Fig. 2C.
Rate increases were compared among categories of taxa (such as photosynthetic and non-photosynthetic), using dN, dS, and the dN/dS ratio (ω) in likelihood ratio tests [36]. These tests were conducted using HYPHY and the category assignments of the branches are those shown in Figure 1. In addition, a previous study [15] indicated that some nonphotosynthetic branches might not have rate increases. Thus, two subsets of the nonphotosynthetic branches were tested: (1) Orobanche, Epifagus and Cistanche branches and (2) Harveya, Hyobanche and Boschniakia branches. Each of these tests used the data set from a single gene and compared two nested hypotheses: H1: the photosynthetic and non-photosynthetic branches share a single value (for one of the parameters dN, dS or ω). H2: the photosynthetic and non-photosynthetic branches have two separate values. If the tree has a significantly higher likelihood under H2, that is taken as evidence that the nonphotosynthetic branches have higher rates. Scatter plots and correlation tests were used to examine the degree of correlation between synonymous and nonsynonymous sites within a gene, and also to see if either class of sites was correlated between genes.
Authors' contributions
NDY and CWD conceived of and designed the study together. NDY did the sequencing, data analyses and drafted the manuscript. CWD provided the genomic DNA samples and provided the conducive laboratory environment, both physical and intellectual, as well as many suggestions for the manuscript.
Acknowledgments
Acknowledgements
We thank R. Olmstead, K. Kirkman, R. Wyatt, J. Armstrong, J. Alison, A. Batten, M. Weatherwax, K. Steiner, L. Musselman, W. Wetschnig, G. Sallé, C. Morden, and J. Palmer for plants or DNAs used in this study; A. Wolfe for some of the rbcL sequences and DNAs; S. Kovalsky-Pond, J. Leebens-Mack and J. Lyons-Weiler for suggestions on analyses; NSF for financial support (DEB-9120258, DBI-9604814) to CWD.
Contributor Information
Nelson D Young, Email: Ned.Young@tufts.edu.
Claude W dePamphilis, Email: cwd3@psu.edu.
References
- Laroche J, Li P, Maggia L, Bousquet J. Molecular evolution of angiosperm mitochondrial introns. Proc Natl Acad Sci USA. 1997;94:5722–5727. doi: 10.1073/pnas.94.11.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Clark LG, Wendel JF, Muse SV. Comparisons of the molecular evolutionary process at rbcLi and ndhF in the grass family (Poaceae) Mol Biol Evol. 1997;14:769–777. doi: 10.1093/oxfordjournals.molbev.a025817. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Johnston MO. Broad-scale analysis contradicts the theory that generation time affects molecular evolutionary rates in plants. Journal of Molecular Evolution. 2003;56:223–233. doi: 10.1007/s00239-002-2395-0. [DOI] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu YL, Palmer JD. Mitochondrial Substitution Rates are Extraordinarily Elevated and Variable in a Genus of Flowering Plants. Proc Natl Acad Sci USA. [DOI] [PMC free article] [PubMed]
- Wernegreen JJ, Moran NA. Evidence for genetic drift in endosymbionts (Buchnera): Analyses of protein-coding genes. Mol Biol Evol. 1999;16:83–97. doi: 10.1093/oxfordjournals.molbev.a026040. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Wernegreen JJ, Moran NA. Intraspecific variation in symbiont genomes: Bottlenecks and the Aphid-Buchnera Association. Genetics. 2001;157:477–489. doi: 10.1093/genetics/157.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Martin W, Nei M. Acceleration of genomic evolution caused by enhanced mutation rate in endocellular symbionts. Proceedings of the National Academy of Sciences, USA. 2002;99:12944–12948. doi: 10.1073/pnas.192449699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG, Savolainen V. Evolutionary rates and species dicersity in flowering plants. Evolution. 2001;55:677–683. doi: 10.1554/0014-3820(2001)055[0677:erasdi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Jobson RW, Albert VA. Molecular rates parallel diversification contrasts between carnivorous plant sister lineages. Cladistics. 2002;18:127–136. doi: 10.1006/clad.2001.0187. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Johnston MO. Male-driven evolution of mitochondrial and chloroplastidial DNA sequences in plants. Molecular Biology and Evolution. 2002;19:938–949. doi: 10.1093/oxfordjournals.molbev.a004151. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch Repair in Replication Fidelity, Genetic Recombination, and Cancer Biology. Annual Review of Biochemistry. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR, Rudd KE. Sequencing and analysis of bacterial genomes. Current Biology. 1996;6:404–416. doi: 10.1016/S0960-9822(02)00508-0. [DOI] [PubMed] [Google Scholar]
- dePamphilis CW, Palmer JD. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990;348:337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]
- dePamphilis CW, Young ND, Wolfe AD. Evolution of plastid gene rps2 in a lineage of hemiparasitic and holoparasitic plants: many losses of photosynthesis and complex patterns of rate variation. Proc Natl Acad Sci USA. 1997;94:7367–7372. doi: 10.1073/pnas.94.14.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden CW, Wolfe KH, dePamphilis CW, Palmer JD. Plastid translation and transcription genes in a non-photosynthetic plant: intact, missing and pseudo genes. EMBO J. 1991;10:3281–3288. doi: 10.1002/j.1460-2075.1991.tb04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Ems SC, Palmer JD. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: Loss or accelerated sequence evolution of tRNA and ribosomal protein genes. Journal of Molecular Evolution. 1992;35:304–317. doi: 10.1007/BF00161168. [DOI] [PubMed] [Google Scholar]
- Young ND, dePamphilis CW. Purifying selection detected in the plastid gene matK and flanking ribozyme regions within a group II intron of nonphotosynthetic plants. Molecular Biology and Evolution. 2000;17:1933–1941. doi: 10.1093/oxfordjournals.molbev.a026295. [DOI] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. Journal of Molecular Evolution. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Wolfe AD, dePamphilis CW. The effect of relaxed functional constraints on the photosynthetic gene rbcL in parasitic plants of Scrophulariales. Molecular Biology and Evolution. 1998;15:1243–1258. doi: 10.1093/oxfordjournals.molbev.a025853. [DOI] [PubMed] [Google Scholar]
- Kuijt J. The biology of parasitic flowering plants. Berkeley, CA, University of California Press; 1969. p. 246. [Google Scholar]
- Backert S, Nielsen BL, Börner T. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997;2:477–483. doi: 10.1016/S1360-1385(97)01148-5. [DOI] [Google Scholar]
- Britt AB. DNA damage and repair in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:75–100. doi: 10.1146/annurev.arplant.47.1.75. [DOI] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL. Chloroplast DNA replication: mechanism, enzymes and replication origins. J Plant Biochem Biotechnol. 1997;6:1–7. [Google Scholar]
- Young ND, Steiner KE, dePamphilis CW. The evolution of parasitism in Scrophulariaceae/Orobanchaceae: plastid gene sequences refute an evolutionary transition series. Ann Mo Bot Gard. 1999;86:876–893. [Google Scholar]
- Wolfe AD, dePamphilis CW. Alternate paths of evolution for the photosynthetic gene rbcL in four nonphotosynthetic species of Orobanche. Plant Mol Biol. 1997;33:965–977. doi: 10.1023/A:1005739223993. [DOI] [PubMed] [Google Scholar]
- Wheeler WC. Sequence alignment, parameter sensitivity, and the phylogenetic analysis of molecular data. Syst Biol. 1995;44:321–331. [Google Scholar]
- Young ND, Healy J. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics. 2003;4:6. doi: 10.1186/1471-2105-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49:369–381. doi: 10.1080/10635159950173889. [DOI] [PubMed] [Google Scholar]
- Farris JS. The retention index and the rescaled consistency index. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogentic analysis using parsimony (*and other methods). Version 4. Sunderland, Massachusetts, Sinauer Associates; 2003. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. 0.901beta ed. Bioinformatics. 2004. [DOI] [PubMed]
- Leebens-Mack J, dePamphilis CW. Power analysis of tests for loss of selective constraint in cave crayfish and non-photosynthetic plant lineages. Mol Biol Evol. 2002;19:1292–1302. doi: 10.1093/oxfordjournals.molbev.a004190. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]