Abstract
Molecular tests revealed influenza D viruses of D/OK lineage widely circulating in farmed animal species in Guangdong Province, southern China. In particular, we found high levels of influenza D virus infection in goats and pigs. We also detected viral RNA in serum specimens and feces of animals with certain severe diseases.
Keywords: influenza, influenza D virus, hemagglutinin-esterase-fusion gene, D/OK lineage, Holstein dairy cattle, yellow cattle, Asian buffalo, American Landrace pigs, hybrid goats, China, viruses, zoonoses
Four types of influenza viruses (A–D) have been confirmed (https://www.cdc.gov/flu/about/viruses/types.htm). The recently discovered influenza D virus is thought to cause respiratory diseases primarily in cattle and to a lesser extent in pigs (1–4). Moreover, serologic evidence for influenza D virus infection in small ruminants and humans has been established (5,6). Since the initial influenza D virus isolation in the United States in 2011 (1), the virus has been reported in China, Mexico, France, Italy, and Japan (7–11). Genetic analysis of the hemagglutinin-esterase-fusion gene demonstrated that these viruses had 2 distinct lineages, represented by D/OK and D/660 (12). Recently, a novel influenza D virus that emerged in Japan has been proposed as the third lineage (11). D/OK lineage–related viruses were previously identified in native Luxi yellow cattle in Shandong Province, northern China (7). Despite good progress in identifying domestic cattle as the primary reservoir of influenza D virus, we know little about prevalence in other animals. We conducted a study to clarify the origin and transmission dynamics of influenza D virus in goats, buffalo, and pigs as well as farmed cattle.
The Study
In 2016, we collected 607 clinical samples from 4 species of animals with different clinical diseases and 250 nasal swab samples from asymptomatic animals (Table) from 16 farms in 4 cities of Guangdong Province: Guangzhou, Qingyuan, Heyuan, and Jiangmen (Figure 1). In addition, we randomly chose 200 archived Holstein dairy cattle serum samples, 40 per year, from 2011–2015 to investigate possible early RNA distribution of influenza D virus in this region. We used the reverse transcription PCR method and subcloning protocol (Technical Appendix). We performed sequence alignment using ClustalW implemented in DNAStar software (DNAStar, Madison, WI, USA), and we conducted phylogenetic analyses based on our obtained sequences and reference truncated sequences (496-bp) of influenza D viruses from GenBank by using MEGA 5.1 software (http://www.megasoftware.net; Technical Appendix Table).
Table. Animal species, location, sample data, and detection rate of influenza D virus, Guangdong Province, China*.
| Animal species and farm | Farm type† | Farm location | No. animals | Age range of animals | Sample type | No. positive/no. samples | Detection rate, % |
|---|---|---|---|---|---|---|---|
| Holstein dairy cattle |
|
|
|
|
|
|
|
| A | Not all-in-all-out | Guangzhou: Tianhe | 2,000 | 3–5 y | Nasal swab | 14/86‡ | 16.3 |
| A | Not all-in-all-out | Guangzhou: Tianhe | 2,000 | 3–5 y | Serum | 10/94‡ | 10.6 |
| B | Not all-in-all-out | Guangzhou: Luogang | 800 | 3–6 y | Nasal swab | 6/70‡ | 8.57 |
| B | Not all-in-all-out | Guangzhou: Luogang | 800 | 3–6 y | Serum | 5/99‡ | 5.05 |
| C |
Not all-in-all-out |
Guangzhou: Tianhe |
175 |
2–5 y |
Nasal swab |
1/50§ |
2 |
| American Landrace pig |
|
|
|
|
|
|
|
| D | Not all-in-all-out | Guangzhou: Huadu | 200 | 10–15 wks | Lung | 4/10‡ | 40 |
| E | All-in-all-out | Heyuan: Yuancheng | 1,000 | 5–5 wks | Nasal swab | 4/10‡ | 40 |
| E | All-in-all-out | Heyuan: Yuancheng | 1,000 | 3–5 wks | Lung | 1/8‡ | 12.5 |
| F | All-in-all-out | Jiangmen: Kaiping | 800 | 8–20 wks | Nasal swab | 3/9‡ | 30 |
| F | All-in-all-out | Jiangmen: Kaiping | 800 | 8–20 wks | Lung | 8/27‡ | 29.6 |
| G |
All-in-all-out |
Heyuan: Dongyuan |
600 |
9–15 wks |
Nasal swab |
1/50§ |
2 |
| Native hybrid white goat |
|
|
|
|
|
|
|
| H | Not all-in-all-out | Guangzhou: Zengcheng | 200 | 0.5–5 y | Serum | 7/25‡ | 28 |
| I |
Not all-in-all-out |
Guangzhou: Luogang |
300 |
2–4 y |
Serum |
20/55¶ |
36.4 |
| Native hybrid black goat |
|
|
|
|
|
|
|
| J | Not all-in-all-out | Qingyuan: Jiangkou | 150 | 1–3 y | Rectal swab | 1/8# | 12.5 |
| K |
Not all-in-all-out |
Jiangmen: Enping |
500 |
1–4 y |
Nasal swab |
0/50§ |
0 |
| Asian buffalo |
|
|
|
|
|
|
|
| L | Not all-in-all-out | Guangzhou: Nansha | 150 | 3–5 y | Serum | 2/26¶ | 7.7 |
| M | Not all-in-all-out | Guangzhou: Panyu | 180 | 3–6 y | Serum | 1/25¶ | 4 |
| N |
Not all-in-all-out |
Qingyuan: Yingde |
400 |
1–4 y |
Nasal swab |
0/50§ |
0 |
| Native yellow cattle |
|
|
|
|
|
|
|
| O | Not all-in-all-out | Qingyuan: Qingxin | 200 | 2–5 y | Nasal swab | 4/55‡ | 7.3 |
| P | Not all-in-all-out | Qingyuan: Fogang | 230 | 1–3 y | Nasal swab | 0/50§ | 0 |
*Feeding type of farms A–G was in captivity (poor biosecurity and high density). Feeding type of farms H–K and N–P was free grazing on the hills in the daytime and in captivity (poor biosecurity and high density) in the nighttime. Feeding type of farms L and M was free grazing in wetland in the daytime and in captivity (poor biosecurity and high density) in the nighttime. †All-in-all-out is a strategy for the control of infectious disease. The barn is emptied of all animals and the accommodation is cleaned and disinfected and then refilled, all on 1 day. ‡These animals had severe respiratory diseases with a 10%–30% mortality rate, mainly characterized by expiratory dyspnea and abdominal respiration. §These animals were asymptomatic. ¶These animals had severe reproductive disorders with a 60%–70% abortion rate. #These animals had severe diarrheal disease, characterized by watery diarrhea, limb weakness, and nearly dying.
Figure 1.
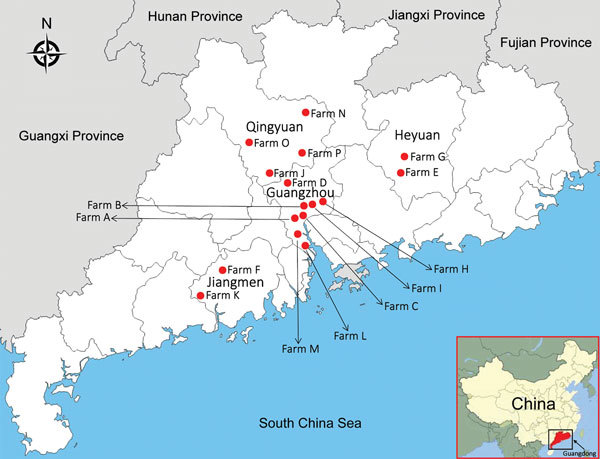
Farm locations for study of influenza D viruses in cattle, goats, buffalo, and pigs, Guangdong Province, China.
After testing by reverse transcription PCR with further sequencing confirmation, we found influenza D virus–positive rates in 230 total nasal swab samples of 12.8% (20/156) for dairy cattle, 7.3% (4/55) for native yellow cattle, and 36.8% (7/19) for pigs. Rates in 324 total serum samples were 7.8% (15/193) for dairy cattle, 5.9% (3/51) for buffalo, and 33.8% (27/80) for goats. The influenza D virus–positive rate was also high (28.9%, 13/45) in swine lung samples. In contrast, we found no or low prevalence (<2%) in asymptomatic animals tested (Table). Moreover, all of the archived serum samples were found to be influenza D virus negative. Interestingly, 1 of 8 rectal swabs of goats with severe diarrhea tested positive (Table). Samples from animals with reproductive problems had a positive rate of 4.3% (5/116) (Table).
Sequence alignment analysis showed that the nucleotide sequences of influenza D viruses found in this study shared high similarity (99%–100%) with previously described sequences from China (7) and low similarity (93.8%–98.8%) with sequences originating from the United States, France, Italy, Mexico, and Japan (1,8–12). Similarly, phylogenetic analysis revealed that all influenza D virus sequences in this study clustered together with previous sequences from China and belonged to the D/OK lineage (Figure 2).
Figure 2.
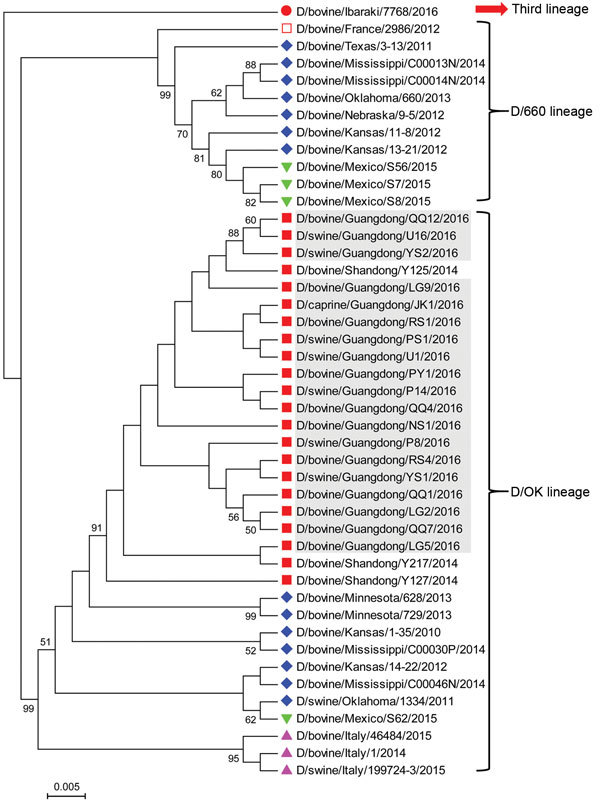
Phylogenetic analysis of viruses from study of influenza D viruses in cattle, goats, buffalo, and pigs in Guangdong Province and neighboring provinces, China, compared with reference viruses. Partial hemagglutinin-esterase-fusion gene sequences (496 bp) were aligned by using ClustalW implemented in DNAStar software (DNAStar, Madison, WI, USA), and the phylogenetic tree was obtained using neighbor-joining method within MEGA 5.1 software (http://www.megasoftware.net). Numbers at nodes are percentages of bootstrap values obtained by repeated analyses (1,000 times) to generate majority consensus tree. Only bootstrap scores of at least 50 were retained. Scale bar indicates 0.5% nucleotide sequence divergence. Gray shading indicates viruses from this study; reference viruses obtained from the United States are marked with ♦; from China, ■; from Italy, ▲; from Mexico, ▼; from France, □; and from Japan ●. Note that D/swine/Guangdong/YS1/2016 and D/swine/Guangdong/YS2/2016 are from the same farm; D/swine/Guangdong/P8/2016 and D/swine/Guangdong/P14/2016 are from the same farm; D/swine/Guangdong/U1/2016 and D/swine/Guangdong/U16/2016 are from the same farm; D/bovine/Guangdong/LG2/2016, D/bovine/Guangdong/LG5/2016 and D/bovine/Guangdong/LG9/2016 are from the same farm; D/bovine/Guangdong/QQ1/2016, D/bovine/Guangdong/QQ4/2016, D/bovine/Guangdong/QQ7/2016 and D/bovine/Guangdong/QQ12/2016 are from the same farm; D/bovine/Guangdong/RS1/2016 and D/bovine/Guangdong/RS4/2016 are from the same farm.
Conclusions
When first discovered, influenza D virus was reported in diseased pigs in the United States (1). Later, it was identified in cattle and swine herds in several other countries, with or without clinical manifestation (7–11). Moreover, antibodies to influenza D virus were detected in goats, sheep, and humans (5–6). Under experimental conditions, influenza D virus replicated and transmitted among ferrets and guinea pigs (13). We confirmed that influenza D virus is widely present in cattle species (dairy cattle, yellow cattle, and buffalo). We also found influenza D virus at a high prevalence (>30%) in pigs and goats (Table), which is in contrast to the low prevalence found in previous investigations (1,5,10). The high prevalence may be caused by poor biosecurity measures and high-density feeding mode practices in China’s animal industry as well as possible cross-species transmission (13). Taken together, our findings expand the host range of influenza D virus and further emphasize the health concern this virus poses to multiple animal species.
Previous studies have shown that influenza D viruses are mainly found in respiratory tract samples (1–4,7,9–12) and that they have played an etiologic role in bovine respiratory diseases (2–4). In this study, we found that influenza D virus RNA was present in cattle and goat serum samples; it was also present in goat rectal swabs, accompanied by peste des petits ruminants virus and caprine kobuvirus (data not shown). The distribution of influenza D virus in our study is not the same as that described under experimental conditions (3).
Influenza viremia, an indicator of disease severity (14), has been detected in 20.9% of severe cases during the acute phase of infection or before host death. Our detection of influenza D virus genome in serum samples from severely diseased animals (Table) implies that the virus could enter transiently into the animal’s circulatory system through capillaries lining the respiratory tract, which further contributes to the possibility of detecting virus in other organs. Similar to previous studies (2,4), we also found that the reverse transcription PCR positive rate was significantly higher (4%–40%) in diseased animals than the rate (<2%) observed in asymptomatic animals (p<0.05), which suggests a potential correlation between the disease severity and presence of influenza D virus. For influenza D virus found in rectal swabs, it might be that animals have swallowed the virus. Another possibility is that, similar to influenza A and B viruses, influenza D virus can replicate within the intestinal tract (15).
We detected influenza D virus in cattle with reproductive disorders. However, we could not determine whether influenza D virus is associated with reproductive problems. Future studies can be designed to investigate these scientific issues.
To date, 2 lineages of influenza D virus (D/OK and D/660) co-circulate in North America and Europe (8–10,12). However, only the D/OK lineage has been found in China, and a potential third lineage was found in Japan (7,11). Our study confirms and further extends the previous observation that D/OK lineage circulates in East Asia. The viral, host, and ecologic factors that shape the observed contrasting phylodynamics of influenza D viruses among different geographic regions warrant further investigation.
In addition, we found different minor genetic variants circulating on the same farm (Figure 2), indicating the ongoing evolution of influenza D viruses in their hosts (7,8,11). In comparing our sequences to the reference sequences from different animal species, we found 4 frequent nucleotide mutations (at positions 136, 231, 263, and 486) (online Technical Appendix Figure 1), which caused 2 amino acid mutations at positions 77 and 88 (online Technical Appendix Figure 2). Interestingly, among 4 nucleotide mutations, 1 unique nucleotide (T at position 486) was originally from the D/660 lineage. Moreover, we found several consistent sequences co-circulating in multiple animal species (online Technical Appendix Figure 1). Our speculation is that homologous recombination among different influenza D viruses and potential cross-species transmission under field conditions are possible, but further study is needed.
In summary, our study investigating the infection status of influenza D virus in different farmed animal species in Guangdong Province provides novel insights into the epidemiology and evolution of this virus. In particular, we document the molecular evidence for influenza D virus infection in goats and buffalo.
RT-PCR method and subcloning protocol used in study of influenza D viruses in cattle, goats, buffalo, and pigs, Guangdong Province and neighboring provinces, China.
Acknowledgments
We are very grateful to Ben M. Hause, Milton Thomas, and Hunter Nedland for their suggestions and English editing when we revised this manuscript.
This work was supported by Guangdong Provincial Department of Science and Technology (Grant no. 2016A040403083), Guangdong Provincial Agricultural Department (Grant no. 2016LM3177), and Ministry of Science and Technology of the People’s Republic of China (Grant no. 2015GA780010). The work was also supported in part by SDSU AES Fund 3AH-477 to F.L. and D.W. S.-L.Z. is sponsored by Guangdong Academy of Agricultural Sciences, Guangzhou, China.
Biography
Dr. Zhai is an associate professor at Animal Disease Diagnostic Center, Institute of Animal Health, Guangdong Academy of Agricultural Sciences. His research interests focus on surveillance and rapid response research of emerging or reemerging animal pathogens. In 2016–17, he is a visiting scholar at the Department of Biology and Microbiology, South Dakota State University.
Footnotes
Suggested citation for this article: Zhai S-L, Zhang H, Chen S-N, Zhou X, Lin T, Liu R, et al. Influenza D virus in animal species in Guangdong Province, Southern China. Emerg Infect Dis. 2017 Aug [date cited]. https://doi.org/10.3201/eid2308.170059
These authors contributed equally to this article.
References
- 1.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:e1003176. 10.1371/journal.ppat.1003176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng TF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol. 2015;89:5340–9. 10.1128/JVI.00064-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson L, Olivier AK, Genova S, Epperson WB, Smith DR, Schneider L, et al. Pathogenesis of influenza D virus in cattle. J Virol. 2016;90:5636–42. 10.1128/JVI.03122-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitra N, Cernicchiaro N, Torres S, Li F, Hause BM. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J Gen Virol. 2016;97:1771–84. 10.1099/jgv.0.000492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quast M, Sreenivasan C, Sexton G, Nedland H, Singrey A, Fawcett L, et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet Microbiol. 2015;180:281–5. 10.1016/j.vetmic.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White SK, Ma W, McDaniel CJ, Gray GC, Lednicky JA. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J Clin Virol. 2016;81:31–3. 10.1016/j.jcv.2016.05.017 [DOI] [PubMed] [Google Scholar]
- 7.Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, et al. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes. 2014;49:493–6. 10.1007/s11262-014-1107-3 [DOI] [PubMed] [Google Scholar]
- 8.Ferguson L, Eckard L, Epperson WB, Long LP, Smith D, Huston C, et al. Influenza D virus infection in Mississippi beef cattle. Virology. 2015;486:28–34. 10.1016/j.virol.2015.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducatez MF, Pelletier C, Meyer G. Influenza D virus in cattle, France, 2011-2014. Emerg Infect Dis. 2015;21:368–71. 10.3201/eid2102.141449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiapponi C, Faccini S, De Mattia A, Baioni L, Barbieri I, Rosignoli C, et al. Detection of influenza D virus among swine and cattle, Italy. Emerg Infect Dis. 2016;22:352–4. 10.3201/eid2202.151439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami S, Endoh M, Kobayashi T, Takenaka-Uema A, Chambers JK, Uchida K, et al. Influenza D virus infection in herd of cattle, Japan. Emerg Infect Dis. 2016;22:1517–9. 10.3201/eid2208.160362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J Virol. 2015;89:1036–42. 10.1128/JVI.02718-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sreenivasan C, Thomas M, Sheng Z, Hause BM, Collin EA, Knudsen DE, et al. Replication and transmission of the novel bovine influenza D virus in a guinea pig model. J Virol. 2015;89:11990–2001. 10.1128/JVI.01630-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse H, To KK, Wen X, Chen H, Chan KH, Tsoi HW, et al. Clinical and virological factors associated with viremia in pandemic influenza A/H1N1/2009 virus infection. PLoS One. 2011;6:e22534. 10.1371/journal.pone.0022534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose R, Daidoji T, Naito Y, Watanabe Y, Arai Y, Oda T, et al. Long-term detection of seasonal influenza RNA in faeces and intestine. Clin Microbiol Infect. 2016;22:813.e1–7. 10.1016/j.cmi.2016.06.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR method and subcloning protocol used in study of influenza D viruses in cattle, goats, buffalo, and pigs, Guangdong Province and neighboring provinces, China.


