Abstract
With the advent of massively parallel sequencing, considerable work has gone into adapting chromosome conformation capture (3C) techniques to study chromosomal architecture at genome-scale. We recently demonstrated that the inactive murine X chromosome adopts a bipartite structure using a novel 3C protocol, termed in situ DNase Hi-C. Like traditional Hi-C protocols, during in situ DNase Hi-C chromatin is chemically crosslinked, digested, end-repaired, and proximity ligated with a biotinylated bridge adaptor. The resulting ligation products are optionally sheared, affinity-purified via streptavidin bead immobilization, and subjected to traditional next-generation library preparation for Illumina paired-end sequencing. Importantly, in situ DNase Hi-C obviates the dependence on a restriction enzyme to digest chromatin, instead relying on the endonuclease DNase I. Libraries generated by in situ DNase Hi-C have a higher effective resolution than traditional Hi-C libraries, making them valuable in cases where high sequencing depth is allowed for, or when hybrid capture technologies are expected to be used. The protocol described here, which involves approximately four days of bench work, is optimized for the study of mammalian cells but can be broadly applicable to any cell or tissue of interest given experimental parameter optimization.
Introduction
The manner in which an incredibly long DNA polymer toplogically organizes itself within a cell or nucleus is crucially linked to higher-order cellular function1,2. This form-function relationship, first realized through early light microscopic studies of higher-order structures like mitotic chromosomes3, the inactive X Barr body4, and polytene chromosomes5, has only become clearer in the face of advancing technologies. Techniques such as fluorescence in situ hybridization (FISH) of chromatin6–8, have provided clear evidence that chromosomes occupy compartments within the nucleus, ultimately leading to the development of correlative models associating biological function (i.e. transcription, splicing, silencing) with particular nuclear locales9,10.
With the advent of genome-scale technologies, high-throughput assays have been developed to characterize nuclear architecture at both increasing scale and resolution. Techniques like DNA adenine methyltransferase identication (DamID)11,12, typically used to map protein-DNA interactions13–15, have been modified to map genome-wide associations between primary sequence and the nuclear lamina16 (i.e. lamina associated domains, or LADs), where silenced domains typically reside. Methods involving the “proximity ligation” of chromatin, now termed chromosome conformation capture (3C)17, have also gained popularity. 3C techniques represent matured versions of early methods that used T4 DNA ligase to quantify the physical proximity of DNA sequences brought together by proteins18,19, and all share a common paradigm: fixation of chromatin within the nucleus via formaldehyde, endonucleolytic digestion of chromatin (normally via restriction enzyme digestion), and re-ligation of physically proximal fragments. The first 3C variants (e.g. 4C, 5C) used specific primers or sets of primers to determine contact frequencies between predefined sites in the genome20,21. Later, massively-parallel versions of 3C, generally termed “Hi-C”, were developed22–24, which leverage paired-end sequencing to generate contact frequency estimates between sequence windows across entire genomes.
Since the advent of 3C techniques, much work has gone into characterizing 3D genome architecture in a wide-variety of biological contexts25–29, including mitotic cell division30, the life cycle of a parasite31, and in mammalian dosage compensation32–35. The vast amount of available Hi-C data has also enabled the discovery of novel “units” of genome topology, including topologically associating domains (TADs)33,36 and chromosomal interacting domains (CIDs)27,37, genomic domains that predominantly self-associate in three-dimensional space. Although the ultimate significance of these domains remains unknown, strong correlations between one-dimensional epigenomic features (e.g., histone marks, DNA methylation, transcription factor binding) and sequence both within and at the borders of these domains suggest that they may play a gene regulatory role.
Although current Hi-C techniques generally allow us to visualize genome-scale chromosome architecture at the resolution of 100 kb to 1 Mb, methodological resolution limitations imposed by incomplete sequencing depth and genome-wide restriction site density have typically precluded identification of topological units at smaller scales, in which local interactions may play crucial gene regulatory roles. The need for fine-scale resolution of these higher-order interactions has only become clearer in the wake of the immense amount of high-resolution, one-dimensional epigenomic data generated by consortia such as ENCODE38 and Roadmap Epigenomics39.
Given the availability of such data, one crucial interest of the gene regulatory field is the potential link between complex gene regulatory programs and dynamic long-range “looping” interactions between distal regulatory elements, features at a scale even smaller than that of TADs and LADs40. Since the earliest realizations that long-range interactions are effectors of gene expression41,42, the gene regulatory field has worked towards completely cataloguing functional DNA looping interactions. In the realm of proximity ligation protocol development, considerable work has gone towards improving the resolution of the Hi-C protocol to the scale of kilobases, where specific regulatory contacts (i.e. enhancer-promoter interactions, CCCTC-binding factor (CTCF)-mediated loops) might be identified.
The protocol presented here complements existing high-resolution Hi-C approaches37,43 by providing another flexible, convenient, and scalable methodology that eschews the use of restriction enzymes. Our approach therefore avoids the theoretical limit in resolution of the standard Hi-C protocol imposed by the occurrence of restriction sites in the genome, given enough sequencing depth and library complexity.
Moving towards fine-scale resolution of 3D contacts
Core methodological improvements to the Hi-C protocol to improve resolution have broadly spanned three primary areas: deeper sequencing36, simplified library preparation protocols43,44, and the use of hybridization capture to enrich for sets of desired loci in a massively parallel fashion45–47. We recently developed a method that unites many of these improvements with additional empirical changes to further increase the effective resolution of Hi-C libraries48. Our method, termed DNase Hi-C, eliminates the reliance on restriction enzymes associated with Hi-C by digesting fixed chromatin with the endonuclease DNase I in the presence of divalent manganese. We demonstrated that DNase Hi-C libraries mitigate many of the biases associated with traditional Hi-C, reducing the effective distance between fragments imposed by 4- and 6-cutter restriction enzymes while improving robustness with respect to G-C content, mappability, and genomic coverage. Furthermore, we also showed that DNase Hi-C may be paired with commercially available hybridization capture kits to visualize long intergenic noncoding RNA (lincRNA) promoters at a previously unprecedented scale of 1 kb without the gross sequencing depth requirements typically associated with high-resolution contact maps.
Motivated by the observation that the vast majority of proximity ligations occur in insoluble chromatin49, and consequent improvements to traditional RE Hi-C using this fact43,44,50, we recently published an improved version of our previously published DNase Hi-C termed in situ DNase Hi-C51. We applied this simplified and robust Hi-C protocol to study the inactive X chromosome in primary mouse brain tissue and an immortalized mouse embryonic kidney cell line, demonstrating for the first time that the murine inactive X chromosome adopts a bipartite conformation. In situ DNase Hi-C represents a considerable improvement over its parent protocol, requiring considerably less hands-on time and lower cellular input requirements51.
Overview of in situ DNase Hi-C
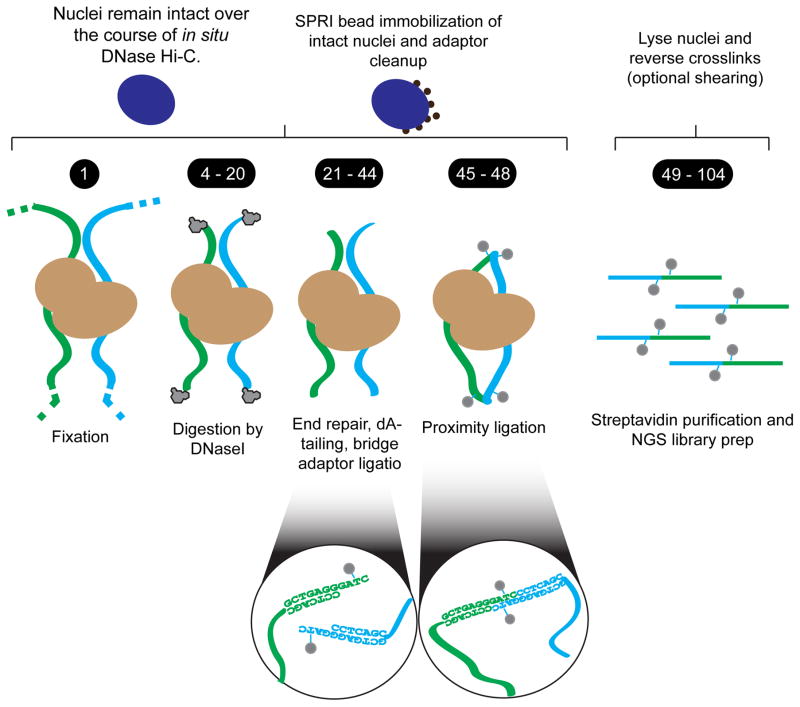
A schematic of the in situ DNase Hi-C protocol is illustrated in Figure 1. Anywhere from 5x105 to 1x107 cells are fixed in formaldehyde to reversibly crosslink in vivo protein-DNA interactions. Fixed cells are then lysed to liberate nuclei, which are treated with the endonuclease DNase I to digest chromatin. Digested chromatin ends are end-repaired and dA-tailed, facilitating the ligation of an exogenous, dT-tailed “bridge” adaptor containing a single biotinylated thymidine, half BamHI restriction site, and 4-base overhang. After clearing out excess adaptors, the free ends of chromatin (now capped with bridge adaptors) are phosphorylated with T4 Polynucleotide Kinase (T4 PNK) and proximity ligated in situ with T4 DNA Ligase I. During all of these steps, nuclei are immobilized against carboxylated paramagnetic beads (commonly referred to as Solid Phase Reversible Immobilization (SPRI) beads18), both providing a scaffold to prevent loss of nuclei during enzymatic reactions and allowing for the simple removal of free DNA and excess bridge adaptor, which adversely affect downstream library preparation.
Figure 1. A schematic overview of in situ DNase Hi-C.
First, fixed cells are lysed and digested with the endonuclease DNase I in the presence of divalent manganese—yielding double stranded breaks. Nuclei are then immobilized on carboxylated paramagnetic beads (i.e. ‘AMPure’ beads) to purify intact nuclei and remove free digested DNA fragments. Chromatin is then end-repaired and dA-tailed in situ, and a biotinylated ‘bridge adaptor’ containing a half BamHI site is ligated onto free chromatin ends. Nuclei are then subjected to phosphorylation and in situ proximity ligation, after which DNA is purified and fragments containing ligation junctions are enriched for via streptavidin beads and on-bead Illumina library prep (optionally following sonication).
Following proximity ligation, nuclei are lysed and crosslinks are reversed with Proteinase K treatment. DNA is then isolated with an isopropanol precipitation, after which fragments are optionally sheared. Ligated DNA fragments harboring the biotinylated bridge adapter are then affinity-purified using streptavidin beads, end-repaired, dA-tailed, and ligated to standard Illumina sequencing adaptors. Finally, ligation products are PCR amplified to generate sequencing libraries. Prior to sequencing, libraries may be treated with a simple BamHI digestion to assess the efficiency of proximity ligation.
Traditional Hi-C vs. in situ DNase Hi-C
In situ DNase Hi-C can be used in any situation where traditional Hi-C would be used. Thanks to a reliance on the endonuclease DNase I, in situ DNase Hi-C eliminates the characteristic restriction enzyme biases that limit resolution in traditional Hi-C libraries while lowering the input cell requirements for library construction. Unlike other Hi-C protocols, in situ DNase Hi-C is the only protocol, to our knowledge, to use paramagnetic carboxylated beads as a tool to immobilize nuclei during in situ enzymatic treatments. This immobilization step not only reduces nuclei loss during the protocol, aiding low-input experiments, but also facilitates the removal of contaminating adaptors and free DNA. Finally, like traditional in situ Hi-C, in situ DNase Hi-C requires considerably less hands-on time for library prep, and more efficiently generates cis (i.e. intrachromosomal) ligation products compared to trans (i.e. interchromosomal) ligation products.
Considering the high sequencing depth required to generate high-resolution genome-wide contact maps, we note that at low resolution, maps generated using in situ DNase Hi-C are practically very similar to those generated using other Hi-C protocols (except in cases where loci may have particularly low restriction site density). In cases where high-resolution (i.e. 1 kb resolution) maps are desired, however, we strongly believe that the relatively unbiased ligation junctions generated through DNase Hi-C present an important alternative to existing methods. This point is particularly relevant when hybrid capture techniques may be applied, as high-resolution, RE independent maps can be generated for a fraction of the cost of genome-scale library sequencing.
Still, we acknowledge that in many cases cost may preclude the use of deep sequencing or hybrid capture. In cases such as these, we suggest more cost-effective solutions using more focused techniques (e.g. 3C, 4C, 5C), albeit at the price of only interrogating interactions among a set number of loci.
In situ DNase Hi-C is broadly applicable to any situation where high-resolution chromatin conformation data or 3D maps are required. We have successfully carried out in situ DNase Hi-C in several immortalized cell lines and primary tissues, generating libraries for the human cell lines K562 and GM12878, as well as mouse embryonic kidney cells and homogenized mouse brain tissue51.
Limitations of the protocol
In situ DNase Hi-C is subject to the same limitations as any bulk Hi-C protocol. First, the protocol requires 5x105 to 1x107 cells to generate sequenceable libraries. Thus, in cases where input might be particularly limited, or where small populations of cells sorted by fluorescence activated cell sorting (FACS), in situ DNase Hi-C may not be appropriate. Second, it is also important to note that while the DNase enzyme is nonspecific when compared to restriction enzymes, it has been shown to exhibit mild sequence bias at cleavage sites52. This must be considered when applying in situ DNase Hi-C to organisms with radical nucleotide content (i.e. low GC content), and when considering the inherent biases within in situ DNase Hi-C maps (as would be done with any Hi-C contact map53).
Experimental design considerations
The in situ DNase Hi-C protocol described here is relatively straightforward, and can be completed over four days, allotting 3 – 6 hours of bench work per day. Still there are several experimental design parameters that should be considered before applying in situ DNase Hi-C to a new cell type of interest. These considerations primarily concern maintaining intact nuclei during the various in situ enzymatic treatments in the protocol. The in situ DNase Hi-C protocol also allows for sequencing-free quality control of libraries, thanks to the integration of half BamH1 sites in the bridge adapter. As discussed below, this allows for easy quantification of the efficiency of proximity ligation in the final in situ DNase Hi-C library.
Although the protocol presented here is robust to many different cell types, different immortalized cell lines may require optimization of formaldehyde crosslinking, DNase I digestion and SDS concentration during digestion. Below we detail our process for optimizing these various parameters:
Formaldehyde concentration
As with other 3C methods and ChIP-seq protocols, formaldehyde fixation is an important component of the in situ DNase Hi-C protocol, promoting proximity ligation of long-range genomic contacts while maintaining the integrity of nuclei during in situ enzymatic steps. Incomplete crosslinking can lead to an underrepresentation of proximity ligation products in Hi-C libraries, and excessive breakage of nuclei can lead to considerable decreases in the ultimate molecular complexity of libraries, and at worst can increase the degree of “spurious” ligations formed. The guidelines for formaldehyde fixation of cells for in situ DNase Hi-C are the same as those for the other 3C-based techniques and ChIP-seq methods. In general, for single-cell suspension cultures (e.g. GM12878 and K562 cells) and monolayer adherent cells (e.g. Hela cells) a standard condition of cross-linking, such as 1% formaldehyde for 10 min at room temperature (RT, 25°C), can be employed. For other cell cultures (e.g. mouse and human embryonic stem cells (ESCs)) and primary tissue cells (e.g. mouse brain cells and plant leaves), for which single-cell suspensions are difficult to obtain, increased formaldehyde concentrations or longer fixation times may be required to ensure efficient crosslinking. For example, both human and mouse ESCs often aggregate to form large clumps in culture. As such, higher concentrations of formaldehyde are generally used in these situations48,54.
Cell lysis and DNase I digestion
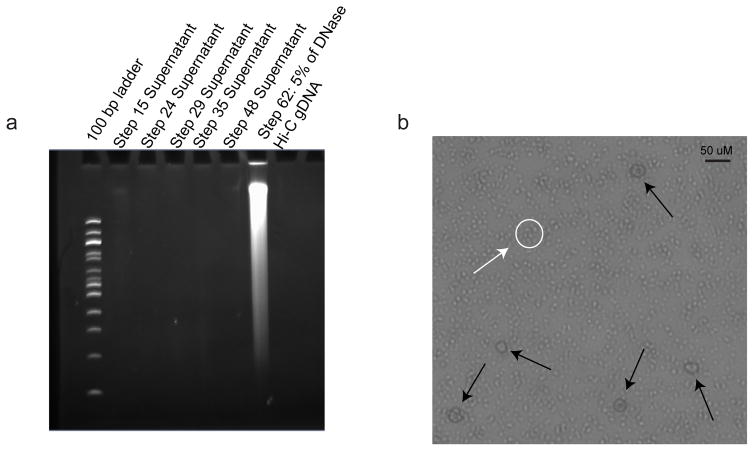
After crosslinking chromatin interactions with formaldehyde, one must render fixed chromatin accessible to enable chromatin fragmentation and other downstream enzymatic reactions. As with restriction digestion-based 3C methods, cell lysis in in situ DNase Hi-C is achieved primarily through SDS treatment. To ensure that nuclei remain intact throughout the multiple enzymatic reactions through the end of nuclear ligation (step 48), the in situ DNase Hi-C protocol employs a relative mild condition (0.3–0.5% SDS treatment for 45 min. at 37°C). During this step, it is crucial to avoid overly lysing nuclei. A simple experiment to determine the extent of nuclear lysis is detailed in Box 1, with expected results shown in Figure 2a. We also note that overly lysed nuclei become apparent during any of the many centrifugation steps in the in situ DNase Hi-C protocol, as no pellet forms. Nuclei should remain intact through proximity ligation, as shown in Figure 2b.
BOX 1. Assessment of nuclear lysis at various steps.
To ascertain whether nuclei remain intact during the protocol perform the following control experiment. Following each enzymatic treatment step (Steps 17, 23, 28, 33, and 47), remove the supernatant and add 10 uL Proteinase K to it. Treat the supernatant overnight at 65°C, and then isopropanol precipitate the DNA by adding 0.1 volumes of 3M sodium acetate, 3 uL GlycoBlue, and 1 volume of 100% isopropanol, mixing, and then incubating mixtures at −80°C for 1 hour. Pellet mixtures at 4°C at 16,000xg, carefully remove the supernatant, and resuspend the pellet in 100 uL ddH2O. Add 10 uL RNase A to each sample and incubate at 37°C for 10 minute, then purify DNA using 1.2 volumes of AMPure XP beads. Resuspend beads in 15 uL ddH2O, and run this out on a 6% TBE gel.
Figure 2. Nuclei remain intact during the in situ DNase Hi-C protocol.
a.) Purified supernatant DNA (see Box 1) from 6 different steps of the DNase Hi-C protocol. Minimal DNA is purified after each enzymatic purification, compared to a large amount of DNA, taken from 5% of the total gDNA yield following nuclear lysis. b.) Phase contrast micrograph (20X magnification) of GM12878 nuclei bound to beads, following proximity ligation (Step 46). Nuclei are highlighted using black arrows, and an example of a clump of carboxylated beads, which are found scattered across the image, is shown circled in white, with an accompanying white arrow.
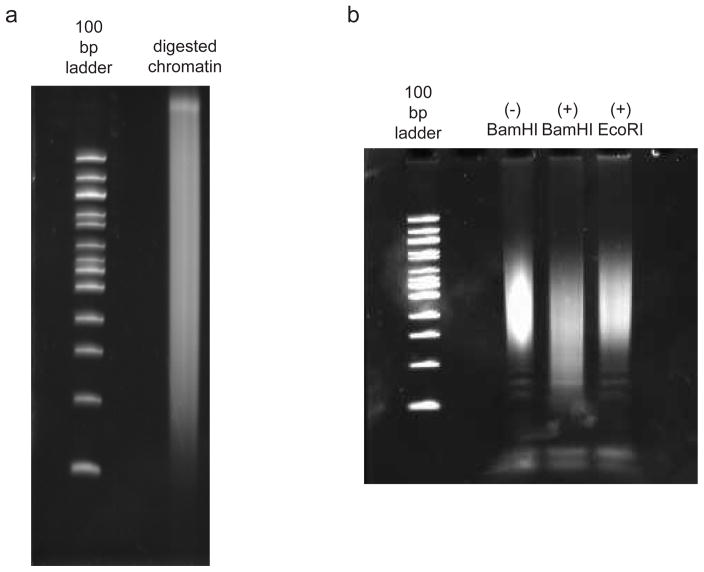
We stress that the required SDS concentration for cell lysis and the amount of DNase I used during the DNase I digestion step can vary depending on the cell type being studied, and the number of nuclei being processed. We recommend carrying out a DNase I and SDS optimization experiment using varying units of DNase I and varying concentrations of SDS when attempting the protocol on new cell types, and examining the DNase I fragmentation pattern following digestion. An example fragmentation pattern is shown in Figure 3a.
Figure 3. Digestion quality controls throughout the in situ DNase Hi-C protocol.
a.) A typical digestion pattern for DNase I-digested fixed chromatin prior to proximity ligation, run on a 6% TBE-PAGE gel. b.) Example of the BamH1 quality control experiment performed on GM12878 in situ DNase Hi-C libraries; in this example, BamH1 shifts the in situ DNase Hi-C library by digesting the reconstituted BamH1 site that forms following proximity ligation of the biotinylated bridge adaptors. Crucially, digestion with another 6-cutter (EcoR1), does not recapitulate this pattern, proving that the BamH1 digestion is specific to proximity ligated fragments. All reactions were run on one 6% TBE-PAGE gel.
The role of paramagnetic carboxylated beads
Paramagnetic carboxylated beads (i.e. AMPure XP beads) have been used in both our standard and in situ DNase Hi-C protocols. As demonstrated in Figure 2, these beads appear to bind to intact nuclei and serve as carriers to pellet the nuclei by low-speed centrifugation. Here, we employ these beads to efficiently remove DNase I and low molecular weight DNA that might escape the nucleus following chromatin digestion, as well as free unligated internal bridge adaptor following bridge adaptor ligation. Furthermore, the beads also aid with visualization of the nuclei pellet throughout the protocol when starting the protocol with fewer than a million cells.
Nuclei treatment
It is crucial that the fixed nuclei remain intact over the course of the DNase Hi-C protocol. To this end, pipetting should be carried out gently to minimize shear forces that may burst nuclei.
BamH1 Digestion Control
A BamH1 digestion test on the final PCR-amplified library can be used to quantify ligation efficiency of the reaction. Lack of a library “shift” (properly digested products shown in Figure 3b) suggests inefficiency in the formation of proximity ligation products, and can be indicative of suboptimal fixation conditions or defective reagents.
Materials
Reagents
-
Cell lines of interest (adherent, suspension, or primary tissue): For example, we have used the human cell line GM12878 (Coriell GM12878) and the Patski cell line in our previous study51.
CAUTION: Cell lines should be regularly checked to ensure that they are authentic and not infected with Mycoplasma.
Pen/Strep Cocktail (Thermo Scientific 15140122)
Fetal Bovine Serum (Thermo Scientific 10437-010)
Cell culture medium: RPMI-1760 w/15% FBS (for GM12878; Thermo Scientific 11875-093) or DMEM w/10% FBS (for Patski; Thermo Scientific 11965118)
Biotinylated Bridge Adaptor 5′: /5Phos/GCTGAGGGA/iBiodT/C (IDT)
Bridge Adaptor 3′T: CCTCAGCT (IDT)
Bridge Adaptor 5′: GCTGAGGGAC (IDT)
Blunt Bridge Adaptor 3′: CCTCAGC (IDT)
SeqAdapt_F: ACACTCTTTCCCTACACGACGCTCTTCCGATC*T (IDT)
SeqAdapt_R: /5Phos/GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (IDT)
SeqPrimer_F: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT (IDT)
SeqPrimer_R: CAAGCAGAAGACGGCATACGAGAT[8 bp barcode] GTGACTGGAGTTCAGACGTGTGCT (IDT)
-
37% (vol/vol) Formaldehyde (Sigma Aldrich F8775)
CAUTION!: Formaldehyde is flammable, can cause skin burns, and is toxic by inhalation. Formaldehyde should be handled using appropriate protective equipment, and be handled in a chemical fume hood.
CRITICAL: Formaldehyde has a limited shelf life; discard solution if it is older than 1 year.
Glycine (Sigma-Aldrich 50046)
NEBuffer 2 (NEB B7002S)
10% (wt/vol) UltraPure SDS (Life Tech 15553-027)
DNase I, RNase-free (supplied with MnCl2 and reaction buffer) (1 U/μL) (Thermo Scientific EN0525)
RNase A, DNase and protease-free (10 mg/ml) (Thermo Scientific EN0531)
Klenow Fragment (10 U/μL) (Thermo Scientific EP0052)
Klenow Fragment (exo–) (5 U/μL) (Thermo Scientific EP0422)
T4 DNA Polymerase (5 U/μL) (Thermo Scientific EP0062)
T4 DNA Ligase (5 U/μL) provided with 50% PEG-4000 (Thermo Scientific EL0012)
T4 Polynucleotide Kinase (10 U/μL) (Thermo Scientific EK0032)
10X T4 DNA Ligase Buffer w/ATP (NEB B0202S)
Agencourt AMPure XP (Beckman Coulter A63880)
2X HotStart PCR ReadyMix (KAPA KK2601)
Fast DNA End Repair Kit (Thermo Scientific K0771)
Proteinase K (Thermo Scientific EO0492)
FastDigest® BamHI (Thermo Scientific FERFD0504)
Dynabeads® MyOne™ Streptavidin C1 (Life Tech 65001)
dNTP Set (100 mM 4x 0.25ml) (Thermo Scientific FERR0181)
100 bp DNA ladder (Thermo Scientific SM0243)
GlycoBlue (Ambion AM9516)
3 M Sodium acetate (pH 5.2) (Cellgro 46-033-CI)
Ethanol (Decon Labs Inc. 2716) CAUTION! Ethanol is flammable, and should be stored and handled under appropriate conditions
Isopropanol (Sigma-Aldrich 437522) CAUTION! Isopropanol is flammable, and should be stored and handled under appropriate conditions
1M UltraPure Tris-HCl pH 8.0 (Life Tech 15568-025)
1M UltraPure Tris-HCl pH 7.5 (Life Tech 15567-027)
Buffer EB (Qiagen 19086)
PEG-8000 (Sigma-Aldrich 89510)
0.5 M EDTA (Cellgro 46-034-CI)
Qubit dsDNA HS kit (Life Tech Q32851)
IGEPAL CA-630 (Sigma-Aldrich I8896-50ML)
Triton X-100 (Sigma-Aldrich X100-5ML)
1X DPBS (Life Tech 14190-250)
Protease Inhibitor Tablets (e.g. Roche 04693116001)
QIAquick PCR Purification Kit (Qiagen 28104)
Rapid DNA Ligation Kit (Thermo Scientific K1422)
5M Sodium Chloride (NaCl) (S5150-1L)
0.25% Trypsin-EDTA (Thermo Scientific 25200056)
Millipore Steriflip Filters (Millipore SCGP00525)
Equipment
Water bath (set to 60°C)
Thermocycler
DynaMag Magnetic Rack (e.g. Life Tech 12321D)
Qubit Fluorometer (e.g. Life Tech Q33216)
0.2 mL PCR tubes (e.g. Fisher 14-230-212)
1.5 mL microcentrifuge tubes (e.g. Fisher 05408129)
6% TBE-PAGE gels (e.g. Life Tech EC6265BOX)
Cell scraper (e.g. Fisher 08-100-241)
50 ml tube (e.g. Fisher 14-432-22)
Cell culture plates (e.g. Sigma CLS430167-100EA)
Microcentrifuge
A computer running Unix/Linux distribution with the software listed below installed.
Software
Python 2.7 (http://www.python.org/)
FastQC version 0.11.3 or higher (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
BWA version 0.5.9 or higher (http://bio-bwa.sourceforge.net/)
samtools version 0.1.18 or higher (http://samtools.sourceforge.net/)
hiclib library (http://mirnylab.bitbucket.org/hiclib/)
matplotlib library (http://matplotlib.org/)
Reagent Setup
2.5 M Glycine: Bring 9.35 g glycine to 50 mL in ddH2O and filter sterilize using Steriflip column. Store at RT for up to 6 months.
80% (vol/vol) Ethanol: Mix 8 mL 100% ethanol with 2 mL ddH2O. Make fresh for every day of experiments.
10% (vol/vol) Triton X-100: Mix 1 mL 100% Triton X-100 with 9 mL ddH2O. Store at RT for up to 6 months.
1X Cell Lysis buffer: Mix 500 μL 1M Tris-HCl pH 8.0, 100 μL 5M NaCl, and 1 mL 10% Igepal CA-630 and bring up to 50 mL in ddH2O. Store at 4°C for up to 6 months.
1X TE Lysis buffer: Mix 2.5 mL 1M Tris-HCl pH 7.0, 100 μL 0.5M EDTA, and 5 mL 10% SDS and bring up to 50 mL in ddH2O. Store at RT for up to 6 months.
2X B&W buffer: Mix 500 μL 1M Tris-HCl pH 8.0, 100 μL 0.5M EDTA, and 20 mL 5M NaCl and bring up to 50 mL in ddH2O. Store at RT for up to 6 months.
0.5X DNase Digestion Buffer: Mix 25 μL 10X DNase digestion buffer with 25 μL 10 mM MnCl2 and 450 μL ddH2O. Use immediately.
6X Stop Solution: Mix 12.5 mL 0.5M EDTA with 12.5 mL 10% SDS and bring to 50 mL in ddH2O. Store at RT for up to 6 months.
AMPure Buffer: Mix 10 g PEG-8000 in 25 mL 5M NaCl and bring to 50 mL with ddH2O. Shake vigorously to mix until PEG-8000 has completely gone into solution. Store at 4°C for up to 6 months
Procedure
Steps 1–2: Adaptor Annealing (Timing: 1h + overnight incubation)
-
1)
Set up the following reactions:
Blunt Bridge Adaptor (40 μM final) Component Amount (μL) Final Concentration 100 μM Bridge Adaptor 5′ 80 40 μM 100 μM Blunt Bridge Adaptor 3′ 80 40 μM 10X NEBuffer 2 20 1X ddH2O 20 Total Volume 200 Biotinylated Bridge Adaptor (40 μM final) Component Amount (μL) Final Concentration 100 μM Biotinylated Bridge Adaptor 5′ 80 40 μM 100 μM Bridge Adaptor 3′T 80 40 μM 10X NEBuffer 2 20 1X ddH2O 20 Total Volume 200 Y-Adaptor (25 μM final) Component Amount (μL) Final Concentration 100 μM SeqAdapt_F 50 25 μM 100 μM SeqAdapt_R 50 25 μM 10X NEBuffer 2 20 1X ddH2O 80 Total Volume 200 -
2)
Anneal mixtures by heating to 98°C for 6 minutes, then allow the tubes to naturally cool to RT overnight.
PAUSE POINT Annealed adapters can be kept at −20°C indefinitely.
Step 3: Cross-linking of cells (Timing: 2h)
[TROUBLESHOOTING 3]
-
3)
Cells should be grown in appropriate culture medium. 2–5 x 106 cells are sufficient for making one DNase Hi-C library. However, we suggest growing, crosslinking, and aliquoting many cells (i.e. 1–5 x 107 cells) to provide replicates if necessary. Below are protocols for handling adherent monolayer cells (option A) or suspension cells (option B):
a) Adherent monolayer cells
Aspirate out media and add 10 ml of serum-free media per 10 cm plate.
Crosslink the cells by adding 280 μl of 37% formaldehyde to obtain 1% final concentration. Mix gently, immediately after addition of formaldehyde.
Incubate cells at RT for exactly 10 min, gently rocking the plates every 2 min.
Quench reaction by adding 0.5 ml of 2.5 M glycine and mixing well.
Incubate for 5 min at RT, then on ice for 15 min to stop cross-linking completely.
Wash cells once with cold 1X PBS.
Treat the cells with 3–5 ml per dish trypsin (0.25%) at 37°C for 5 min.
Add 5 ml fresh medium with serum.
Scrape the cells from the plates with a cell scraper and transfer to a 50 ml tube (combine all the cells from all the dishes to one tube).
Centrifuge the cross-linked cells at 800xg for 10 min.
Discard the supernatant by aspiration and wash the cross-linked cells with 1 x PBS once.
-
Aliquot the cells into 1.5 ml microtubes (2.5 million cells per tube).
PAUSE POINT: Cells can be snap-frozen in liquid nitrogen and stored for at least one year at −80°C, or one can continue with cell lysis.
b) Suspension cells
Gently pellet the cells by spinning at 300xg for 10 min at RT.
Discard the supernatant.
Resuspend the pellet in 10 ml of fresh culture medium without serum. Break cell clumps by pipetting up and down.
Crosslink the cells by adding 280 μl of 37% formaldehyde (1% final concentration). Mix quickly by inverting the tube several times.
Incubate at RT for exactly 10 min. Gently invert the tube every 2 min.
Add 0.5 ml of 2.5 M glycine to quench the cross-linking reaction, mix well.
Incubate for 5 min at RT, then on ice for 15 min to stop cross-linking completely.
Centrifuge the cross-linked cells at 800xg for 10 min at 4°C.
Discard the supernatant by aspiration and wash the cross-linked cells with 1X PBS once.
Split the cross-linked cell suspension into aliquots of 2.5 x 106 cells (in 1.5 ml microtubes).
Centrifuge the cross-linked cells at 800xg for 10 min at RT.
-
Discard the supernatant by aspiration.
PAUSE POINT: Cells can be snap-frozen in liquid nitrogen and stored for up to 1.5 years at −80°C, or one can continue with cell lysis.
Steps 4 – 20: Cell lysis and chromatin digestion with DNase I (Timing: 1.5 h)
-
4)
Resuspend one cross-linked cell aliquot (0.5–2.5 x 106 cells) in 0.4 ml of ice-cold cell lysis buffer containing protease inhibitor.
CRITICAL STEP: Add 1 tablet protease inhibitor to 10 mL of ice-cold lysis buffer immediately prior to lysis. We recommend using lysis buffer with freshly added protease inhibitor for all experiments.
-
5)
Incubate on ice for 10 min.
-
6)
Centrifuge for 60 seconds at 2,500xg at RT.
[TROUBLESHOOTING 7]
-
7)
Discard the supernatant and resuspend the pellet in 100 μl of 0.5X DNase I digestion buffer containing 0.2% SDS.
CRITICAL STEP: For larger cell inputs (i.e. 3–5 x 106), we recommend using 200 μl 0.5X DNase I digestion buffer instead.
-
8)
Incubate at 37°C for 30 min.
-
9)
Add 100 μl of 0.5X DNase I digestion buffer containing 2% Triton X-100 and 4 μl RNase A, mix well.
-
10)
Incubate at 37°C for 10 min.
-
11)
Add 1.5 units of DNase I and mix well.
-
12)
Incubate at RT for 4 min.
-
13)
Add 40 μl of 6X Stop Solution, mix well.
[TROUBLESHOOTING 14]
-
14)
(Optional) To determine the efficacy of DNase I digestion, take 20 μl of lysed cells from the previous step and add to a new tube. Add 70 μl 1X TE lysis buffer and 10 μl Proteinase K (20 mg/ml). Incubate for 30 minutes at 65°C. Purify DNA using a Qiaquick PCR purification kit. Check the quality of chromatin digestion by running the samples out on a 6% TBE-PAGE gel. The sample is properly digested if one sees a large smear of DNA fragments between ~100 bp and 1 kb (see Figure 3a). We recommend characterizing DNase I digestion efficiency when performing the protocol on a new cell type. In the event of over-digestion or under-digestion of chromatin, we recommend optimizing the concentration of SDS in the digestion reaction, amount of DNase I used, or digestion time.
-
15)
Centrifuge for 60 seconds at 2,500xg at RT.
-
16)
Discard the supernatant and resuspend the pellet in 150 μl water.
-
17)
Add 300 μl AMPure XP beads; mix thoroughly by pipetting up and down.
-
18)
Incubate at RT for 5 min and place the tube in a DynaMag magnet for 2 min.
-
19)
Discard the supernatant and wash the beads twice with 1 ml of freshly prepared 80% ethanol. Briefly spin down the beads and remove the residual ethanol.
-
20)
Resuspend the beads in 169 μl of water, and proceed immediately to the next step.
Steps 21 – 30: Chromatin End Repair and dA-tailing (Timing: 2.5 h)
-
21)
Prepare the End-Repair reaction as follows:
Reagents (add in this order) Volume (μL) Final Concentration Nuclei w/ beads 169 10X T4 ligase buffer w/ ATP 20 1X 10 mM dNTPs 5 0.25 mM T4 DNA Polymerase (3U/ μl) 3 0.045 U / μL Klenow (10U/ μl) 3 0.15U / μL Total Volume 200 -
22)
Incubate at RT for 1 h.
-
23)
Add 5 μl 10% SDS to stop the reaction.
-
24)
Centrifuge for 60 seconds at 2,500xg at RT.
-
25)
Aspirate and resuspend the pellet in 135 μl water.
-
26)
Prepare the dA-Tailing reaction as follows:
Reagents (add in this order) Volume (μL) Final Concentration Nuclei w/ beads 135 10X NEBuffer 2 20 1X 10 mM dATP 10 0.5 mM 10% Triton X-100 20 1% Klenow (exo-) (5U/ μl) 15 0.375U / μL Total Volume 200 -
27)
Incubate the resulting mixture at 37°C for 1 hr.
-
28)
Add 5 μl 10% SDS to stop reaction.
-
29)
Centrifuge for 60 seconds at 2,500xg at RT.
-
30)
Aspirate and resuspend the pellet in 30 μl nuclease-free water.
Steps 31 – 44: Ligation of Biotin-labeled Bridge adaptors (Timing: Overnight, followed by 0.5 h)
-
31)
Prepare the adaptor ligation reaction as follows:
Reagents (add in this order) Volume (μl) Final Concentration Nuclei w/ beads 30 Annealed Bridge Adaptor w/ Biotin (40 μM) 20 8 μM Annealed Blunt Adaptor w/o Biotin (40 μM) 20 8 μM 10X T4 ligase buffer w/ ATP 10 1X PEG-4000 (50%) 10 5% 10% Triton X-100 5 0.5% T4 DNA Ligase (5 U/ μl) 5 0.25U / μL Total Volume 100 -
32)
Incubate at 16°C overnight.
PAUSE POINT: Reaction should be allowed to incubate overnight.
-
33)
(Optional) To examine the efficacy of the above end-repair, dA-tailing and adaptor ligation reactions, take 3 μl of nuclei from the step 30 to perform a control ligaion reaction with the Illumina Y-adaptor as below:
Reagents (add in this order) Volume (μl) Final Concentration Nuclei w/ beads 3 Illumina Y-adaptor (50 μM) 1 2.5 μM Water 10 10X T4 ligase buffer w/ ATP 2 1X PEG-4000 (50%) 2 5% 10% Triton X-100 1 0.5% T4 DNA Ligase (5 U/ μl) 1 0.25U / μL Total Volume 20 After incubation at 16°C for overnight, add 70 μl 1X TE lysis buffer and 10 μl Proteinase K (20 mg/ml). Incubate for 30–60 min at 65°C. Purify genomic DNA using a QiaQuick PCR purification kit. Check the ligation efficiency by carrying out qPCR with Illumina PCR primers. If upstream end-repair and dA-tailing steps are efficient, one should see amplification before 10 PCR cycles using 10 ng genomic DNA as template. We recommend this quality control step when performing the protocol on a new cell type. In the event of inefficiency of these steps, we recommend optimizing the concentration of SDS in the cell lysis step, or the amount of DNase I digestion used.
-
34)
Add 5 μl of 10% SDS to stop the reaction.
-
35)
Centrifuge for 60 seconds at 2,500xg at RT.
-
36)
Resuspend the pellet in 200 μl nuclease-free water.
-
37)
Add 165 μl AMPure buffer; mix thoroughly by pipetting up and down.
-
38)
Incubate at RT for 5 min, and place the tube in a DynaMag magnet for 2 min.
-
39)
Discard the supernatant and wash the beads once with 1 ml of freshly prepared 80% ethanol. Briefly spin down the beads and remove the residual ethanol.
CRITICAL STEP: We recommend diluting fresh 80% ethanol before every experiment.
-
40)
Resuspend the pellet in 200 μl water.
-
41)
Add 165 μl of AMPure bead buffer; mix thoroughly by pipetting up and down.
-
42)
Incubate mixture at RT for 5 min, then place tube in DynaMag magnet for 2 min.
-
43)
Discard the supernatant and wash the beads twice with 500 μl of 80% ethanol. Briefly spin down the beads and remove residual ethanol as completely as possible, then air-dry the beads for no more than 2 min.
-
44)
Resuspend the nuclei-bead mixture in 80 μl of nuclease-free water.
Steps 45 – 46: In situ phosphorylation (Timing: 1.25 h)
-
45)
Prepare the PNK reaction as follows:
Reagents (add in this order) Volume (μL) Final Concentration Nuclei w/ beads 80 10X T4 ligase buffer w/ ATP 10 1X PNK (10 U/ μl) 10 1U / μL Total Volume 100 -
46)
Incubate at 37°C for 1 hr.
Steps 47 – 48: In situ ligation (Timing: 4.25 h)
-
47)
Add the following reaction to the above tube:
Reagents (add in this order) Volume (μl) Final Concentration H2O 794 10X T4 ligase buffer 100 1X T4 DNA ligase (5 U/ μl) 6 0.03U / μL Total Volume 1 mL -
48)
Incubate at RT for 4 hr. For a micrograph of nuclei after this stage, see Figure 2b.
Steps 49 – 62: Cross-linking reversal, isopropanol precipitation and DNA purification (Timing: Overnight, followed by 2.5 h)
-
49)
Centrifuge for 60 seconds at 2,500xg at RT.
-
50)
Resuspend the pellet in 400 μl 1X NEBuffer 2.
-
51)
Add 40 μl 10% SDS.
-
52)
Add 40 μl of 20 mg/ml Proteinase K.
-
53)
Incubate overnight at 60°C.
-
54)
Add 3 μl GlycoBlue, 50 μl 3M sodium acetate, pH 5.2 and 550 μl of isopropanol.
-
55)
Incubate mixture at −80°C for 2 hours.
-
56)
Centrifuge mixture for 30 min. at 4°C at maximum speed in a microcentrifuge.
-
57)
Resuspend the DNA pellets in each tube with 100 μl nuclease-free water.
-
58)
Add 100 μl AMPure XP beads, mix well.
-
59)
Incubate mixture at RT for 5 min, and place the tube in a DynaMag magnet for 2 min.
-
60)
Discard the supernatant and wash the beads twice with 1 ml of 80% ethanol. Briefly spin down the beads and remove residual ethanol as completely as possible, then air-dry the beads for no more than 2 min.
-
61)
Resuspend the beads in 130 μl nuclease-free water.
-
62)
Incubate beads at RT for 1 min. Collect beads via DynaMag magnet and transfer eluent to fresh 1.5 mL tube. At this point, determine the concentration of the recovered DNA with a spectrophotometer. A typical yield is 3–5 μg if starting with 2.5x106 cells.
PAUSE POINT: Purified DNA can be stored indefinitely at −20°C.
Steps 63–65: DNA Sonication (Timing: 0.5 h)
CRITICAL At this point, purified DNA may be sonicated to shear large fragments to the 100–500 bp range or taken directly to sequencing library prep. Sonication promotes a less biased representation of fragment ends at the cost of additional prep time and loss of material. The protocol here is suitable for Covaris sonicators. If sonication is not desired, skip to step 66.
-
63)
Transfer DNA to Covaris microtube.
-
64)
Shear the DNA to a size of 100 – 500 bp using a sonicator. For a Covaris instrument use the following parameters:
Duty Cycle 15% Peak Incident Power 450 Cycles per Burst 200 Set Mode Frequency sweeping Continuous degassing Process time 80 s Number of cycles 5 -
65)
Transfer 130 μL sonicated DNA to a 1.5 ml tube.
PAUSE POINT: Eluted DNA may be stored indefinitely at −20°C.
Steps 66 – 74: Biotin pull-down (Timing: 0.5 h)
-
66)
Wash 30 μl of MyOne C1 beads twice with 100 μl 1X B&W buffer, once with 100 μl 2X B&W buffer, then resuspend in 100 μl 2X B&W buffer.
-
67)
Add 100 μl eluted DNA to resuspended streptavidin beads and mix well.
-
68)
Incubate the sample for 20 min at RT on a rotator.
-
69)
Place tube in DynaMag magnet for 1 min and discard the supernatant.
-
70)
Wash beads once with 300 μl 0.5X TE Lysis Buffer plus 300 μl 0.5X B&W buffer.
-
71)
Wash beads twice with 600 μl 1X B&W buffer.
-
72)
Wash beads once with 600 μl 1X NEBuffer 2.
-
73)
Wash beads once with 600 μl EB buffer.
-
74)
Resuspend beads in 170 μl of EB buffer.
PAUSE POINT: Resuspended beads may be stored at −20°C indefinitely or 4°C for short-term storage.
Steps 75 – 84: End Repair and dA-tailing (Timing: 1.5 h)
-
75)
Set up the end-repair reaction with the Fast DNA End Repair Kit as follows:
Reagents (add in this order) Volume (μl) Final Concentration Purified DNA 170 10X Reaction buffer 20 1X End-repair enzyme mix 10 Total Volume 200 -
76)
Incubate at 18°C for 10 min.
-
77)
Add 200 μl of Ampure buffer, mix thoroughly by pipetting up and down.
-
78)
Incubate at RT for 5 min and place the tube in a DynaMag-Spin magnet for 2 min.
-
79)
Discard the supernatant and wash the beads twice with 500 μl of 80% ethanol. Briefly spin down the beads, remove the residual ethanol as completely as possible, and air-dry the beads for 5 min.
-
80)
Resuspend beads in 21.5 μl water.
-
81)
Set up the dA-tailing reaction as follows:
Reagents (add in this order) Volume (μl) Final Concentration End-repaired DNA w/ beads 21.5 10X NEBuffer 2 3 1X 10 mM dATP 3 1 mM Klenow (exo-) (5 U/ μl) 2.5 0.42U / μL Total Volume 30 -
82)
Incubate at 37°C for 30 min.
-
83)
Wash beads twice with 400 μl 1X B&W buffer.
-
84)
Wash beads twice with 400 μl EB buffer and resuspend in 30 μl EB buffer.
CRITICAL STEP: Proceed immediately to adaptor ligation.
Steps 85 – 95: Ligation of sequencing adaptors (Timing: 1 h)
-
85)
Immediately resuspend beads in the following reaction mixture:
Reagents (add in this order) Volume (μl) Final Concentration dA-tailed DNA w/ beads 30 5X Thermo Rapid Ligation Buffer 10 1X Y-Adaptor (2.5 μM) 6 0.3 μM T4 DNA ligase (5 U/ μl) 4 0.4U / μL Total Volume 50 -
86)
Incubate at RT for 30 min.
PAUSE POINT: The ligation reaction in step 86 can also be performed at 16°C overnight.
-
87)
Add 5 μl of 0.5 M EDTA to stop the reaction. Add 145 μl of ddH2O to bring up the volume to 200 μl and mix thoroughly by pipetting up and down.
-
88)
Add 200 μl of AMPure buffer to each tube and mix thoroughly by pipetting up and down.
-
89)
Incubate at RT for 5 min and then place the tubes in a DynaMag magnet for 2 min.
-
90)
Discard the supernatant and wash the beads twice with 500 μl of 80% ethanol. Briefly spin down the beads and remove residual ethanol as completely as possible, then air-dry the beads for no more than 2 min.
-
91)
Resuspend beads in 200 μl ddH2O and add 165 μl of AMPure buffer
-
92)
Mix thoroughly by pipetting up and down.
-
93)
Incubate at RT for 5 min, and place the three tubes in a DynaMag magnet for 2 min.
-
94)
Discard the supernatant and wash the beads twice with 0.5 ml of 80% ethanol. Briefly spin down the beads and remove the residual ethanol as completely as possible, then air dry the beads for 5 min.
-
95)
Resuspend the beads in each tube with 50 μl of EB.
Steps 96 – 104: Library amplification (Timing: 2.5 h)
CRITICAL: Optimization of input amount and PCR cycle number is integral to obtaining a sufficiently diverse in situ DNase Hi-C library. We recommend running several “pilot” PCR reactions with various bead input amounts and various cycle numbers and running these “pilot” libraries on a 6% TBE-PAGE gel to ensure that library overamplification is not occurring.
-
96)
To determine the number of PCR cycles necessary to generate ample PCR products for sequencing—importantly, without over-amplification—set up trial PCR reactions with 10, 12, or 14 cycles, and 2.5 or 5 μl of DNA-bound streptavidin beads as follows:
Reagents (add in this order) Volume (μl) End-repaired DNA w/ beads 2.5 / 5 2X HotStart ReadyMix 10 1X 10 μM SeqPrimer_F 1 1 μM 10 μM SeqPrimer_R 1 1 μM ddH2O up to 20 Total Volume 20 Using the following PCR program:
Cycle number Denature Anneal Extend 1 95°C, 3 min. 2–6 98°C, 20 sec. 60°C, 20 sec. 72°C, 1 min. 7–17* 98°C, 20 sec. 65°C, 20 sec. 72°C, 1 min. *Use optimized cycle number -
97)
Run 2 μl of each PCR reaction on a 6% TBE-PAGE gel to determine the appropriate number of cycles and amount of input beads for each PCR reaction. PCR products should run from ~300 bp to ~1 kbp, with the vast majority of the fragments with a size of 300bp–600bp, as shown in Figure 3b. Presence of products much larger than 1 kbp (i.e. will not migrate on a 6% TBE-PAGE gel) indicates overamplification, and should be avoided by reducing PCR cycle number or volume of beads used.
-
98)
Aliquot remaining beads into 20 μl PCR reaction and amplify the remaining beads using multiple PCR reactions at the optimized cycle and input parameters.
-
99)
Pool all PCR reactions into one 1.5 mL microcentrifuge tube.
-
100)
Purify library by adding 0.8X volumes of AMPure XP beads.
-
101)
Incubate mixture at RT for 5 min and place tube in a DynaMag magnet for 2 min.
-
102)
Discard the supernatant and wash the beads twice with 1 ml of 80% ethanol. Briefly spin down the beads and remove residual ethanol as completely as possible, then air-dry the beads for no more than 2 min.
-
103)
Resuspend beads in 25 μl EB buffer and incubate at RT for 1 min.
-
104)
Place resuspended beads on DynaMag magnet and transfer supernatant containing eluted DNA to fresh 1.5 mL tube.
Steps 105 – 109: Quality control of DNase Hi-C library by BamHI digestion (Timing: 1.25 h)
-
105)
Quantitate amount of dsDNA in library using Qubit dsDNA HS kit as per manufacturer’s protocols.
-
106)
Digest a small aliquot of the final DNase Hi-C library (50 – 100 ng) with BamHI to estimate the portion of molecules with valid biotinylated junctions as follows:
Reagents (add in this order) Digest (-) Control 10X Fast digestion buffer 1 μl 1 μl DNase Hi-C product 1–2 μl (50–100 ng) 1–2 μl (50–100 ng) Fast digestion BamHI 1 μl 0 μl Water to 10 μl to 10 μl -
107)
Incubate at 37°C for 30 min.
-
108)
Run the entire volume of the reaction on a 6% TBE-PAGE gel. Digested libraries should demonstrate a marked shift in library size distribution, as shown in Figure 3b. If libraries pass this QC metric, proceed to Illumina sequencing.
-
109)
(Optional) Hybrid capture experiments may be carried out according to manufacturer’s protocols provided with the Agilent SureSelect system.
Steps 110 – 120: Mapping, Normalization, and Visualization of Hi-C Contact Maps. TIMING dependent on volume of data)
-
110)
Copy the output fastq sequencing files generated by the Illumina sequencer to the storage on the Linux computer.
[TROUBLESHOOTING 111]
-
111)
Open a terminal on the computer and enter after the $ sign the commands described in the following steps. First, run FastQC to investigate the sequencing qualities, in which “L1_1” and “L1_2” correspond to the fastq sequence files for read 1 and read 2, respectively.
$ fastqc --extract -f fastq L1_1.fq L1_2.fq
-
112)
Obtain reference genome sequences. For instance, the mouse mm9 reference sequences can be downloaded from the UCSC Genome browser using the command below.
$ wget “http://hgdownload.cse.ucsc.edu/goldenPath/mm9/bigZips/chromFa.tar.gz” $ tar -xzvf chromFa.tar.gz $ gunzip -c chr*.fa.gz > mm9.fa
CRITICAL STEP: If the in situ DNase Hi-C data are from female cells, do not include chrY.
-
113)
Run BWA to generate index files for the reference genome.
$ bwa index -a bwtsw -p mm9 mm9.fa
-
114)
Run BWA to map each end of the pair-ended reads to the reference genome separately.
$ bwa aln mm9 L1_1.fq > L1_1.sai $ bwa samse mm9 L1_1.sai mm9.fa > L1_1.sam $ bwa aln mm9 L1_2.fq > L1_2.sai $ bwa samse mm9 L1_2.sai mm9.fa > L1_2.sam
CRITICAL STEP: The two ends of the reads should be mapped separately.
-
115)
Run samtools to exact high-quality (MAPQ>=30) and uniquely mapped reads.
$ samtools view -S -F 4 L1_1.sam | awk ‘$5>=30 && $12==“XT:A:U”‘ | cut –f 1-4 | sort -k1,1 > L1_1.mapped $ samtools view -S -F 4 L1_2.sam | awk ‘$5>=30 && $12==“XT:A:U”‘ | cut –f 1-4 | sort -k1,1 > L1_2.mapped
-
116)
Join mapped loci pairs if both ends are successfully mapped.
$ join L1_1.mapped L1_2.mapped > L1.mapped
-
117)
Remove PCR duplicates.
$ cut –f 2-7 L1.mapped | awk ‘BEGIN{OFS=“\t”;} {if($2<$5){print $0;} else if($2>$5){print $4,$5,$6,$1,$2,$3;} else if($3<=$6){ print $0;} else{print $1,$2,$6,$2,$5,$3;}}’ | sort –u > L1.unique -
118)
Parse the mapped contacts loci pairs to generate the Hi-C contact map at a given resolution.
-
119)
Run ICE53 to normalize the contact matrix using the Mirny lab’s hiclib library (https://bitbucket.org/mirnylab/hiclib).
-
120)
Visualize the contact map.
Troubleshooting
| Step | Problem | Possible reasons | Solution |
|---|---|---|---|
| 3 | Low percentage of long-range contacts in sequencing library or BamHI digest does not shift library | Inefficient or incomplete formaldehyde crosslinking | For new cell types, optimizing the amount of formaldehyde used for crosslinking may be necessary. |
| 7 | Nuclear pellet disappears during in situ enzymatic treatments | Overtreatment of fixed nuclei with SDS | Reduce the amount of SDS used in the cell lysis. |
| 14 | gDNA digestion efficiency is poor | Undertreatment of fixed nuclei with SDS; inadequate amount of DNase I used for digestion | Optimization of the appropriate SDS and DNase I amounts may be necessary. We recommend performing the protocol through Step 38 for a variety of SDS concentrations (i.e. 0.1% – 0.5%) and DNase I amounts (i.e. 1U – 8U). |
| 111 | FastQC metrics are poor | High duplication rate in library (e.g. Fewer than 60% unique sequences); low quality sequencing run (e.g. total percentage of bases with q > 30 is less than 85%) | To maximize library complexity, make sure to set up several PCR reactions in Step 114. Issues with sequencing runs themselves may be difficult to diagnose and may require outside help. |
Timing
Day 1: Steps 1 – 32: Fixation; cell lysis; chromatin digestion, end repair, and adaptor ligation; ~6 h
Day 2: Steps 33 – 53: Adapter cleanup; in situ phosphorylation and ligation; crosslink reversal; ~6.5 h
Day 3: Steps 54 – 65: DNA purification and sonication; 2.5 – 3.5 h
Day 4: Steps 66 – 86: Biotin pulldown, end repair/dA tailing, and adaptor ligation of Hi-C fragments; ~3 h
Day 5: Steps 87 – 109: Library amplification, Bam HI quality check, and sequencing; ~4 h for amplification and quality check; up to several days / weeks for sequencing, instrumentation depending.
Day 6 and beyond: Steps 110 – 120: Data analysis time depends on sequencing depth and available compute resources.
Anticipated Results
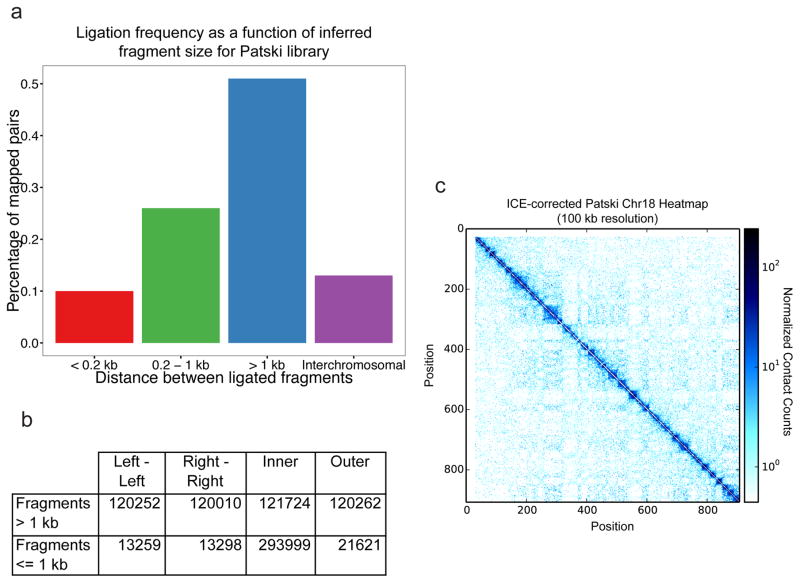
We recommend QCing all libraries that pass the BamHI digestion test (typical results, including a negative control EcoRI digest, shown in Figure 3b) by sequencing at low depth first to ensure that the libraries are sufficiently complex for your desired application. We also recommend quantifying the length-classes of sequenced ligation pairs in libraries; in situ DNase Hi-C libraries should demonstrate an enrichment for pairs mapping with long-range (i.e. > 1 kb) distances between them (example distributions shown in Figure 4a). Furthermore, we recommend quantifying the relative numbers of different ligation pairs (i.e. “in-facing,” “out-facing,” “left,” and “right”) in libraries (a typical example is shown in Figure 4b). Corrected matrices generated from valid in situ DHC libraries should be analogous to the example shown in Figure 4c, with large scale structures (i.e. TADs) clearly visible even at 100 kb resolution.
Figure 4. In situ DNase Hi-C results for the mouse embryonic kidney Patski cell line.
a.) In situ DNase Hi-C reads (950,206 downsampled reads from data published in Deng, Ma et al51 (using the mouse Patski cell line, rather than GM12878) demonstrate an enrichment for long-range (i.e. > 1 kb) intrachromosomal read pairs expected of Hi-C libraries. b.) Expected breakdown of mate orientations for read pairs in in situ DNase Hi-C data. For intrafragment distances > 1 kb, a roughly 25% split should be observed for each orientation class. c.) Normalized heat map generated from data published in Deng, Ma et al (GEO Accession: GSE68992) for mouse chromosome 18 at 100 kb resolution. The dataset used to generate this heatmap contained 60,666,200 uniquely mapped, high-quality read pairs.
We have observed that the relative fraction of interchromosomal ligation pairs in in situ DNase Hi-C libraries is largely cell-type specific, but highly reproducible—in line with previously published in situ results43,50. This is evident in Supplementary Figure 1, which compares fractions of various ligation pairs between the Patski cell line, and three replicates of the human lymphoblastoid cell line GM12878. When considering gold-standards for in situ DNase Hi-C experiments, we typically look to the abundance of “long-range” ligation pairs in our libraries, which typically make up > 40% of uniquely mapped read pairs.
Using this modified DHC protocol, we have shown that the inactive murine X chromosome adopts a bipartite structure, consistent with results obtained using traditional Hi-C both in an analogous murine system35 and human lymphoblastoid cells43. These results suggest that the in situ DHC protocol produces signal comparable to existing Hi-C protocols while ultimately providing a less-biased empirical method for generating higher-resolution 3D maps of chromatin structure.
Supplementary Material
Acknowledgments
The authors thank S. Kasinathan and members of the Shendure lab for helpful comments on the manuscript. This work was funded by National Institutes of Health (NIH) Director’s Pioneer Award (1DP1HG007811Q14 to J.S.) and an NIH National Human Genome Research Institute (NHGRI) Genome Training Grant (5T32HG000035 to V.R.).
Footnotes
Author Contributions
V.R. and Z.D. developed the protocol. V.R., D.A.C., R.J.H., R.Q., and Z.D. performed experiments and optimized the protocol. W.M. and W.S.N. devised the processing pipeline for in situ DNase Hi-C data. X.D., C.A.B., C.M.D., W.S.N., J.S., and Z.D. supervised research. V.R., J.S., and Z.D. wrote the manuscript, with input from all authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 2.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 3.Rieder CL, Khodjakov A. Mitosis through the microscope: advances in seeing inside live dividing cells. Science. 2003;300:91–96. doi: 10.1126/science.1082177. [DOI] [PubMed] [Google Scholar]
- 4.BARR ML, BERTRAM EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 5.Hochstrasser M, Sedat JW. Three-dimensional organization of Drosophila melanogaster interphase nuclei. I. Tissue-specific aspects of polytene nuclear architecture. J Cell Biol. 1987;104:1455–1470. doi: 10.1083/jcb.104.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manuelidis L. Individual interphase chromosome domains revealed by in situ hybridization. Hum Genet. 1985;71:288–293. doi: 10.1007/BF00388453. [DOI] [PubMed] [Google Scholar]
- 7.Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schardin M, Cremer T, Hager HD, Lang M. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum Genet. 1985;71:281–287. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- 10.Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]
- 11.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 12.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 13.Orian A, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wit E, Greil F, van Steensel B. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Research. 2005;15:1265–1273. doi: 10.1101/gr.3198905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izzo A, et al. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 17.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 18.Mukherjee S, Erickson H, Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. 1988;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- 19.Cullen KE, et al. Science. 1993;261 doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 20.de Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 21.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Research. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman-Aiden E, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le TBK, et al. High-Resolution Mapping of the Spatial Organization of a Bacterial Chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ay F, et al. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Research. 2014;24:974–988. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Splinter E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giorgetti L, et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minajigi A, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349:aab2276–aab2276. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh THS, et al. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabo PJ, et al. Integrative analysis of 111 reference human epigenomes. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine M, Cattoglio C, Tjian R. Looping Back to Leap Forward: Transcription Enters a New Era. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffith J, Hochschild A, Ptashne M. DNA loops induced by cooperative binding of [lambda] repressor. 1986;322:750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- 42.Müller HP, Sogo J, Schaffner W. An enhancer stimulates transcription in Trans when attached to the promoter via a protein bridge. 1989;58:767–777. doi: 10.1016/0092-8674(89)90110-4. [DOI] [PubMed] [Google Scholar]
- 43.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 46.Schoenfelder S, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Research. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahlén P, et al. Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol. 2015;16:156. doi: 10.1186/s13059-015-0727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma W, et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Meth. 2015;12:71–78. doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gavrilov AA, et al. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Research. 2013;41:3563–3575. doi: 10.1093/nar/gkt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagano T, et al. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 2015;16:175. doi: 10.1186/s13059-015-0753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng X, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He HH, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Meth. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Meth. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Wit E, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.