Abstract
We have analyzed several cases of Beckwith–Wiedemann syndrome (BWS) with Wilms' tumor in a familial setting, which give insight into the complex controls of imprinting and gene expression in the chromosome 11p15 region. We describe a 2.2-kbp microdeletion in the H19/insulin-like growth factor 2 (IGF2)-imprinting center eliminating three target sites of the chromatin insulator protein CTCF that we believe here is necessary, but not sufficient, to cause BWS and Wilms' tumor. Maternal inheritance of the deletion is associated with IGF2 loss of imprinting and up-regulation of IGF2 mRNA. However, in at least one affected family member a second genetic lesion (a duplication of maternal 11p15) was identified and accompanied by a further increase in IGF2 mRNA levels 35-fold higher than control values. Our results suggest that the combined effects of the H19/IGF2-imprinting center microdeletion and 11p15 chromosome duplication were necessary for manifestation of BWS.
Keywords: insulin-like growth factor 2, H19, differentially methylated region
Genomic imprinting is an important component of the regulation of human gene expression, and a number of genetic diseases can be caused by errors in imprinting. The imprinting of a gene is thought to be regulated by epigenetic modifications, such as CpG methylation, which are coordinated by imprinting centers (ICs). The mechanism of this regulation appears to be complex but of potential importance in developing therapeutic interventions for disorders caused by imprinting errors.
Beckwith–Wiedemann syndrome (BWS), a congenital overgrowth condition, in which ≈5% of children develop embryonal tumors [predominantly Wilms' tumor (WT)] is associated with escape from epigenetic silencing [loss of imprinting (LOI)] of genes on chromosome 11p15. The three most frequent findings identified are (i) loss-of-function alterations in the CDKN1C gene (reviewed in ref. 1), (ii) alterations affecting the tight regulation of 11p15 gene expression [e.g., insulin-like growth factor 2 (IGF2)-LOI], observed in cases with uniparental paternal disomy (patUPD) (2), and (iii) alterations smaller than patUPDs that cluster to two ICs implicated in the control of epigenetically regulated genes in 11p15.5. The proximal IC is termed IC2 or KvDMR1 [KvLQT1-associated differentially methylated region (DMR) 1]. Hypomethylation of this sequence mediates silencing of the KCNQ1 and CDKN1C genes by permitting transcription of the antisense transcript KCNQ1OT1 (3, 4). The distal IC, IC1, is a DMR positioned between H19 and IGF2. The expression of these two reciprocally imprinted genes is dependent on the same enhancers located distal of H19 (5). Access to these enhancers is regulated by binding of CTCF (CCCTC-binding factor) to the unmethylated maternal IC1 allele, thus creating a functional chromatin boundary and blocking the interaction of IGF2 promoters and enhancers. IC1-methylation (on the paternal chromosome) prevents CTCF binding, thus allowing IGF2 expression (6, 7). Human IC1 contains seven CTCF target sites (CTSs) (Fig. 1). Methylation aberrations of IC1 have been reported to occur in a graded fashion, and pathologic conditions are associated with aberrant methylation resulting in IGF2-LOI (8, 9) or H19-LOI (10).
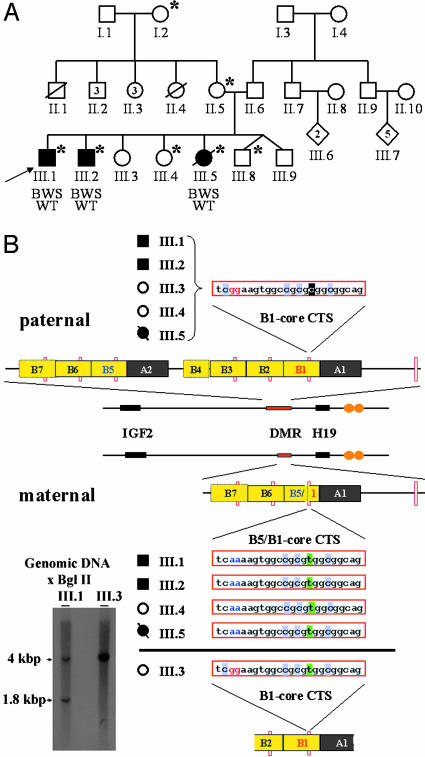
Fig. 1.
IC1 structure in the three-generation BWS family with WT. (A) Pedigree of a large three-generation family with three children affected by BWS and WT. *, Those individuals who inherited the same 11p15 maternal haplotype, along with the IC1 deletion described in this report. The proband (III.1) is indicated by an arrow. (B) Organization of the paternal and maternal IC1 region in members of the BWS/WT pedigree. The integral DMR status on the paternal haplotype of individuals III.1, III.2, III.3, III.4, and III.5 is shown, with B repeats represented by yellow boxes and A repeats denoted by gray boxes. The locations of the CTCF-binding sites are denoted by small orange hatches. The core CTS sequence is shown, with the blackened C or green T, denoting a paternal or maternal single-nucleotide polymorphism (SNP), respectively. Nucleotides specific for the B1 and B5 sequences are marked in red and blue, respectively. The microdeletion generates a chimeric CTS sequence derived from B1 and B5 repeats in the maternal haplotypes of III.1, III.2, III.4, and III.5. The heterozygous nature of the deletion in III.1 was verified by Southern blot analysis (Left).
Small deletions can give insight into the function of an imprinted region and have been extremely informative in understanding Prader–Willi syndrome and Angelman syndrome, where paternally or maternally transmitted 15q deletions, respectively, lead to different complex syndromes (for review see ref. 11). However, in BWS, small deletions that include chromosomal ICs have not been identified until very recently. One reported microdeletion includes the entire KCNQ1OT1 gene and when maternally inherited, causes BWS by silencing of CDKN1C (12). Another microdeletion was described for IC1 (13). In two families, each with one child that suffered from BWS, an inherited 1.8-kbp microdeletion in the H19 DMR eliminated two CTSs. Maternal transmission of the deletion correlated with hypermethylation of the H19 DMR, biallelic IGF2 expression, H19 silencing, and the BWS phenotype.
We report on a microdeletion for chromosome 11p15 IC1 that occurs in a familial setting in a sibship with three cases of BWS and WT. We describe a 2.2-kbp microdeletion in the H19/IGF2-IC, eliminating three target sites of the chromatin insulator protein CTCF, which we believe is necessary but not sufficient to cause BWS and WT. Maternal inheritance of the deletion is associated with IGF2-LOI and up-regulation of IGF2 mRNA. However, in at least one affected family member, a second genetic lesion, duplication of maternal 11p15, was identified, which was absent in unaffected family members, suggesting that the combined effects of the deletion and chromosome duplication were necessary for the BWS disease phenotype to manifest.
Materials and Methods
BWS/WT Family Members. Informed written consent was obtained from the adult individuals, and consent was obtained from parents for minors. The ethical Review Board of Germany approved all study protocols.
Allelotyping, 11p Marker Analysis, and Southern Blotting Analysis. Genomic DNA was isolated from peripheral blood lymphocytes by using the Wizard genomic DNA purification kit (Promega). Then 42 polymorphic microsatellite markers spanning the entire short arm of chromosome 11 were PCR amplified and analyzed with an ABI 310 capillary sequencing machine (Applied Biosystems). Southern analysis for the IC1 region was performed by using 10 μg of BglII-digested genomic DNA, hybridized with a PCR-generated probe detecting the B7 repeat (primers were B7F, 5′-GGAGAACCAAGCATTAATGCG-3′, and B7R, 5′-AGTCATGACCACTGCAGAAC-3′). Genomic DNA of all coding exons of WT1, CDKN1C, and CTCF were sequenced by using an ABI PRISM 377 DNA analyzer (Applied Biosystems). Primer sequences are available on request.
Bisulfite Sequencing. Two micrograms of genomic DNA from peripheral blood lymphocytes were cleaved with TliI, BsiWI, and BsmBI (New England Biolabs) overnight at 55°C, purified, and treated with the CpGenome DNA modification kit according to the manufacturer's instructions (Invitrogen). Bisulfite-treated DNA samples were PCR amplified as described (8). Purified PCR products were cloned into a T-vector (Promega), and 16–20 independent clones per patient were sequenced.
FISH. IGF2 cosmid clone RPCI-11-46C8 (IGF2 genomic sequence GenBank accession no. AC132217.15) was nick translated with Cy3- or SpectrumGreen-dUTP according to the manufacturer's instructions (Roche). Metaphase spreads were prepared from fibroblasts (14), and FISH was performed together with a CEP11-SpectrumGreen probe (Vysis, Downers Grove, IL) as described (15). Posthybridization washes were done at 71°C in 0.4× SSC and 0.3% Nonidet P-40 for 2 min, and in 2× SSC and 0.1% Nonidet P-40 for 2 min. Images were analyzed by using cytovision 3.52 software (Applied Imaging, San Jose, CA).
Quantitative Genomic PCR. Relative quantification by real-time PCR was carried out by using a Roche LightCycler system and SYBR Green mix (Roche) with 3.5 mM MgCl2 in 10-μl reactions. Cycling conditions were 95°C for 10 min, followed by up to 50 cycles of 95°C (5 sec), 65°C (2 sec), and 72°C (3 sec). Primers for amplification of IC1 (IC1 genomic sequence GenBank accession no. AF125183) with and without the microdeletion were forward (0.6 μM), 5′-CGGTTGTAAGTGTGGACTCAA-3′ or 5′-TGGTTGTAGTTGTGGAATCGG-3′, respectively, and reverse (0.4 μM), 5′-CTGTGGATAATGCCCGACCT-3′.
Quantitative RT-PCR of IGF2 Alleles. Total RNA (4 μg), random hexamer, and Moloney murine leukemia virus reverse transcriptase (Invitrogen) were used for cDNA production. Forward primers for isoform-specific cDNA-PCR amplification were IGF2-3F, 5′-GGACAATCAGACGAATTCTCC-3′ (isoform 3) and IGF2-4F, 5′-CTTCTCCTGTGAAAGAGACTTC-3′ (isoform 4) and for isoform independent amplification IGF2-8F, 5′-TCCTGGAGACGTACTGTGCTA-3′. General reverse primer was IGF2-9R, 5′-CGGGGATGCATAAAGTATGAG-3′.
Relative quantitative analysis was performed by using real-time PCR with the cDNA samples prepared as described above. Amplifications were done in 10- or 20-μl reactions with the SYBR Green mix (Roche), 3.5 mM MgCl2, and 0.6 μM primers (IGF2-3F, -4F, IGF2-7R: 5′-CAGCAATGCAGCACGAGGCGAAGGC-3′, IGF2-8F, -9R) with cycling conditions: 95°C for 10 min and up to 55 cycles of 95°C (4 sec), 64°C (7 sec), and 72°C (40 or 10 sec for isoform-independent or isoform-specific amplification, respectively). cDNA concentrations were calculated in respect to equal cDNA starting amounts relative to GAPDH and pyruvate dehydrogenase amplifications.
Results
Chromosome 11 Genotyping of Familial BWS with WT. We analyzed a large, three-generation family with three children affected by BWS (Fig. 1A) presenting with the cardinal features of this syndrome: overgrowth (90th to 95th percentile), macroglossia, umbilical hernia, hepatomegaly, and WT. Microsatellite marker analysis excluded patUPD for 11p in this family (data not shown). A single maternal 11p15 haplotype was found to be common to all affected (III.1, III.2, and III.5) and two unaffected children (III.4 and III.8) (data not shown). This result suggested that the genetic lesion responsible for BWS in this family resides on the maternal chromosome bearing this haplotype. In addition, transmission of the same haplotype to both affected and unaffected children suggests that at least one additional factor underlies the phenotypic variation among the siblings who inherit this chromosome. The BWS/WT individuals in the sibship had an unaltered CDKN1C-coding sequence and retained differential IC2 methylation, suggesting that the genetic lesion responsible for their BWS might lie elsewhere in 11p15.5 (data not shown). We therefore undertook a detailed analysis of the IC1 region.
Identification of a Microdeletion Within IC1. Initially we sought to distinguish the maternal and paternal copies of the IC1 region. We identified four SNPs in the B1/A1 region (GenBank accession no. AF125183) of IC1, which were informative in the sibship. Genotyping revealed that a single 11p15.5 haplotype had been transmitted from the maternal grandmother to the mother and to all three affected and two unaffected children. Direct sequencing of the DMR from the inherited maternal haplotype revealed that this haplotype carried a deletion in IC1 (Fig. 1B). This deletion, generated by recombination between the B1 and B5 repeats, eliminates one copy of repeat A and three and one-half copies of repeat B, abolishing three CTSs (Fig. 1B). Southern blot analysis confirmed the presence of the heterozygous 2.2-kbp microdeletion (Fig. 1B). The deletion was not present in the genomic DNA of the father, the maternal grandfather, and two unaffected children, as well as in 100 control chromosomes, excluding it as a common polymorphism (data not shown).
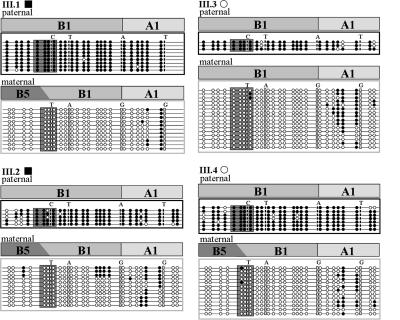
We also assessed the methylation status of the B1, fused B5/B1, and A1 repeats of the IC1 region in four individuals of this sibship. Sequencing of bisulfite-treated patient DNA indicated that methylation was restricted to the paternal allele (Fig. 2). Because this pattern represents the normal epigenetic state for the paternally transmitted allele, it further supports the view that the genetic lesions in the maternally derived allele were likely to be responsible for the BWS in this sibship.
Fig. 2.
Methylation status of IC1 in children III.1, III.2, III.3, and III.4. Allele-specific methylation was analyzed by bisulfite sequencing of IC1 fragments, including the core CTS in the B1 and B5/B1 repeats (boxed). Paternal and maternal clones, identified by four SNPs (CTAT and TAGG), are grouped. •, methylated CpGs; ○, transformed unmethylated CpGs. Allele-specific paternal methylation was detected for the complete B1, as well as the fused B5/B1 sequences, of all four DNA samples.
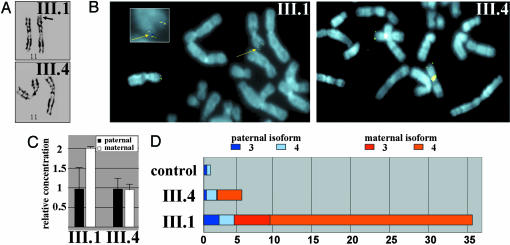
Chromosomal Duplication of the Maternal IGF2 Locus. Our findings implicate the microdeletion described above in the etiology of the BWS/WT phenotype. However, the microdeletion is inherited from the maternal grandmother and mother without the occurrence of BWS and WT in either of these individuals and is also transmitted to one of the siblings in the third generation without BWS (III.4) (Fig. 1A). We considered the possibility that an additional genetic event was necessary to manifest the symptoms of BWS in this sibship. We were able to obtain primary fibroblast cell cultures and primary lymphocytes from two of the siblings, one affected and the other unaffected (III.1 and III.4), both of whom had inherited the same maternal 11p haplotype containing the IC1 microdeletion. Karyotypic analysis revealed an additional lesion in the affected sibling. High-resolution GTG banding of metaphase chromosome spreads showed a prolonged 11p15 subtelomeric region on one chromosome 11 of the proband. This finding was not present on either chromosome 11 of the healthy sibling (Fig. 3A). FISH analysis was therefore carried out to examine the DNA sequences included in the duplication. A genomic probe for IGF2 detected a duplication of the distal part of 11p in fibroblasts (Fig. 3B). The presence of the duplication was also apparent when a genomic real-time PCR quantitation assay was used. The presence of the IC1 microdeletion enabled discrimination between the maternal and paternal 11p15.5 copies in this assay, leading to results consistent with a partial trisomy of 11p15 with a 2:1 (maternal:paternal) ratio (Fig. 3C). The occurrence of the distal 11p15 duplication in III-1 suggested that this genomic alteration might account for the BWS of all three affected siblings. Were this the case, the most likely mechanism that would account for the occurrence of BWS in all three affected siblings would be maternal gonadal mosaicism.
Fig. 3.
Chromosome analysis, genomic quantitation of IC1 alleles, and quantitative RT-PCR analysis of allelic IGF2 expression. (A) Metaphase spreads of primary fibroblasts from an affected individual (III.1) and unaffected individual (III.4) were analyzed. High-resolution GTG banding revealed a prolonged 11p15 subtelomeric region (black arrow) on one chromosome 11 of the proband. (B) FISH analysis with a genomic probe for IGF2 shows two signals on one chromosome 11 of the proband (yellow arrow), indicating a duplication of distal 11p. (Inset) An enlarged view of 11p to illustrate the duplication (yellow arrow). (C) Quantitation of IGF2 gene dosage by using real-time PCR quantification. (D) Quantitative RT-PCR analysis, haplotype-specific expression, and promoter usage of the IGF2 gene. The relative IGF2 mRNA amounts are represented by horizontal bars, with colored segments denoting contribution of the parental haplotypes. As well, the relative contribution to IGF2 expression by the P3 or P4 promoters is indicated by darker and lighter shading, respectively. Allelic contribution to expression levels was determined by sequencing cloned transcripts of all individuals heterozygous for a SNP in IGF2 exon 9.
The IC1 Microdeletion Is Associated with IGF2 Overexpression. To determine whether the genomic alterations affect transcription of the neighboring IGF2 gene, we examined IGF2 mRNA levels in the established fibroblasts. This analysis revealed a significant up-regulation (≈5-fold compared with a normal fibroblast control) of the IGF2 transcript in the unaffected sibling (III.4). However, the increase in IGF2 mRNA levels in the fibroblasts derived from the proband was considerably higher than in the unaffected sibling: a total of ≈35-fold higher than in the normal fibroblast control (Fig. 3D). IGF2 is transcribed from four alternative promoters (P1–P4), generating mRNA isoforms differing in their 5′ untranslated regions, with P3 and P4 responsible for the majority of transcripts in most tissues. Although transcripts from promoter P1 normally are expressed biallelically, promoters P2, P3, and P4 are maternally silent (16). Splice version-specific real-time cDNA amplification revealed that IGF2 overexpression in the fibroblasts was primarily the consequence of enhanced usage of the P4 promoter, and in the case of the fibroblasts from the affected child, to a lesser extent, of the P3 promoter. We found the maternal and paternal IGF2 alleles to contribute almost equally to expression of IGF2 in the fibroblast from the unaffected child, whereas expression of IGF2 in fibroblasts from the BWS child was predominantly (83%) of maternal origin (Fig. 3D).
Discussion
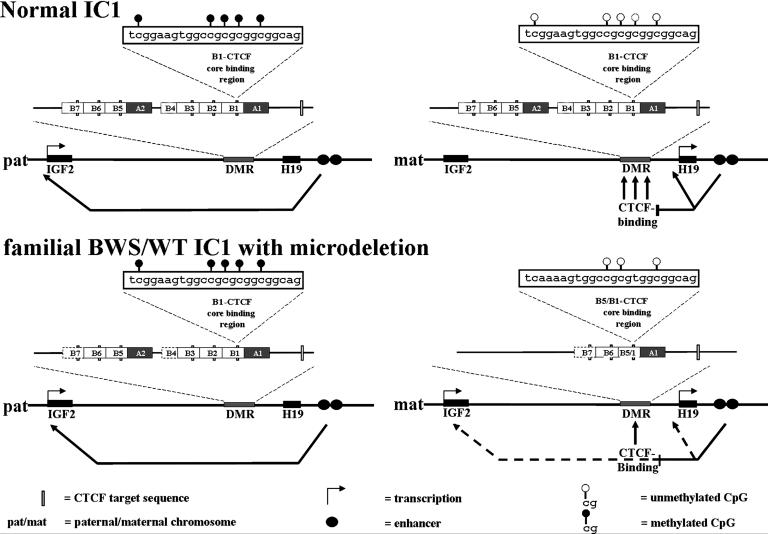
A key feature of the BWS/WT family characterized herein is that the maternal IGF2 copy appears to have been “paternalized” by the IC1 microdeletion, leading to activation of the normally silent maternal IGF2 allele. In general, BWS cases with partial chromosome 11 duplications are of paternal origin (patUPD), whereas duplication of maternal 11p15 is associated with growth retardation, but not with BWS (17). The occurrence of the microdeletion thus gives some insight into the control mechanisms that regulate imprinting through IC1 (Fig. 4). Although we found no methylation changes associated with the IC1 microdeletion, deletion of the three CTSs results in loss of insulation by CTCF on the maternal allele (schematically represented in Fig. 4). An additional structural difference in the deleted allele that might also contribute to the loss of CTCF insulation is the hybrid B5/B1 sequence, which differs from the normal B1 sequence at its 5′ end (Fig. 4).
Fig. 4.
Schematic representation of the IC1 microdeletion effects on IGF2 expression. (Upper) The current enhancer insulation model (7). In the analyzed BWS/WT family (Lower), the microdeletion results in LOI of IGF2, although differential methylation is maintained.
A number of previous studies have addressed the relationship of IC1 to expression levels of IGF2 and H19. Takai and coworkers (10) have suggested that methylation/demethylation of the wrong IC1-CTS could adversely affect expression of either IGF2 or H19 and that only B1 is differentially methylated and all other B elements are hypermethylated on both alleles (10). Methylation aberrations in the B1-CTS have been described for WT (8), osteosarcoma (18), and colorectal (19) and bladder cancer (10), which argue for a central role of the A1/B1-B4 cluster in regulating IC1 function. Cui and coworkers (9, 20) found differential methylation in the A2/B5-B7 repeat cluster (affecting the CTS in B7) in addition to the A1/B1-B4 cluster as well as B7 hypomethylation associated with colorectal cancer.
An IC with functional properties similar to the human IC1 has been demonstrated in the mouse. The mouse IC1 contains four CpG-rich CTS but has significant architectural differences from the human IC1. Recent findings from Fedoriw and coworkers (21) indicate that the murine CTCF can protect the H19 DMR from de novo methylation during oocyte growth and is required for normal preimplantation development. Reduced CTCF levels result in hypermethylation of the CTS. A similar observation was made when a deletion abolished a dyad-Oct-binding sequence, resulting in a maternal IC1 methylation and subsequent LOI of IGF2 (22). Because the murine IC1 region has a different architecture compared with the human IC1, the function of the putative human dyad-Oct-binding sequence in A2 still remains to be determined. Dyad-Oct binding in human A1 and A2 may protect the two clusters of CTS from regional hypermethylation. The microdeletion described in the BWS patients in the present study erases three CTSs (CTS3, CTS4, and CTS5 in repeats B5, B3, and B2) and a putative dyad-Oct-binding sequence in A2. Fusion of the B1/B5 repeats generates a CTS, similar in sequence to the original B5 repeat, which is differentially methylated, like the original B1 repeat. Dyad-Oct-binding in A1 therefore seems to work properly in maintaining differential methylation in the neighboring CTS, although the deletion of the putative dyad-Oct-binding sequence in A2 might result in hypermethylation of the B6 and B7 repeats. The observed activation of the maternal IGF2 locus without hypermethylation of CTS6 argues for a pivotal role of additional IC1-CTS in generating a functional CTCF block. Although the deletion described by Sparago et al. (13) is smaller and encompasses fewer CTS than our familial BWS case, they find that the retained fourth and sixth CTS (in B3 and B1) are hypermethylated. For the murine IC, Kanduri and coworkers (23) demonstrated that nucleosome positioning sites optimize the fidelity of the interaction of CTCF with its target sites in the H19 DMR. Therefore alteration of CTS6 methylation might reflect differences in nucleosome positioning with signals in the retained repeats B5–B7 because a set of strong nucleosome positioning sites have been described for this part of the H19 DMR (24).
The impact on BWS and WTs may be thought of in terms of the effects of the IC microdeletion and 11p15 duplication on IGF2 expression levels (Fig. 4). Transgenic mice that overexpress Igf2 develop features of BWS, including prenatal and neonatal overgrowth, organomegaly, and macroglossia. The overgrowth phenotype is dependent on the Igf2 mRNA level. Although a 2-fold increase in murine Igf2 dose is sufficient to generate some of the somatic overgrowth features of BWS (25), biallelic IGF2 expression in humans does not necessarily give identical results (26). In mice a combination of two events, inactivation of Cdkn1c and LOI of Igf2 (resembling the patUPD situation in BWS patients) is necessary to produce a BWS-like phenotype (27). Additional arguments in favor of IGF2 as a central player in BWS/WT pathogenesis come from studies of sporadic BWS cases (28) and 40–60% of WTs (29), where biallelic expression of IGF2 is observed. A variety of other pediatric tumors, including the BWS-associated rhabdomyosarcoma and neuroblastoma (reviewed in ref. 30), also show IGF2 overexpression. Finally, the enhanced IGF2 levels reported in cases of congenital fibrosarcoma and congenital mesoblastic nephroma are of special interest, because molecular studies implicate the presence of an intact IGF signaling axis in malignant transformation (31).
However, loss of imprinting alone does not necessarily raise IGF2 levels sufficiently to cause BWS. LOI of IGF2 without pathologic effects has in fact been reported in peripheral blood leukocytes in ≈10% of the normal population (26). In the case of the familial BWS described here, LOI of IGF2 leads to enhanced IGF2 mRNA levels in the fibroblasts of the unaffected child with the IC1 microdeletion, yet this LOI does not result in BWS. The differences in phenotype between siblings with and without 11p15 duplication are consistent with a threshold model in which IGF2 expression levels that exceed a threshold are critical in the etiology of BWS and its associated tumors. The IC1 deletion, according to this model, would drive pathogenesis as a first hit by influencing the expression level of IGF2 (Fig. 4). Increased IGF2 levels may contribute to the BWS phenotype and predispose to WTs by leading to unscheduled stimulation of the insulin receptor. This in turn would activate the phosphatidylinositol 3-kinase/Akt/mTOR pathway, a signal transduction cascade implicated in regulating cell size and proliferation. The further understanding of the complex pattern of control exerted through the chromosome 11p15 ICs in BWS should be helpful in ultimately designing therapeutic intervention for this disorder and its associated tumors.
Acknowledgments
We thank the BWS family members for their participation and Drs. J. Spranger and G. Wildhardt for helpful discussions and support. We are grateful to M. Holl, J. Truebenbach, S. Schambach, M. Engel, C. Schueller, and P. Garneau for expert technical assistance and J. Loesow for editorial support. J.P. is a Canadian Institute of Health Research Senior Investigator. This work was supported by a grant from the Canadian Institute of Health Research to J.P. and Deutsche Forschungsgemeinschaft Schwerpunktprogramm Grant 1129 (project PR688/1) to D.P. and B.Z.
Author contributions: D.P. and B.Z. designed research; D.P., T.E., B.G.-R., M.O., D.R., S.F., and R. L. performed research; R.L. contributed new reagents/analytic tools; D.P., T.E., B.G.-R., C.S., E.L., P.S., D.E.H., J.P., and B.Z. analyzed data; D.P., T.E., D.E.H., J.P., and B.Z. wrote the paper; and M.K., I.B., and J.L. examined patients and provided contact to the family to enable us to obtain the sample materials.
Abbreviations: BWS, Beckwith–Wiedemann syndrome; IC, imprinting center; LOI, loss of imprinting; WT, Wilms' tumor; DMR, differentially methylated region; IGF2, insulin-like growth factor 2; CTS, CTCF target site; patUPD, uniparental paternal disomy; SNP, single-nucleotide polymorphism.
References
- 1.Weksberg, R., Smith, A. C., Squire, J. & Sadowski, P. (2003) Hum. Mol. Genet. 12, Spec. no. 1, R61–R68. [DOI] [PubMed] [Google Scholar]
- 2.Rainier, S., Johnson, L. A., Dobry, C. J., Ping, A. J., Grundy, P. E. & Feinberg, A. P. (1993) Nature 362, 747–749. [DOI] [PubMed] [Google Scholar]
- 3.Smilinich, N. J., Day, C. D., Fitzpatrick, G. V., Caldwell, G. M., Lossie, A. C., Cooper, P. R., Smallwood, A. C., Joyce, J. A., Schofield, P. N., Reik, W., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 8064–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soejima, H., Nakagawachi, T., Zhao, W., Higashimoto, K., Urano, T., Matsukura, S., Kitajima, Y., Takeuchi, M., Nakayama, M., Oshimura, M., et al. (2004) Oncogene 23, 4380–4388. [DOI] [PubMed] [Google Scholar]
- 5.Banet, G., Bibi, O., Matouk, I., Ayesh, S., Laster, M., Kimber, K. M., Tykocinski, M., de Groot, N., Hochberg, A. & Ohana, P. (2000) Mol. Biol. Rep. 27, 157–165. [DOI] [PubMed] [Google Scholar]
- 6.Bell, A. C. & Felsenfeld, G. (2000) Nature 405, 482–485. [DOI] [PubMed] [Google Scholar]
- 7.Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M. & Tilghman, S. M. (2000) Nature 405, 486–489. [DOI] [PubMed] [Google Scholar]
- 8.Frevel, M. A., Sowerby, S. J., Petersen, G. B. & Reeve, A. E. (1999) J. Biol. Chem. 274, 29331–29340. [DOI] [PubMed] [Google Scholar]
- 9.Cui, H., Niemitz, E. L., Ravenel, J. D., Onyango, P., Brandenburg, S. A., Lobanenkov, V. V. & Feinberg, A. P. (2001) Cancer Res. 61, 4947–4950. [PubMed] [Google Scholar]
- 10.Takai, D., Gonzales, F. A., Tsai, Y. C., Thayer, M. J. & Jones, P. A. (2001) Hum. Mol. Genet. 10, 2619–2626. [DOI] [PubMed] [Google Scholar]
- 11.Nicholls, R. D. & Knepper, J. L. (2001) Annu. Rev. Genomics Hum. Genet. 2, 153–175. [DOI] [PubMed] [Google Scholar]
- 12.Niemitz, E. L., DeBaun, M. R., Fallon, J., Murakami, K., Kugoh, H., Oshimura, M. & Feinberg, A. P. (2004) Am. J. Hum. Genet. 75, 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparago, A., Cerrato, F., Vernucci, M., Ferrero, G. B., Silengo, M. C. & Riccio, A. (2004) Nat. Genet. [DOI] [PubMed]
- 14.Lemieux, N., Dutrillaux, B. & Viegas-Pequignot, E. (1992) Cytogenet. Cell Genet. 59, 311–312. [DOI] [PubMed] [Google Scholar]
- 15.Winterpacht, A., Endele, S., Enklaar, T., Fuhry, M. & Zabel, B. (1996) Cytogenet. Cell Genet. 75, 132–135. [DOI] [PubMed] [Google Scholar]
- 16.Ekstrom, T. J., Cui, H., Li, X. & Ohlsson, R. (1995) Development (Cambridge, U.K.) 121, 309–316. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, A. M., Thomas, N. S., Cockwell, A., Stecko, O., Kerr, B., Temple, I. K. & Clayton, P. (2002) Hum. Genet. 111, 290–296. [DOI] [PubMed] [Google Scholar]
- 18.Ulaner, G. A., Vu, T. H., Li, T., Hu, J. F., Yao, X. M., Yang, Y., Gorlick, R., Meyers, P., Healey, J., Ladanyi, M. & Hoffman, A. R. (2003) Hum. Mol. Genet. 12, 535–549. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa, H., Chadwick, R. B., Peltomaki, P., Plass, C., Nakamura, Y. & de La, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui, H., Onyango, P., Brandenburg, S., Wu, Y., Hsieh, C. L. & Feinberg, A. P. (2002) Cancer Res. 62, 6442–6446. [PubMed] [Google Scholar]
- 21.Fedoriw, A. M., Stein, P., Svoboda, P., Schultz, R. M. & Bartolomei, M. S. (2004) Science 303, 238–240. [DOI] [PubMed] [Google Scholar]
- 22.Hori, N., Nakano, H., Takeuchi, T., Kato, H., Hamaguchi, S., Oshimura, M. & Sato, K. (2002) J. Biol. Chem. 277, 27960–27967. [DOI] [PubMed] [Google Scholar]
- 23.Kanduri, M., Kanduri, C., Mariano, P., Vostrov, A. A., Quitschke, W., Lobanenkov, V. & Ohlsson, R. (2002) Mol. Cell. Biol. 10, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davey, C., Fraser, R., Smolle, M., Simmen, M. W. & Allan, J. (2003) J. Mol. Biol. 325, 873–887. [DOI] [PubMed] [Google Scholar]
- 25.Sun, F. L., Dean, W. L., Kelsey, G., Allen, N. D. & Reik, W. (1997) Nature 389, 809–815. [DOI] [PubMed] [Google Scholar]
- 26.Sakatani, T., Wei, M., Katoh, M., Okita, C., Wada, D., Mitsuya, K., Meguro, M., Ikeguchi, M., Ito, H., Tycko, B., et al. (2001) Biochem. Biophys. Res. Commun. 283, 1124–1130. [DOI] [PubMed] [Google Scholar]
- 27.Caspary, T., Cleary, M. A., Perlman, E. J., Zhang, P., Elledge, S. J. & Tilghman, S. M. (1999) Genes Dev. 13, 3115–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reik, W., Brown, K. W., Schneid, H., Le Bouc, Y., Bickmore, W. & Maher, E. R. (1995) Hum. Mol. Genet. 4, 2379–2385. [DOI] [PubMed] [Google Scholar]
- 29.Vu, T. H., Chuyen, N. V., Li, T. & Hoffman, A. R. (2003) Cancer Res. 63, 1900–1905. [PubMed] [Google Scholar]
- 30.Toretsky, J. A. & Helman, L. J. (1996) J. Endocrinol. 149, 367–372. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, K. B., Tognon, C. E., Garnett, M. J., Deal, C. & Sorensen, P. H. (2002) Oncogene 21, 5684–5695. [DOI] [PubMed] [Google Scholar]