Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is caused by the cooperation of multiple oncogenic lesions1,2. We used exome sequencing on 67 T-ALLs to gain insight into the mutational spectrum in these leukemias. We detected protein-altering mutations in 508 genes, with an average of 8.2 mutations in pediatric and 21.0 mutations in adult T-ALL. Using stringent filtering, we predict seven new oncogenic driver genes in T-ALL. We identify CNOT3 as a tumor suppressor mutated in 7 of 89 (7.9%) adult T-ALLs, and its knockdown causes tumors in a sensitized Drosophila melanogaster model3. In addition, we identify mutations affecting the ribosomal proteins RPL5 and RPL10 in 12 of 122 (9.8%) pediatric T-ALLs, with recurrent alterations of Arg98 in RPL10. Yeast and lymphoid cells expressing the RPL10 Arg98Ser mutant showed a ribosome biogenesis defect. Our data provide insights into the mutational landscape of pediatric versus adult T-ALL and identify the ribosome as a potential oncogenic factor.
T-ALL is a genetically heterogeneous leukemia that is caused by the accumulation of multiple oncogenic lesions, which have been identified through characterization of chromosomal aberrations or via candidate gene sequencing4–7. In addition, recent whole-genome sequencing of 12 immature early T-cell precursor (ETP) ALLs revealed several new oncogenic drivers in this T-ALL subtype8. To discover new disease-driving genes in pediatric and adult T-ALL, we performed exome sequencing on 67 diagnostic T-ALL samples, 39 matched remission samples and 17 T-ALL cell lines (Supplementary Tables 1–3 and Supplementary Note).
To identify somatic mutations, we limited our initial analysis to the 39 paired samples acquired in the same individual at diagnosis and remission. To assess the performance of variant calling, 185 predicted single-nucleotide variations (SNVs) were validated by Sanger sequencing. This set was used to determine the filtering strategy with the best sensitivity and specificity. Different parameters were tested as filters, including sequencing coverage of the variant nucleotide, variant allele frequency, variant quality and presence in repeat regions. Finally, ranging thresholds of the somatic score, as calculated by SomaticSniper9, were used as a filter (Supplementary Fig. 1). Removing variants with a somatic score below 70 resulted in 89% sensitivity and 96% specificity. Using this filter, a second batch of SNVs was selected for testing with capillary sequencing, which confirmed 80% (67 of 84) of the predicted SNVs.
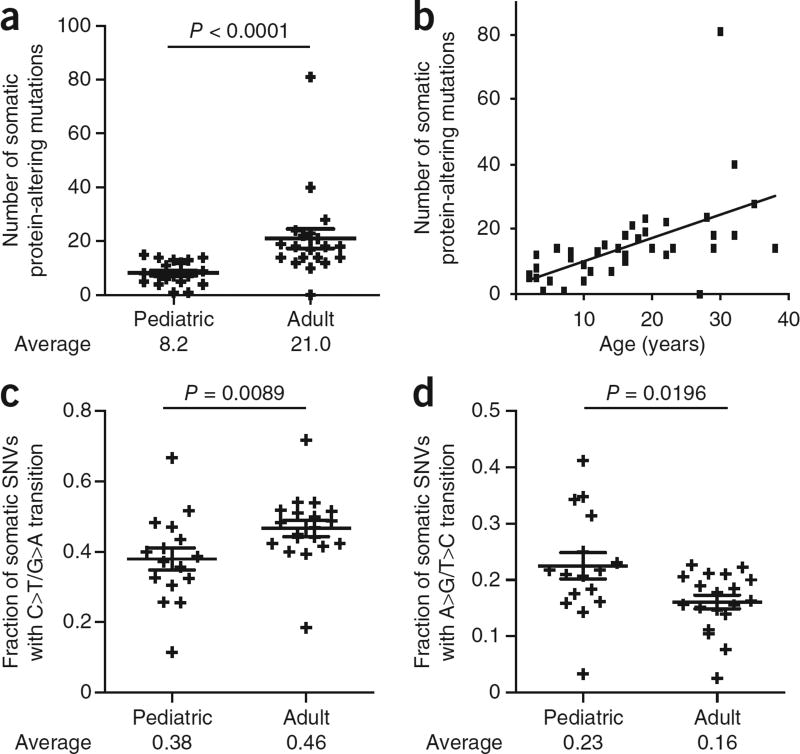
We identified 1,810 somatic SNVs and 1,248 insertion and/or deletions (indels) in the 39 diagnostic-remission pairs. Excessively high numbers of somatic indels were present in three samples, possibly owing to defective DNA repair. These indels were excluded from candidate gene detection. One-fourth of the somatic mutations were protein altering, with the majority being missense mutations (413); the rest were frameshift indels (55), in-frame indels (30), nonsense coding (32) or splice-site mutations (39). On average, each sample contained 14.7 somatic protein-altering SNVs and indels (Supplementary Table 4). Notably, adults (age of >15 years) showed 2.5 times more somatic protein-altering mutations than children (21.0 versus 8.2; P < 0.0001), and there was a clear correlation between the age of the affected individual and mutation number (Fig. 1a,b and Supplementary Fig. 2). However, disease outcome was not linked to mutation number (Supplementary Fig. 3). Notably, a larger fraction of somatic SNVs in adults were cytosine-to-thymine transitions, and adults had a lower fraction of adenine-to-guanine transitions than children (Fig. 1c,d).
Figure 1.
Correlation between the age of the affected individual and mutation number and type. (a) Plot showing the number of protein-altering somatic mutations in pediatric (≤15 years) and adult (≥16 years) individuals with T-ALL. Averages and s.e.m. are shown. The P value tested whether there was a significantly different mutation number in adults versus children and was calculated using the two-tailed Wilcoxon signed-rank test. Pediatric cases, n = 19; adult cases, n = 20. (b) Dot plot representing the number of protein-altering somatic mutations versus age of the affected individual. (c,d) Plots showing the fraction of somatic SNVs that were C>T/G>A transitions (c) or A>G/T >C transitions (d) in pediatric and adult individuals with T-ALL. Averages and s.e.m. are shown. Samples with fewer than ten somatic SNVs were excluded from this analysis. The reported P values test whether there was a significant difference between adults and children and was calculated using the two-tailed Wilcoxon signed-rank test. Pediatric cases, n = 16; adult cases, n = 19.
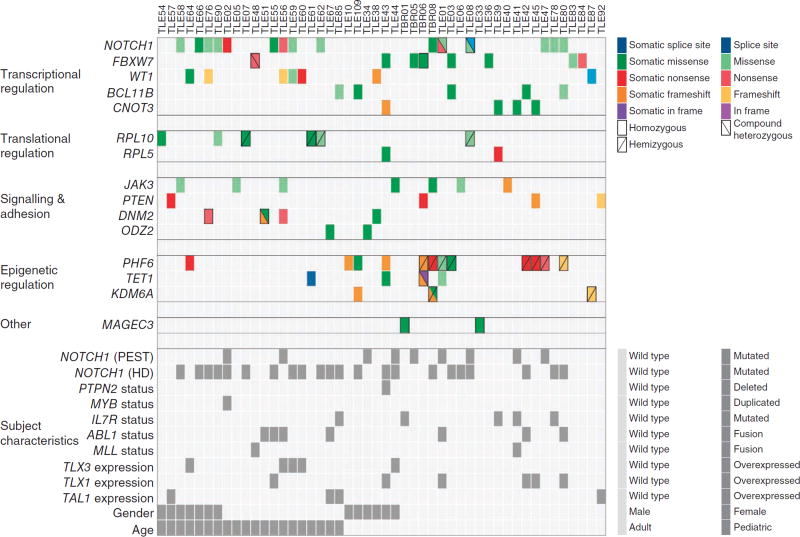
Protein-altering mutations occurred across 508 genes (Supplementary Table 5). To distinguish driver mutations from passenger ones, we only considered genes that were mutated in at least two samples and were significantly more mutated than the local background mutation rate, as calculated by Genome MuSiC10 (Supplementary Table 6). We identified 15 candidate driver genes meeting these two criteria (Fig. 2 and Table 1) and 11 additional genes that were recurrently but not significantly mutated (Supplementary Fig. 4). Of the 15 candidate driver genes, 8 were known drivers in T-ALL, and 7 were new. Reassuringly, we found further mutations in many of the 15 candidate driver genes across the 28 additional diagnosis samples and 17 cell lines that were sequenced (Fig. 2, Supplementary Fig. 5 and Supplementary Table 7).
Figure 2.
Overview of mutations in 15 identified candidate T-ALL driver genes in 67 samples from affected individuals. Top, mutations in 15 candidate T-ALL driver genes are shown across the set of cases. For clarity, only affected individuals harboring mutations in at least 1 of the 15 genes are included. Each type of mutation is indicated by color, and symbols indicate whether the mutation was homozygous, hemizygous or compound heterozygous. Mutations with no indication are heterozygous. All mutations shown here were validated by Sanger sequencing. Bottom, the characteristics of the relevant individuals (identified by Sanger sequencing, karyotyping or gene expression analysis) are shown. Mutations in NOTCH1 were hard to identify by exome sequencing owing to low capture efficiency and resulting low sequence coverage of NOTCH1. Detailed descriptions of the mutations shown in this figure are provided in Supplementary Tables 5 and 7–9.
Table 1.
Significantly and recurrently mutated genes in T-ALL
| Gene | Number of subjects with mutations |
Gene function | Associated age group |
Associated pathologies with genomic alterations |
|---|---|---|---|---|
| NOTCH1 | 29/67 (43.3%)a | Transmembrane receptor, releases intracellular NOTCH1 transcriptional enhancer upon activation | None | T-ALL25, CLL26,27, lung cancer28, head and neck cancer29,30, breast cancer31,32 |
| FBXW7 | 8/67 (11.9%) | Part of ubiquitin ligase complex targeting cyclin E, MYC and NOTCH1 | None | T-ALL33, various cancer types34 |
| WT1 | 7/67 (10.4%) | Zinc-finger transcription factor | None | T-ALL35, AML36, Wilms tumor |
| BCL11B | 5/67 (7.5%) | Zinc-finger transcription factor | None | T-ALL37 |
| CNOT3 | 8/211 (3.8%) | Part of the CCR4-NOT complex that regulates gene expression | Adult | |
| RPL10 | 11/211 (5.2%) | Ribosomal protein of the 60S ribosomal subunit | Pediatric | Autism38,39 |
| RPL5 | 4/211 (1.9%) | Ribosomal protein of the 60S ribosomal subunit | None | Diamond Blackfan anemia11 |
| JAK3 | 7/67 (10.4%) | Kinase involved in cytokine receptor signaling | None | T-ALL8,40, various myeloid and lymphoid malignancies |
| PTEN | 4/67 (6.0%) | Phosphatase antagonizing PI3K function | None | T-ALL18, various cancer types |
| DNM2 | 4/67 (6.0%) | Microtubule-associated GTPase | None | T-ALL8, Charcot-Marie-Tooth disease41, centronuclear myopathy41 |
| ODZ2 | 2/67 (3.0%) | May function as a cellular signal transducer | None | |
| PHF6 | 12/67 (17.9%) | Plant homeodomain-like finger (PHF) family protein that may regulate transcription | Adult | T-ALL42, AML43, Borjeson-Forssman-Lehmann syndrome44 |
| TET1 | 4/67 (6.0%) | Epigenetic regulator converting methylcytosine (5mC) to 5-hydroxymethylcytosine (5-hmC) | None | t(10;11)(q22;23) (MLL-TET1 fusion) in AML and B-ALL45–47 |
| KDM6A | 3/67 (4.5%) | Histone demethylase for Lys27 of histone H3 | None | Various cancer types48,49, Kabuki syndrome50 |
| MAGEC3 | 2/67 (3.0%) | Gene function unknown, only expressed in normal testis and in various tumor types | None | Diffuse large B-cell lymphoma51 |
CLL, chronic lymphocytic leukemia; AML, acute myeloid leukemia; B-ALL, B-cell acute lymphoblastic leukemia.
Mutations in NOTCH1 were hard to identify by exome sequencing due to poor capture efficiency and resulting low sequence coverage of NOTCH1. The reported mutation number reflects NOTCH1 mutations detected by complementary Sanger sequencing.
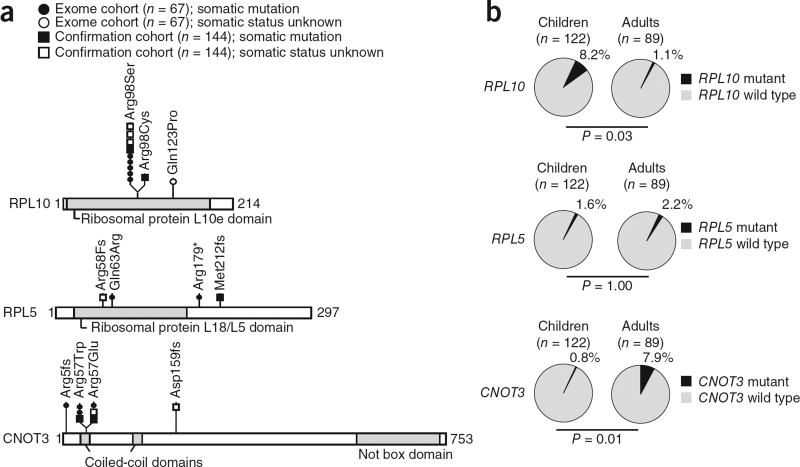
Adult samples showed 2.7 times more mutations in candidate driver genes than children (1.9 versus 0.7; P = 0.0034) (Supplementary Fig. 2). Moreover, mutations in FBXW7, CNOT3, PHF6, KDM6A and MAGEC3 were mainly present in adults, whereas RPL10 mutations were almost exclusively found in children (Figs. 2 and 3, Table 1 and Supplementary Tables 8–10). Notably, our list of candidate driver genes included RPL5 and RPL10, two genes encoding ribosomal proteins that occupy neighboring positions in the 60S ribosomal complex (Supplementary Fig. 6), with five exome samples carrying the same somatic mutation encoding a p.Arg98Ser alteration in RPL10. Also, the CNOT3 gene showed evidence of a mutational hotspot, with three affected individuals carrying a substitution at Arg57 (Fig. 3a). Screening for mutations in these 3 genes in an independent confirmation cohort of 144 T-ALLs identified additional mutations in each of these genes (Fig. 3a and Supplementary Tables 8 and 9), resulting in total mutation frequencies of 3.8% (8/211) for CNOT3 and 7.1% (15/211) for RPL5 and RPL10. Adding the results from the confirmation cohort consolidated the association between CNOT3 mutations and adult age (P = 0.01), and CNOT3 was found to be mutated in 7 of 89 (7.9%) adult T-ALLs. In contrast, RPL10 mutations were associated with young age (P = 0.03), and 10 of 122 (8.2%) pediatric cases had RPL10 mutations (Fig. 3b). Mutations in CNOT3, RPL10 or RPL5 were not associated with any of the major molecular subgroups in T-ALL or with NOTCH1 mutations (Supplementary Tables 8–10).
Figure 3.
Overview of mutations in RPL10, RPL5 and CNOT3. (a) Schematics of RPL10, RPL5 and CNOT3 protein structures with the positions of the alterations detected in 211 T-ALL samples indicated. The somatic status of the mutations is indicated. The characteristics of the individuals with RPL10, RPL5 or CNOT3 mutations are reported in Supplementary Tables 8 and 9. (b) Pie charts reporting mutation frequencies detected in adult versus pediatric individuals with T-ALL. All reported P values tested whether there was a significant difference in mutation frequency in adults versus children and were calculated using the unpaired t test.
Ribosomal defects have been identified in inherited hematopoietic disorders (called ribosomopathies) that result in anemia and a propensity to develop leukemia11. Mutations in RPL5 have previously been associated with Diamond Blackfan anemia and were studied in much detail in that context11, but mutation of RPL10 has not been described in any disease. Notably, loss of RPL22, another 60S ribosomal protein, was also recently identified in T-ALL12, and, in our exome cohort, we also detected one affected individual with a frameshift mutation in RPL22 (Supplementary Table 5).
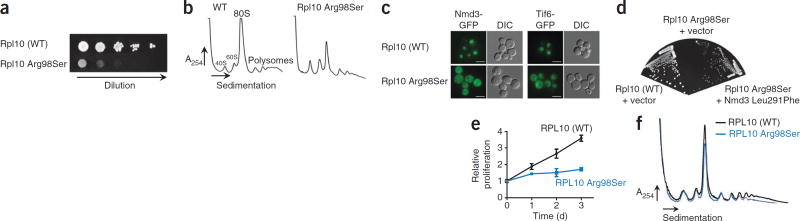
RPL10 is located on the X chromosome, and 7 of 11 mutant cases were males carrying the mutation in nearly all leukemia cells. Moreover, the single RPL10-mutated female from whom we had RNA available expressed only the mutant allele in tumor cells (Supplementary Fig. 7). To confirm that these RPL10 mutations were not random passenger mutations but alter RPL10 function, we engineered yeast cells expressing wild-type Rpl10, Rpl10 Arg98Ser, Rpl10 Arg98Cys or Rpl10 His123Pro as the sole copy of Rpl10. Rpl10 has been intensively studied13 and is highly conserved in yeast, with Arg98 being unchanged from yeast to humans (Supplementary Fig. 8a). Notably, the Arg98 and His123 residues are closely apposed in a β hairpin near the peptidyltransferase center, the catalytic core of the ribosome (Supplementary Fig. 8b,c). In yeast, expression of the Rpl10 mutants impaired proliferation and caused a ribosome biogenesis defect, evidenced by the altered ratio of mature 80S to free subunits and the reduced presence of polysomes (Fig. 4a,b and Supplementary Fig. 9). In addition, Nmd3 and Tif6 showed aberrant accumulation in the cytoplasm in cells expressing Rpl10 Arg98Ser (Fig. 4c and Supplementary Fig. 9), indicating that this alteration impaired the release of the 60S export adaptor Nmd3 as well as the subunit anti-association factor Tif6. The deleterious effects of the Rpl10 mutants were partially suppressed by expressing Nmd3 Leu291Phe (Fig. 4d), a mutant with weakened affinity for the ribosome14, and by increasing the dosage of the Nmd3 gene (Supplementary Fig. 9). These data indicate that the Rpl10 mutants affect the release of Nmd3 from the ribosome. Retention of Nmd3 and Tif6 on pre-60S subunits blocks ribosome assembly, and the resulting depletion of Nmd3 from the nucleus reduces the export of new ribosome subunits15. We also tested the effect of the expression of human RPL10 Arg98Ser, the most frequent RPL10 alteration, in lymphoid cells. In these cells, expression of RPL10 Arg98Ser also resulted in proliferation and ribosome biogenesis defects (Fig. 4e,f).
Figure 4.
Cellular effects of the RPL10 p.Arg98Ser alteration. (a,b) The growth of yeast cells expressing wild-type (WT) Rpl10 or Rpl10 Arg98Ser was compared by plating tenfold serial dilutions (a), and polysome profiles were obtained (b). A254, absorbance at 254 nm. (c) The fluorescence of Nmd3-GFP and Tif6-GFP was examined in cells expressing wild-type Rpl10 or Rpl10 Arg98Ser. Scale bars, 5 µm. DIC, differential interference contrast. In the case of Nmd3, cells also contained leptomycin B (LMB)-sensitive Crm1, and Nmd3-GFP localization was examined after treatment with LMB to trap Nmd3 in the nucleus. (d) Yeast cells expressing wild-type Rpl10 or Rpl10 Arg98Ser were transformed with empty vector or vector expressing Nmd3 Leu291Phe. Tenfold serial dilutions were grown. (e) Proliferation curves of mouse B cells (Ba/F3) expressing wild-type RPL10 or RPL10 Arg98Ser. Error bars, s.d. of measurements in triplicate. (f) Polysome profiling in Ba/F3 cells expressing human wild-type RPL10 or RPL10 Arg98Ser.
In the context of CNOT3, some of the mutations we identified were clearly truncating, whereas another group of mutations seemed to represent missense mutations affecting Arg57. Analysis of mRNA expression, however, showed that, also in the cases with Arg57 alteration, the mutant transcripts were not or were only weakly expressed (Supplementary Fig. 7). This is most likely caused by splicing defects, as the mutations are located at the splice donor site of exon 5. The mutations in CNOT3 thus suggest that this gene acts as a tumor suppressor in T-ALL. CNOT3 is part of the CCR4-NOT complex that regulates gene expression transcriptionally and post-transcriptionally16. Cnot3 also mediates self-renewal in mouse embryonic stem cells, where Cnot3 shares many target genes with Myc17, a known oncogene in T-ALL. To investigate the effect of loss of CNOT3 in tumor formation, we used an established Drosophila eye cancer model. We used the sensitized model in which the Notch ligand Delta is overexpressed in the developing eyes. These flies have larger eyes but by themselves do not develop tumors3,7,18,19, and this model is relevant for T-ALL, given the central role of NOTCH1 signaling in this disease1. Reduction of Not3 expression in this genetic background resulted in a marked increase in tumor incidence from 8% of the eyes with control RNA interference (RNAi) to 46% up to 90% with three different Not3 RNAi lines and one line with a P-element insertion in Not3 (Fig. 5). These data support the notion that reduction in Not3 expression is sufficient to transform sensitized cells.
Figure 5.
Reduced Not3 expression promotes tumor development in a Drosophila sensitized background. (a,b) Sensitized flies overexpressing the Notch ligand Delta in the eye were crossed to one of three different Drosophila Not3 RNAi fly lines (v105990, v37545, v37547), to the 15271 line with a P-element insertion in Not3 or to control RNAi flies (with an RNAi construct against the white (w) gene). Shown are quantitative (a) and qualitative (b) representations of the eye tumor burden in different genotypes. ***, tumor incidence in this cross was significantly different from that in the control cross (P < 0.001) as analyzed by two-tailed Fisher’s exact test. Scale bars in b, 200 µm.
Using whole-exome sequencing, we describe clear differences between pediatric and adult T-ALL and identify a spectrum of driver mutations that function in various cellular processes. One notable observation is that a subset of T-ALL cases have accumulated mutations that affect the function of the ribosome, and it is currently unclear what advantage this might provide to the cancer cells. This is, however, very similar to recent findings of deregulated splicing in myelodysplasia and chronic lymphocytic leukemia20–23 and might indicate that cancer cells have mechanisms to overcome defects in these basic processes. Indeed, cancer cells might compensate for the deleterious effects of mutations affecting the ribosome by acquiring additional mutations, similar to the suppressive effect of the Nmd3 Leu291Phe alteration that we describe in the yeast model (Fig. 4d). Alternatively, the mutations affecting the ribosome might downregulate the hyperactive translation machinery in cancer cells24, which could be beneficial for the fitness of these cells. Our data shed light on the diversity of mutations that are implicated in T-ALL development and on the differences between adult and pediatric T-ALL.
ONLINE METHODS
Leukemia samples and cell lines
T-ALL samples were collected at various institutions. All patients gave their informed consent, and all samples were obtained according to the guidelines of the local ethical committees. This study was approved by the ethical committee of the University Hospital Leuven. The Ba/F3 mouse pro-B-cell line and all human T-ALL cell lines were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ).
Sequencing
Genomic DNA samples from affected individuals and cell lines were sonicated to give a fragment size of 250 bp on a Bioruptor UCD-300 instrument (Diagenode). Library preparation was carried out using the Truseq DNA sample prep kit (Illumina), and capture was performed with SeqCapEZ Exome v2.0 (Nimblegen) or Agilent SureSelect 38 Mb (Agilent) capture reagents. Captured samples were sequenced on a HiSeq 2000 (Illumina) operated in paired-end 2 × 100 bp mode. Reads from each sample were aligned to the reference genome version hg19 using the Burrows-Wheeler Aligner (BWA)52. Alignment files were processed further with the Genome Analysis Toolkit (GATK) before variant calling. Duplicate removal, local realignment around known indels and base quality recalibration were performed as described elsewhere53. Variants were called using the variant quality score recalibration (VQSR) protocol in GATK. Variant calling was performed for each sample separately, and somatic variants were identified by selecting the variants observed at diagnosis that were absent at remission. The somatic score for each variant was calculated with the Somatic Sniper54 algorithm using the processed alignment files for diagnosis and remission samples. Indel calling was performed with DINDEL55 using the calibrated bam files for each sample separately. Somatic indel detection was performed by identifying the high-quality indels in the diagnosis sample (minimum read depth of 15 at the variant site and 15% of the reads supporting the variant allele) and then filtering out the ones observed in the corresponding remission sample (minimum read depth of 1).
Sanger validation of results
All predicted somatic variants shown in Figure 2 were tested and confirmed by conventional Sanger sequencing. These and additional SNVs and indels validated by Sanger sequencing are reported in Supplementary Tables 5 and 7. Overall, we performed Sanger sequencing validation for 219 SNVs, of which 202 were confirmed (92.2% validation rate), and of 78 indels, of which 61 were confirmed (78% validation rate). Analysis of the chromatograms from Sanger sequencing was performed using CLC Main Workbench 6 (CLC Bio). Predicted somatic variants that were not confirmed by Sanger sequencing are not shown in Figure 2 but are reported in Supplementary Table 5 and are indicated as false positives. The entire coding sequences of the RPL5, RPL10 and CNOT3 genes were PCR amplified, and Sanger sequencing was performed on whole-genome amplified DNA from an independent set of 144 individuals with T-ALL. Primer sequences are provided in Supplementary Table 11. Mutation detection was performed using Mutation Surveyor v4.0.4 (Softgenetics). Detected variants were confirmed on original, non-amplified material and were tested for their somatic origin using DNA from remission samples, if available.
Statistical testing
Throughout this study, statistical significance was defined as P < 0.05. The statistical testing methods used are described in the figure and table legends.
Drosophila experiments
The fly line 15271 (y1; P{SUPor-P}l(2)NC136KG10496/CyO; ry506) with a P-element insertion in the Not3 gene was obtained from the Bloomington Stock Center56. Not3 RNAi fly lines v37547 (w1118; P{GD4068}v37547), v37545 (w1118; P{GD4068}v37545) and v105990 (w1118; P{KK102144}VIE-260B) were purchased from the Vienna Drosophila RNAi Center (VDRC)57. Control RNAi flies (w; Sp/CyO; UAS-w dsRNA) and the Cy8 sensitized fly (w; eyeless-Gal4; UAS-Dl/CyO) were described previously3. All fly strains were crossed to Cy8 virgin females. From the performed crosses, the F1 progeny obtained with the following genotypes were selected: Cy8 × 15271 (eyeless-Gal4; UAS-Dl/P{SUPor-P}l(2)NC136KG10496; ry506/+), Cy8 × v37547 (eyeless-Gal4; UAS-Dl/+; P{GD4068}v37547/+), Cy8 × v37545 (eyeless-Gal4; UAS-Dl/+; P{GD4068}v37545/+), Cy8 × v105990 (eyeless-Gal4; UAS-Dl; P{KK102144}VIE-260B) and Cy8 × control (eyeless-Gal4; UAS-Dl/CyO; UAS-w dsRNA/+).
All flies were raised at 25 °C on standard fly food. To analyze the tumor burden, each eye was scored separately in the selected F1 progeny. Eyes were counted as hyperplastic when the eye showed at least one fold.
Yeast experiments
Codon 98 of yeast (Saccharomyces cerevisiae) RPL10 in the centromeric LEU2 vector pAJ2522 was changed from AGA to either TCT or TGT, and codon 123 was changed from CAC to CCA by site-specific mutagenesis. Wild-type and mutant constructs were introduced into the Rpl10 deletion strain AJY1437 (MATα rpl10::KanMX lys2Δ0 met15Δ0 his3Δ1 leu2Δ0 ura3Δ0 pAJ392 - Rpl10 URA3 CEN) by plasmid shuffle or the conditional glucose-repressible Rpl10 strain AJY3373 (MATα KanMX-GAL1-RPL10 his3Δ1 leu2Δ0 ura3Δ0) and assayed for growth by plating tenfold serial dilutions onto selective medium. To test suppression of Rpl10 mutants by mutations in Nmd3, empty vector or vector expressing wild-type Nmd3 or Nmd3 Leu291Phe was introduced into the indicated strains.
Polysome profiles were analyzed as described2. Constructs expressing wild-type Rpl10 or Rpl10 mutants were introduced into AJY1837, which contains a glucose-repressible Rpl10 gene (GAL-Rpl10), the LMB-sensitive Crm1 allele encoding the Thr539Cys variant and Nmd3-GFP, and AJY2766, containing GAL-Rpl10 and Tif6-GFP. Cultures were grown in selective medium containing galactose. Glucose was added to repress the expression of wild-type genomic Rpl10 for 2 h, and LMB was added to a final concentration of 0.1 µg/ml for 30 min to block Nmd3 shuttling. Images were captured using a Nikon E800 microscope fitted with a 100× Plan Apo objective and a Photometrics CoolSNAP ES camera controlled by NIS-Elements AR 2.10 software. Images were prepared using Adobe Photoshop 7.0.
Experiments in mammalian cells
Wild-type RPL10 cDNA (ENST00000344746) was PCR amplified from human thymus cDNA and cloned into the BglII and EcoRI sites of pMSCV-GFP. The mutation encoding the p.Arg98Ser alteration was introduced by mutagenesis of the wild-type RPL10 construct. Mouse lymphoid pro-B cells (Ba/F3) were transduced with pMSCV retroviral constructs expressing wild-type RPL10 or the Arg98Ser mutant according to standard methods. To mimic the situation in individuals with T-ALL, where only mutant and not wild-type RPL10 is expressed in leukemia cells (Supplementary Fig. 7), Ba/F3 cell experiments were performed with concurrent knockdown of endogenous Rpl10. Delivery of small interfering RNA (siRNA) into the cells was performed by electoporation on a Gene Pulser Xcell machine (Bio-Rad) using exponential decay, 250 V and 950 µF. siRNAs against mouse Rpl10 were used at a concentration of 400 nM during electroporation and were obtained from Integrated DNA Technologies (IDT). Cell proliferation was measured on a Guava flow cytometer (Millipore) at the indicated time points after electroporation.
For polysome profiling, Ba/F3 cells were homogenized in a solution with 100 mM Tris-HCl, pH 7.5, 100 mM NaCl, 100 mM MgCl2, 1% DOC/Triton X-100, 1 mM dithiothreitol, 10 µl/ml Protease Inhibitor Cocktail (Sigma), 10 µl/ml Phosphatase Inhibitor Cocktail I (Sigma), 10 µl/ml Phosphatase Inhibitor Cocktail II (Sigma) and 30 U/ml RNasin supplemented with 100 µg/ml cycloheximide. After 5 min of incubation on ice, the extract was centrifuged for 5 min at 12,000 g at 4 °C. The supernatant was loaded onto a 10–60% sucrose gradient and sedimented by centrifugation at 4 °C for 150 min at 37,000 rpm in a Beckman SW41 rotor.
Supplementary Material
Acknowledgments
This work was supported by grants from KU Leuven (concerted action grant to J. Cools and P.V. and PF/10/016 SymBioSys to J. Cools and S.A.), FWO-Vlaanderen (G.0546.11 to J. Cools, P.V., S.A. and A.U. and G.0704.11N to S.A.), the Foundation against Cancer (SCIE2006-34 to J. Cools and 2010-154 to S.A.), a European Research Council (ERC) starting grant (J. Cools), the Interuniversity Attraction Poles (IAP) granted by the Federal Office for Scientific, Technical and Cultural Affairs, Belgium (J. Cools and P.V.), a grant from the Ministry of Health, Cancer Plan (J. Cools, P.V. and S.A.), a grant from the French program Carte d’Identité des Tumeurs (CIT, Ligue Contre le Cancer) and from Canceropole d’Ile de France (J.S.), and a grant from the US National Institutes of Health (NIH; GM53655 to A.W.J. and S.P.). K.D.K. is a postdoctoral researcher, and P.V. is a senior clinical investigator of FWO-Vlaanderen.
Footnotes
Accession codes. Sequence and variant data are available from the European Genome-phenome Archive (EGA) under accession EGAS00001000296, and somatic variants are available through a BioMart interface at http://lcbmart.aertslab.org/.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
All authors contributed to the writing of the manuscript. K.D.K., Z.K.A., N.L., C.B., B.A.H. and A.W.J. designed and performed experiments and analyzed data. C.V. and J.Y. performed and analyzed Not3 Drosophila experiments. S.P. performed and analyzed Rpl10 yeast studies. R.L. performed and analyzed polysome profiling experiments. T.G., V.G., E.G., M.P., I.L., G.H., E.C., R.V., B.S., K.J., N.M. and I.W. performed experiments and analyzed data. H.V., B.C., J. Cloos, J.S., A.U. and P.V. collected samples and analyzed data. S.A. and J. Cools supervised the project, designed experiments and analyzed data.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 2.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Invest. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferres-Marco D, et al. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 4.De Keersmaecker K, Marynen P, Cools J. Genetic insights in the pathogenesis of T-cell acute lymphoblastic leukemia. Haematologica. 2005;90:1116–1127. [PubMed] [Google Scholar]
- 5.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N. Engl. J. Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 6.Homminga I, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson DE, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dees ND, et al. MuSiC: identifying mutational significance in cancer genomes. Genome. Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao S, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120:3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofer A, Bussiere C, Johnson AW. Mutational analysis of the ribosomal protein Rpl10 from yeast. J. Biol. Chem. 2007;282:32630–32639. doi: 10.1074/jbc.M705057200. [DOI] [PubMed] [Google Scholar]
- 14.Hedges J, et al. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo KY, et al. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collart MA, Panasenko OO. The Ccr4-not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 17.Hu G, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes. Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bossuyt W, et al. The atonal proneural transcription factor links differentiation and tumor formation in Drosophila. PLoS Biol. 2009;7:e40. doi: 10.1371/journal.pbio.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 21.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 23.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 25.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 26.Fabbri G, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westhoff B, et al. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. USA. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao X, et al. Somatic mutations in the Notch, NF-κB, PIK3CA, and Hedgehog pathways in human breast cancers. Genes Chromosom. Cancer. 2012;51:480–489. doi: 10.1002/gcc.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 35.Tosello V, et al. WT1 mutations in T-ALL. Blood. 2009;114:1038–1045. doi: 10.1182/blood-2008-12-192039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King-Underwood L, Pritchard-Jones K. Wilms’ tumor (WT1) gene mutations occur mainly in acute myeloid leukemia and may confer drug resistance. Blood. 1998;91:2961–2968. [PubMed] [Google Scholar]
- 37.De Keersmaecker K, et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat. Med. 2010;16:1321–1327. doi: 10.1038/nm.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klauck SM, et al. Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism. Mol. Psychiatry. 2006;11:1073–1084. doi: 10.1038/sj.mp.4001883. [DOI] [PubMed] [Google Scholar]
- 39.Chiocchetti A, et al. Mutation and expression analyses of the ribosomal protein gene RPL10 in an extended German sample of patients with autism spectrum disorder. Am. J. Med. Genet. A. 2011;155:1472–1475. doi: 10.1002/ajmg.a.33977. [DOI] [PubMed] [Google Scholar]
- 40.Kalender Atak Z, et al. High accuracy mutation detection in leukemia on a selected panel of cancer genes. PLoS ONE. 2012;7:e38463. doi: 10.1371/journal.pone.0038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durieux AC, Prudhon B, Guicheney P, Bitoun M. Dynamin 2 and human diseases. J. Mol. Med. 2010;88:339–350. doi: 10.1007/s00109-009-0587-4. [DOI] [PubMed] [Google Scholar]
- 42.Van Vlierberghe P, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat. Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Vlierberghe P, et al. ETV6 mutations in early immature human T cell leukemias. J. Exp. Med. 2011;208:2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lower KM, et al. 1024C>T (R342X) is a recurrent PHF6 mutation also found in the original Börjeson-Forssman-Lehmann syndrome family. Eur. J. Hum. Genet. 2004;12:787–789. doi: 10.1038/sj.ejhg.5201228. [DOI] [PubMed] [Google Scholar]
- 45.Ono R, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer. Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 46.Lorsbach RB, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 47.Burmeister T, et al. The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia: results from the GMALL study group. Blood. 2009;113:4011–4015. doi: 10.1182/blood-2008-10-183483. [DOI] [PubMed] [Google Scholar]
- 48.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lederer D, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larson DE, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albers CA, et al. Dindel: accurate indel calls from short-read data. Genome. Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bellen HJ, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.