Abstract
Recurrent spontaneous abortion (RSA) occurs in 1–5% of parturients. The sustained therapy and research for RSA is expensive, which is a serious issue faced by both patients and doctors. The aim of the present study was to detect protein expression profiles in the serum of RSA patients and healthy controls, and to identify potential biomarkers for this disease. A 1,000-protein microarray consisting of a combination of Human L-507 and L-493 was used. The microarray data revealed that eight serum protein expression levels were significantly upregulated and 143 proteins were downregulated in RSA patients compared with the healthy controls. ELISA individually validated 5 of these 151 proteins in a larger cohort of patients and control samples, demonstrating a significant decrease in insulin-like growth factor-binding protein-related protein 1 (IFGBP-rp1)/IGFBP-7, Dickkopf-related protein 3 (Dkk3), receptor for advanced glycation end products (RAGE) and angiopoietin-2 levels in patients with RSA. Sensitivity and specificity analyses were calculated by a receiver operating characteristics curve, and were revealed to be 0.881, 0.823, 0.79 and 0.814, with diagnostic cut-off points of 95.44 ng/ml for IFGBP-rp1, 32.84 ng/ml for Dkk3, 147.27 ng/ml for RAGE and 441.40 ng/ml for angiopoietin-2. The present study indicated that these four proteins were downregulated in RSA samples and may be useful as biomarkers for the prediction and diagnosis of RSA. Subsequent studies in larger-scale cohorts are required to further validate the diagnostic value of these markers.
Keywords: recurrent spontaneous abortion, serum biomarker, antibody array
Introduction
Recurrent spontaneous abortion (RSA), also referred to as recurrent miscarriage, habitual abortion or recurrent pregnancy loss, is defined by more than three consecutive miscarriages prior to 20 gestational weeks (1,2). RSA occurs in 1–5% of women during pregnancy (3). The cause of RSA remains unknown; thus, continuing clinical and laboratory investigations are required (4,5). Previous studies have reported that various etiologic factors are involved in certain RSA cases; including chromosome abnormalities, endocrine diseases, uterine abnormalities, placental anomalies, hormonal problems, thrombophilia, infections, nutritional disorders, autoimmune disease and anatomy (6–8). The etiology of RSA remains to be fully elucidated despite numerous studies investigating the above factors. Early prediction of the potential risk of RSA is required to increase live birth rates in patients with RSA (9).
Biomarkers are currently widely used to refine diagnoses, predict disease and monitor the effects of treatment (10). It is established that the human proteome regulates cellular function and determines the phenotype; thus, the identification of relevant proteins is likely to reveal reliable biomarkers for predicting disease (11). A range of potential biomarkers for RSA have been previously reported. Stortoni et al (12) reported that expression levels of thrombomodulin were reduced by 45% in patients with RSA compared with healthy individuals, as determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Bao et al (13) determined by RT-qPCR, western blot analysis and immunohistochemistry that serum Dickkopf-related protein (Dkk) 1 levels were increased in RSA patients compared with controls. Additional studies are required to validate these potential biomarkers and their prognostic value. Identifying novel RSA biomarkers may improve the diagnosis, safety and efficacy of current therapies for RSA. As one of the most intensely studied protein families in biomedical science, cytokines have been widely investigated as potential disease biomarkers (14). The introduction of high-throughput and high-specificity detection of complex proteins at picomolar and femtomolar quantities, and antibody arrays, are now widely used for mining complex proteomes (15), facilitating simultaneous screening of numerous secreted signal proteins in complex biological samples (16). However, to the best of our knowledge, no previous study has identified serum RSA biomarkers using antibody array technology. Therefore, the present study used a RayBio® Label-Based (L-Series) Human Antibody Array 1000 Membrane kit (RayBiotech, Inc., Norcross, GA, USA) to identify reliable biomarkers for the prediction of RSA.
Patients and methods
Patients and controls
From January 2014 to March 2015, a total of 60 Chinese patients with a history of RSA were recruited as the patient group from the Department of Traditional Chinese Medicine at the Beijing Obstetrics and Gynecology Hospital. They had normal endocrine levels, and their partners had normal spermatogenesis and sperm function. ‘Blood stasis’ syndrome (BSS, also known as Xueyu zheng in Chinese) is characterized in traditional medicine as ‘pain that occurs in a fixed location, dark-purple face or tongue, bleeding, blood spots under the skin, and an astringent pulse’ among other features (17). The concept of blood stasis has been interpreted, changed and developed systematically since ancient times (18). All 60 RSA patients exhibited the ‘blood stasis’ features described above at the time of study. Patient characteristics, including age at diagnosis, gravidities, number of child births and timing of spontaneous abortion, are summarized in Table I. For the control group, 20 Chinese females who had experienced full-term pregnancies were recruited from the Department of Traditional Chinese Medicine at the Beijing Obstetrics and Gynecology Hospital.
Table I.
Characteristics of 60 patients with recurrent spontaneous abortion.
| Characteristic | Value |
|---|---|
| Age at diagnosisa | 30±2.8 |
| Graviditiesa | 3±0.5 |
| No. of childbirths | |
| 1b | 5 (8%) |
| 0b | 55 (92%) |
| Spontaneous abortionsa | 3±0.5 |
| 2b | 1 (2%) |
| 1b | 12 (20%) |
| 0b | 47 (78%) |
Data are expressed as the mean ± standard deviation.
Data are expressed as the number of patients (% of total).
Ethical approval and sample collection
All participants signed informed consent forms prior to participation. The present study was approved by the Ethics Committee of the Beijing Obstetrics and Gynecology Hospital, Capital Medical University (Beijing, China; approval no. 2014-KY-001). Whole blood samples were collected from each participant. Serum was collected following blood centrifugation at 550 × g for 10 min at 4°C, and stored at −80°C. The sera of 23 RSA patients and 10 healthy subjects were pooled into 6 samples followed by standard processing (19). The samples included those that could be classified as ‘blood stasis’ 1, 2 and 3, ‘non-blood stasis’ 1, 2, and 3, and controls 1–6. The order of mixing is presented in Table II. All mixtures were obtained by mixing equal volumes of sera.
Table II.
Pooling of serum samples.
| Pooled serum sample | Original sample |
|---|---|
| Patients with RSA | |
| Blood stasis group 1 (thrombus 1) | A1, A2, A3, A4 |
| Blood stasis group 2 (thrombus 2) | A5, A6, A7, A8 |
| Blood stasis group 3 (thrombus 3) | A9, A10, A11, A12 |
| Non-blood stasis group 1 (non-thrombus 1) | C1, C2, C3, C4 |
| Non-blood stasis group 2 (non-thrombus 2) | C5, C6, C7, C8 |
| Non-blood stasis group 3 (non-thrombus 3) | C9, C10, C11 |
| Control | |
| 1 | E1, E2, E3 |
| 2 | E4, E5, E6 |
| 3 | E7 |
| 4 | E8 |
| 5 | E9 |
| 6 | E10 |
RSA, recurrent spontaneous abortion.
Antibody array assay
The 12 samples described above were assayed for the relative expression of 1,000 human proteins. The target proteins included cytokines, chemokines, adipokines, growth factors, angiogenic factors, proteases, soluble receptors and soluble adhesion molecules. A RayBio® Label-Based (L-Series) Human Antibody Array 1,000 Membrane kit (consisting of a combination of Human L-507 and L-493) was used for protein detection in accordance with the manufacturer's protocol. The signals were scanned at a wavelength of 532 nm using an InnoScan 300 Microarray Scanner (Innopsys, Carbonne, France; resolution, 10 µm) and analyzed using RayBio Analysis Tool software (AAH-BLG-1-SW and AAH-BLG-2-SW; RayBiotech, Inc.).
Detection of protein levels by ELISA
As determined by microarray analysis, serum markers with significant differences in expression levels between patients and healthy individuals were detected in 60 patients and 20 controls using ELISA kits (ELH-TRAPPIN2, ELH-IGFBPRP1, ELH-RAGE, ELH-DKK3 and ELH-Angiopoietin-2; RayBiotech, Inc.) according to the manufacturer's protocol. Trappin-2, insulin-like growth factor-binding protein-related protein 1 (IGFBP-rp1)/IGFBP-7, receptor for advanced glycation end products (RAGE), Dkk3, and angiopoietin-2 levels were detected. Serum samples were incubated at room temperature. Following washing with wash buffer, a prepared biotinylated antibody was added into the microplate to capture the target protein. Following this, horseradish peroxidase-conjugated streptavidin was used to bind with biotin from the biotinylated antibody. Finally, 1-Step 3,3′,5,5′-tetramethylbenzidine-ELISA substrate solution was added followed by stop solution, and absorbance was measured at a wavelength of 450 nm by absorbance microplate reader ELx800 (BioTek Instruments, Inc., Winooski, VT, USA).
Statistical analysis and bioinformatics
All array data analyses were performed using RayBio Analysis Tool software. Biostatistics and bioinformatics analysis included discriminatory protein analysis and data mining cluster analysis. Statistical differences between two groups were determined by Student's t-test. Fold change values of proteins were used as indicators of relative expression levels. Data mining cluster analysis was used to identify potential biomarkers by clustering all relevant proteins according to the similarity of their expression profiles using Cluster software version 3.0 (http://cluster2.software.informer.com/3.0). ELISA data was analyzed using SigmaPlot software version 12.0 (Systat Software, Inc., San Jose, CA, USA). T- and F-tests were used to analyze ELISA quantification. The receiver operating characteristics curve (ROC) method was used to assess sensitivity and specificity of potential biomarkers using SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Analysis of antibody microarrays
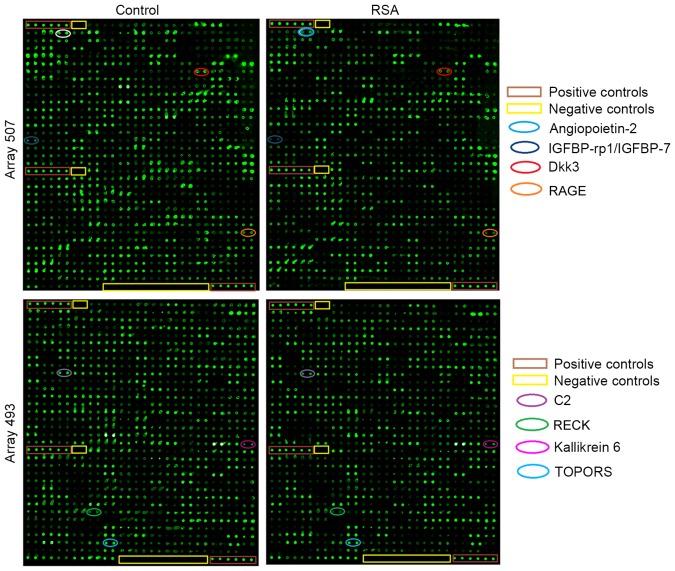
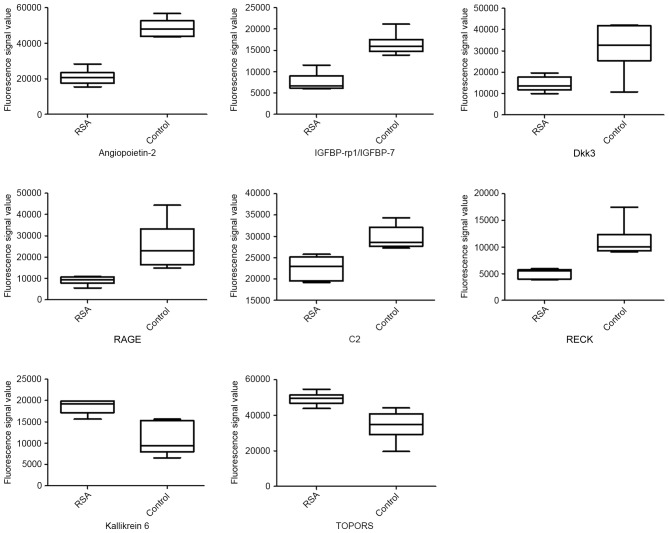
A total of 1,000 proteins were measured in the serum mixture using the microarray. The spectra of 1,000 proteins from eight samples are presented in Fig. 1. The results demonstrated that 151 proteins had significantly different expressions between the two groups. Of these differential proteins, eight were significantly upregulated, and 143 proteins were downregulated in RSA patients compared with controls (Table III). Fig. 2 presents are boxplots of the fluorescence signal values of eight differential proteins, selected for signal strength, fold changes and clinical significance. Serum mixture samples were arranged by similarities in the abundance of these 151 markers in the sera clustering algorithm, which produced two clusters that contained patients and healthy individuals (Fig. 3).
Figure 1.
Protein spectra from RayBio L-Series Human 507 (507 proteins) and 493 (493 proteins) antibody arrays. Representative images from human antibody arrays demonstrating the reactivity of pooled serum samples to arrays L series (1,000 proteins) in healthy controls and RSA patients. Each protein was measured in duplicate. A total of eight of significantly different factors on the microarrays are marked in elliptical boxes. IGFBP-rp1/IGFBP-7, insulin-like growth factor-binding protein-related protein 1/insulin-like growth factor-binding protein 7; Dkk3, Dickkopf-related protein 3; RAGE, receptor for advanced glycation end products; RSA, recurrent spontaneous abortion; TOPORS, topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase; C2, complement C2; RECK, reversion-inducing-cysteine rich protein with kazal motifs.
Table III.
A total of 151 proteins with significantly different expression levels between patients with RSA and controls.
| RSA | Control | RSA vs. control | |||||
|---|---|---|---|---|---|---|---|
| Target | Mean | SD | Mean | SD | F-test | t-test P-value | Fold-change |
| A2M | 4,650.558 | 373.053 | 6,374.645 | 269.687 | 0.494 | 0.000 | 0.730 |
| ADAMTS-10 | 7,862.456 | 1,306.692 | 13,925.383 | 550.378 | 0.081 | 0.000 | 0.565 |
| ADAMTS-15 | 2,999.115 | 479.112 | 5,705.655 | 1,924.875 | 0.008 | 0.017 | 0.526 |
| ADAMTS-5 | 558.209 | 250.695 | 1,466.103 | 483.403 | 0.176 | 0.002 | 0.381 |
| ADAMTS-L2 | 20,775.755 | 2,676.424 | 27,565.213 | 1,305.789 | 0.141 | 0.000 | 0.754 |
| ALK | 7,976.603 | 1,428.122 | 13,199.048 | 2,632.151 | 0.206 | 0.002 | 0.604 |
| Angiopoietin-2 | 20,789.029 | 4,292.976 | 48,570.398 | 4,977.529 | 0.753 | 0.000 | 0.428 |
| ApoC2 | 14,305.316 | 2,066.666 | 19,754.508 | 2,309.965 | 0.813 | 0.002 | 0.724 |
| ApoH | 4,898.886 | 462.884 | 7,760.884 | 1,065.437 | 0.091 | 0.000 | 0.631 |
| ApoM | 1,502.749 | 273.114 | 3,924.876 | 654.067 | 0.078 | 0.000 | 0.383 |
| APP | 12,633.899 | 1,318.521 | 19,428.245 | 5,101.616 | 0.010 | 0.021 | 0.650 |
| Axl | 1,510.055 | 1,117.320 | 4,936.198 | 495.598 | 0.099 | 0.000 | 0.306 |
| BAF57 | 1,181.342 | 265.140 | 1,906.967 | 361.301 | 0.513 | 0.003 | 0.619 |
| BAFF R/TNFRSF13C | 130.169 | 152.078 | 1,362.334 | 162.415 | 0.889 | 0.000 | 0.096 |
| Bax | 15,217.862 | 3,242.826 | 38,315.362 | 7,712.389 | 0.080 | 0.000 | 0.397 |
| BDNF | 13,455.044 | 6,688.395 | 25,250.436 | 5,906.534 | 0.792 | 0.009 | 0.533 |
| β 2M | 1,320.861 | 133.263 | 1,910.310 | 294.761 | 0.106 | 0.001 | 0.691 |
| BIK | 311.407 | 134.485 | 1,179.513 | 269.458 | 0.153 | 0.000 | 0.264 |
| BMP-3 | 10,702.264 | 3,851.243 | 28,110.700 | 15,617.736 | 0.008 | 0.041 | 0.381 |
| BMP-3b/GDF-10 | 124.436 | 41.431 | 347.417 | 122.473 | 0.033 | 0.005 | 0.358 |
| BMP-4 | 901.991 | 2,56.578 | 1,767.609 | 343.897 | 0.536 | 0.001 | 0.510 |
| BMPR-IB/ALK-6 | 23,780.461 | 9648.386 | 62,589.402 | 6,422.893 | 0.393 | 0.000 | 0.380 |
| BTC | 11,944.615 | 4,921.916 | 21,760.107 | 3,620.813 | 0.517 | 0.003 | 0.549 |
| C2 | 22,604.021 | 2,719.955 | 29,647.219 | 2,700.060 | 0.988 | 0.001 | 0.762 |
| C5/C5a | 12,737.991 | 1,525.086 | 22,749.974 | 7,304.236 | 0.004 | 0.019 | 0.560 |
| Calsyntenin-1 | 1,911.570 | 607.215 | 4,505.218 | 691.477 | 0.782 | 0.000 | 0.424 |
| CD40/TNFRSF5 | 1,101.031 | 337.593 | 3,397.425 | 678.353 | 0.152 | 0.000 | 0.324 |
| Chordin-Like 1 | 1,391.942 | 585.338 | 3,128.589 | 566.356 | 0.944 | 0.000 | 0.445 |
| CNTF R α | 77,532.324 | 10,164.182 | 114,914.734 | 12,924.497 | 0.611 | 0.000 | 0.675 |
| Contactin-1 | 3,977.884 | 438.734 | 6,093.532 | 1,458.926 | 0.020 | 0.015 | 0.653 |
| Cripto-1 | 8,176.343 | 2,831.476 | 18,724.736 | 4,786.781 | 0.274 | 0.001 | 0.437 |
| CRTH-2 | 34,856.201 | 6,547.851 | 55,742.907 | 8,503.753 | 0.580 | 0.001 | 0.625 |
| CXCR4 (fusin) | 3,702.606 | 1,090.358 | 11,095.207 | 6,134.514 | 0.002 | 0.031 | 0.334 |
| Dkk-3 | 14,387.266 | 3,452.455 | 31,716.357 | 11,437.946 | 0.020 | 0.012 | 0.454 |
| DLL4 | 1,759.891 | 629.238 | 3,330.977 | 898.310 | 0.453 | 0.006 | 0.528 |
| EDAR | 1,240.627 | 223.958 | 4,773.756 | 1,971.378 | 0.000 | 0.007 | 0.260 |
| EGF R/ErbB1 | 11,026.167 | 1,733.072 | 15,758.956 | 2,371.519 | 0.508 | 0.003 | 0.700 |
| EG-VEGF/PK1 | 29,916.922 | 4,877.762 | 51,349.446 | 16,211.012 | 0.020 | 0.022 | 0.583 |
| EMAP-II | 10,666.342 | 2,600.714 | 19,708.795 | 3,896.837 | 0.395 | 0.001 | 0.541 |
| EphB4 | 1,962.256 | 406.898 | 5,161.814 | 945.891 | 0.088 | 0.000 | 0.380 |
| ErbB2 | 4,267.172 | 1,130.288 | 15,536.343 | 9,241.970 | 0.000 | 0.030 | 0.275 |
| ESAM | 2,854.186 | 495.592 | 5,990.106 | 2,581.532 | 0.002 | 0.030 | 0.476 |
| FAM3B | 1,270.946 | 447.324 | 4,455.137 | 885.200 | 0.160 | 0.000 | 0.285 |
| FGF R4 | 3,992.796 | 1,049.190 | 12,794.810 | 7,288.004 | 0.001 | 0.031 | 0.312 |
| FGF R5 | 2,826.176 | 1,013.809 | 7,878.562 | 1,003.601 | 0.983 | 0.000 | 0.359 |
| FGF-19 | 1,641.777 | 369.438 | 4,442.636 | 778.147 | 0.128 | 0.000 | 0.370 |
| FGF-9 | 6,413.361 | 1,443.488 | 22,180.535 | 5,185.520 | 0.014 | 0.000 | 0.289 |
| FGFR1 | 11,978.622 | 1,527.435 | 16,683.655 | 4,343.023 | 0.039 | 0.045 | 0.718 |
| FGFR2 | 13,703.293 | 2,914.828 | 23,307.209 | 7,653.038 | 0.054 | 0.017 | 0.588 |
| Ficolin-3 | 3,374.750 | 486.123 | 8,120.462 | 4,212.321 | 0.000 | 0.040 | 0.416 |
| Follistatin-like1 | 2,481.289 | 721.297 | 7,152.976 | 1,161.224 | 0.319 | 0.000 | 0.347 |
| Galectin-1 | 1,435.877 | 634.069 | 3,694.218 | 803.285 | 0.616 | 0.000 | 0.389 |
| Galectin-3BP | 10,655.461 | 1,406.056 | 13,522.242 | 1,910.307 | 0.517 | 0.014 | 0.788 |
| Gas1 | 4,530.505 | 1,493.529 | 7,110.508 | 1,228.339 | 0.678 | 0.008 | 0.637 |
| GASP-1/WFIKKNRP | 85,406.876 | 7,073.932 | 128,207.327 | 24,054.577 | 0.018 | 0.006 | 0.666 |
| GATA-3 | 7,625.978 | 591.277 | 13,276.727 | 2,655.282 | 0.005 | 0.003 | 0.574 |
| GCP-2/CXCL6 | 847.677 | 514.092 | 2,243.977 | 1,012.301 | 0.163 | 0.013 | 0.378 |
| GLO-1 | 997.491 | 290.387 | 2,106.667 | 501.997 | 0.255 | 0.001 | 0.473 |
| Glucagon | 61,025.896 | 9,853.803 | 128,506.268 | 35,031.652 | 0.015 | 0.004 | 0.475 |
| GluT2 | 11,546.660 | 704.259 | 37,514.070 | 2,261.457 | 0.023 | 0.000 | 0.308 |
| Glypican 3 | 1,389.140 | 249.379 | 2,554.322 | 649.947 | 0.056 | 0.002 | 0.544 |
| Glypican 5 | 15,529.644 | 2,266.574 | 25,519.026 | 7,257.053 | 0.023 | 0.018 | 0.609 |
| GPX1 | 2,008.854 | 557.930 | 4,383.148 | 782.893 | 0.475 | 0.000 | 0.458 |
| GPX3 | 3,009.541 | 1,283.488 | 5,083.531 | 927.399 | 0.493 | 0.009 | 0.592 |
| GRP78 | 2,266.940 | 302.999 | 4,387.312 | 1,150.085 | 0.011 | 0.005 | 0.517 |
| Hemopexin | 725.265 | 151.625 | 3,871.951 | 913.350 | 0.001 | 0.000 | 0.187 |
| HRG-α | 5,321.121 | 1,385.473 | 7,078.008 | 834.906 | 0.291 | 0.024 | 0.752 |
| HSP10 | 26,971.939 | 3,802.952 | 43,781.370 | 10,058.305 | 0.052 | 0.003 | 0.616 |
| I-309 | 4,283.951 | 2,116.728 | 13,856.485 | 3,913.389 | 0.204 | 0.000 | 0.309 |
| IBSP | 7,712.034 | 986.133 | 10,133.874 | 1,761.061 | 0.229 | 0.015 | 0.761 |
| IGFBP-4 | 1,453.473 | 684.730 | 4,146.757 | 1,511.595 | 0.107 | 0.003 | 0.351 |
| IGFBP-rp1/IGFBP-7 | 7,523.924 | 2,135.058 | 16,380.247 | 2,502.068 | 0.736 | 0.000 | 0.459 |
| IGF-II | 65,869.138 | 5,912.640 | 94,321.230 | 10,392.485 | 0.241 | 0.000 | 0.698 |
| IL-1 ra | 361,872.941 | 34,398.268 | 563,878.996 | 45,414.321 | 0.011 | 0.011 | 0.642 |
| IL-13 R α1 | 6,360.695 | 1,012.833 | 24,083.997 | 13,484.474 | 0.000 | 0.023 | 0.264 |
| IL-17C | 2,345.533 | 541.033 | 7,048.395 | 748.914 | 0.493 | 0.000 | 0.333 |
| IL-18 R β/AcPL | 1,935.580 | 858.981 | 5,832.686 | 1,358.705 | 0.337 | 0.000 | 0.332 |
| IL-29 | 8,733.803 | 1,800.279 | 24,866.993 | 5,813.232 | 0.022 | 0.001 | 0.351 |
| IL-31 | 1,970.735 | 687.942 | 5,958.295 | 784.836 | 0.779 | 0.000 | 0.331 |
| IL-31 RA | 1,182.894 | 372.281 | 3,207.266 | 506.214 | 0.516 | 0.000 | 0.369 |
| IL-33 | 2,441.589 | 430.891 | 3,847.273 | 1,061.515 | 0.070 | 0.013 | 0.635 |
| IL-6 R | 3,540.308 | 909.773 | 6,566.421 | 1,881.755 | 0.137 | 0.005 | 0.539 |
| IL-8 | 25,002.761 | 6,204.334 | 34,829.214 | 6,706.528 | 0.869 | 0.025 | 0.718 |
| Kallikrein 14 | 2,979.489 | 615.746 | 5,825.747 | 1,585.646 | 0.058 | 0.002 | 0.511 |
| LBP | 3,409.164 | 463.893 | 13,342.910 | 8,052.946 | 0.000 | 0.029 | 0.256 |
| LIF R α | 13,797.957 | 2,293.325 | 27,604.197 | 8,923.296 | 0.010 | 0.012 | 0.500 |
| LIF | 332.827 | 242.463 | 1,628.345 | 875.661 | 0.014 | 0.014 | 0.204 |
| LIGHT/TNFSF14 | 2,092.869 | 917.268 | 6,360.354 | 1,227.569 | 0.538 | 0.000 | 0.329 |
| Lipocalin-1 | 20,832.310 | 4,151.253 | 34,570.502 | 9,172.736 | 0.107 | 0.007 | 0.603 |
| Livin | 2,707.067 | 795.904 | 4,194.195 | 745.424 | 0.889 | 0.007 | 0.645 |
| LRG1 | 11,113.403 | 863.592 | 28,581.729 | 14,531.460 | 0.000 | 0.032 | 0.389 |
| Lymphotoxin β R/TNFRSF3 | 788.596 | 380.913 | 3,291.572 | 764.370 | 0.153 | 0.000 | 0.240 |
| M-CSF | 28,394.735 | 5,351.774 | 41,766.017 | 11,016.348 | 0.139 | 0.023 | 0.680 |
| Midkine | 3,267.246 | 433.600 | 5,032.121 | 1,617.032 | 0.012 | 0.044 | 0.649 |
| MIF | 3,951.056 | 885.814 | 6,986.334 | 1,120.792 | 0.618 | 0.000 | 0.566 |
| MIP-1α | 50,222.493 | 12,359.813 | 94,756.426 | 25,311.008 | 0.142 | 0.003 | 0.530 |
| MMP-11/Stromelysin-3 | 54,153.970 | 12,843.620 | 69,764.345 | 7,386.564 | 0.250 | 0.027 | 0.776 |
| MMP-16/MT3-MMP | 11,330.829 | 2,524.054 | 26,764.023 | 11,916.806 | 0.004 | 0.024 | 0.423 |
| MMP-8 | 52,067.139 | 3,933.472 | 70,211.908 | 7,728.075 | 0.165 | 0.000 | 0.742 |
| MSP α Chain | 13,201.992 | 2,412.896 | 25,409.389 | 4,525.441 | 0.194 | 0.000 | 0.520 |
| NEP | 1,581.827 | 658.480 | 3,958.249 | 666.439 | 0.980 | 0.000 | 0.400 |
| NM23-H1/H2 | 550.675 | 189.223 | 2,651.125 | 1,022.967 | 0.002 | 0.004 | 0.208 |
| Orexin B | 43,718.747 | 7,996.118 | 74,138.593 | 18,347.514 | 0.092 | 0.004 | 0.590 |
| Osteoactivin/GPNMB | 2,895.689 | 312.735 | 5,540.870 | 1,872.568 | 0.001 | 0.017 | 0.523 |
| PD-1 | 2,591.490 | 389.734 | 4,141.350 | 1,038.516 | 0.051 | 0.007 | 0.626 |
| PDGF-C | 9,264.920 | 2,881.398 | 24,377.899 | 9,157.178 | 0.024 | 0.008 | 0.380 |
| PDGF-D | 2,883.630 | 396.751 | 5,130.131 | 907.360 | 0.093 | 0.000 | 0.562 |
| PDX-1 | 2,270.140 | 363.742 | 3,981.057 | 824.869 | 0.097 | 0.001 | 0.570 |
| PEPSINOGEN I | 2,439.074 | 824.492 | 6,093.801 | 1,335.963 | 0.313 | 0.000 | 0.400 |
| Persephin | 2,515.448 | 1,011.303 | 4,523.807 | 838.775 | 0.691 | 0.004 | 0.556 |
| PGRP-S | 12,316.910 | 730.024 | 16,966.564 | 1,180.649 | 0.315 | 0.000 | 0.726 |
| PIM2 | 1,865.699 | 437.191 | 4,879.191 | 1,260.593 | 0.036 | 0.001 | 0.382 |
| PKM2 | 2,655.795 | 1,099.697 | 4,528.436 | 478.749 | 0.092 | 0.003 | 0.586 |
| RAGE | 9,020.357 | 1,947.254 | 25,325.305 | 10,739.884 | 0.002 | 0.013 | 0.356 |
| RANK/TNFRSF11A | 2,020.058 | 587.942 | 4,020.679 | 833.497 | 0.462 | 0.001 | 0.502 |
| RECK | 5,084.724 | 891.920 | 11,081.323 | 3,186.035 | 0.014 | 0.005 | 0.459 |
| RELT/TNFRSF19L | 31,871.125 | 4,886.989 | 61,866.224 | 17,568.797 | 0.014 | 0.007 | 0.515 |
| ROBO4 | 66,732.387 | 11,925.740 | 100,118.428 | 17,349.652 | 0.430 | 0.003 | 0.667 |
| S100A10 | 1,189.482 | 653.171 | 4,146.007 | 336.156 | 0.171 | 0.000 | 0.287 |
| S100A4 | 4,024.174 | 328.979 | 5,366.717 | 482.437 | 0.421 | 0.000 | 0.750 |
| S100A6 | 3,485.252 | 541.367 | 6,023.360 | 1,041.092 | 0.178 | 0.000 | 0.579 |
| Serpin A8 | 1,755.946 | 304.447 | 3,270.617 | 704.644 | 0.089 | 0.001 | 0.537 |
| Serpin A9 | 1,258.179 | 327.453 | 2,710.239 | 999.220 | 0.029 | 0.015 | 0.464 |
| Smad 1 | 1,028.273 | 540.825 | 3,720.431 | 748.020 | 0.494 | 0.000 | 0.276 |
| Smad 7 | 34,565.690 | 2,075.517 | 55,551.432 | 18,891.474 | 0.000 | 0.042 | 0.622 |
| Smad 8 | 6,886.663 | 2,599.832 | 15,373.512 | 4,688.229 | 0.221 | 0.003 | 0.448 |
| SOST | 3,117.271 | 461.906 | 6,218.660 | 1,701.605 | 0.012 | 0.006 | 0.501 |
| Spinesin | 20,626.782 | 4,848.625 | 38,828.941 | 4,444.642 | 0.853 | 0.000 | 0.531 |
| Syndecan-1 | 1,772.888 | 379.277 | 3,778.232 | 1,215.352 | 0.023 | 0.008 | 0.469 |
| Thrombospondin-4 | 10,921.569 | 2,002.073 | 23,198.652 | 9,994.362 | 0.003 | 0.029 | 0.471 |
| TIM-1 | 2,132.294 | 259.727 | 4,438.018 | 697.608 | 0.049 | 0.000 | 0.480 |
| TIMP-3 | 4,008.775 | 1,259.727 | 6,988.724 | 1,853.036 | 0.417 | 0.009 | 0.574 |
| TRADD | 127,030.588 | 28,309.796 | 222,266.652 | 75,285.718 | 0.051 | 0.016 | 0.572 |
| TRAIL R2/DR5/TNFRSF10B | 17,190.463 | 4,094.960 | 32,861.735 | 13,104.866 | 0.023 | 0.032 | 0.523 |
| Trappin-2 | 5,865.048 | 978.164 | 19,388.585 | 11,625.715 | 0.000 | 0.036 | 0.303 |
| TROY/TNFRSF19 | 2,324.349 | 786.973 | 5,012.391 | 1,945.216 | 0.069 | 0.011 | 0.464 |
| TRPC1 | 2,436.721 | 681.450 | 6,693.017 | 2,511.517 | 0.012 | 0.008 | 0.364 |
| TSLP | 1,853.969 | 303.884 | 5,110.938 | 2,119.578 | 0.001 | 0.013 | 0.363 |
| TSLP R | 1,905.470 | 965.346 | 6,488.242 | 1,625.517 | 0.277 | 0.000 | 0.294 |
| Ubiquitin+1 | 17,529.029 | 5,002.370 | 34,561.192 | 16,122.797 | 0.023 | 0.049 | 0.507 |
| uPA | 52,463.526 | 6,879.542 | 94,616.706 | 25,331.681 | 0.012 | 0.008 | 0.554 |
| VEGF-D | 24,467.055 | 5,668.534 | 52,501.169 | 6,836.652 | 0.691 | 0.000 | 0.466 |
| WISP-1/CCN4 | 2,330.112 | 1,166.842 | 5,516.779 | 2,727.490 | 0.086 | 0.025 | 0.422 |
| GASP-2/WFIKKN | 71,246.693 | 3,344.127 | 31,422.325 | 19,817.529 | 0.001 | 0.004 | 2.267 |
| IL-1 F5/FIL1δ | 30,158.276 | 6,083.967 | 13,083.494 | 8,056.469 | 0.553 | 0.002 | 2.305 |
| IL-28A | 71,058.941 | 7,420.008 | 30,883.742 | 18,108.416 | 0.072 | 0.001 | 2.301 |
| Kallikrein 6 | 18,588.432 | 1,695.500 | 10,786.937 | 3,766.885 | 0.104 | 0.001 | 1.723 |
| NGF R | 42,876.389 | 1,905.084 | 1,8740.985 | 13,128.177 | 0.001 | 0.006 | 2.288 |
| NrCAM | 73,805.167 | 6,435.403 | 5,1415.054 | 8,782.973 | 0.511 | 0.001 | 1.435 |
| TOPORS | 49,264.099 | 3,557.276 | 3,4240.228 | 8,383.960 | 0.083 | 0.002 | 1.439 |
| VEGF R2 (KDR) | 37,152.074 | 4,541.513 | 1,2904.085 | 9,743.845 | 0.119 | 0.000 | 2.879 |
SD, standard deviation; RSA, recurrent spontaneous abortion.
Figure 2.
Boxplots of differential serum proteins between RSA patients and controls. P<0.05, RSA vs. control for all presented proteins. ANG-2, angiopoietin 2; IGFBP-rp1/IGFBP-7, insulin-like growth factor-binding protein-related protein 1/insulin-like growth factor-binding protein 7; Dkk3, Dickkopf-related protein 3; RAGE, receptor for advanced glycation end products; RSA, recurrent spontaneous abortion; TOPORS, topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase; C2, complement C2; RECK, reversion-inducing-cysteine rich protein with kazal motifs.
Figure 3.
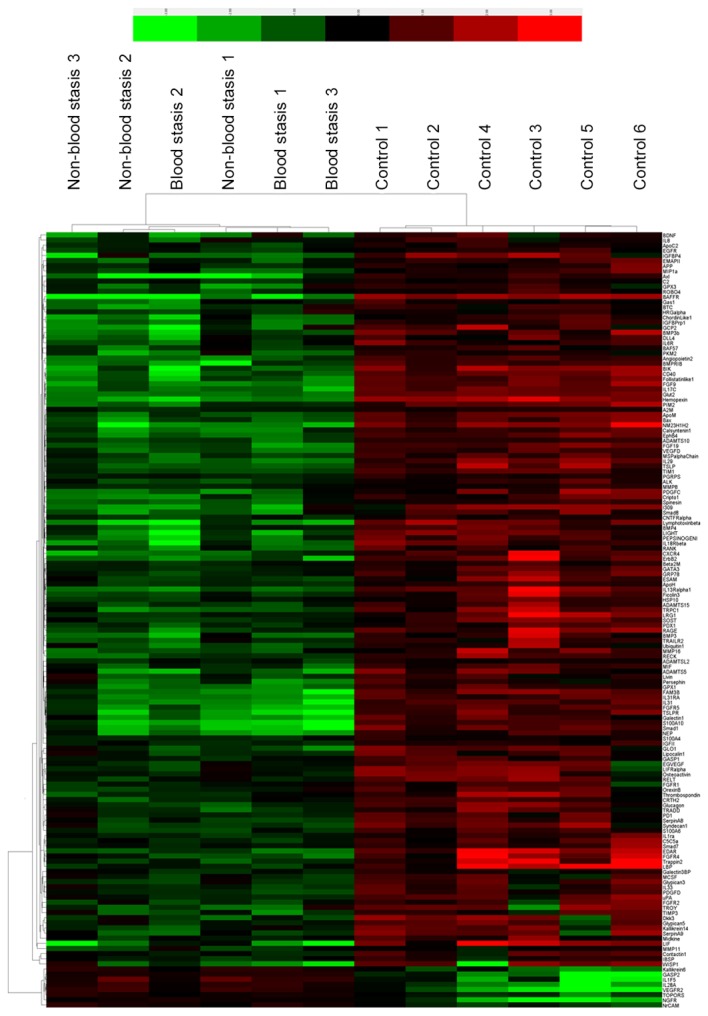
Cluster map of protein expression levels of the 151 proteins in 12 mixed serum samples. The signal values of the 151 proteins from microarray analyses were used to prepare the cluster map. Red shades indicate higher expression levels, green shades indicate lower expression levels, and black shades indicate median expression levels.
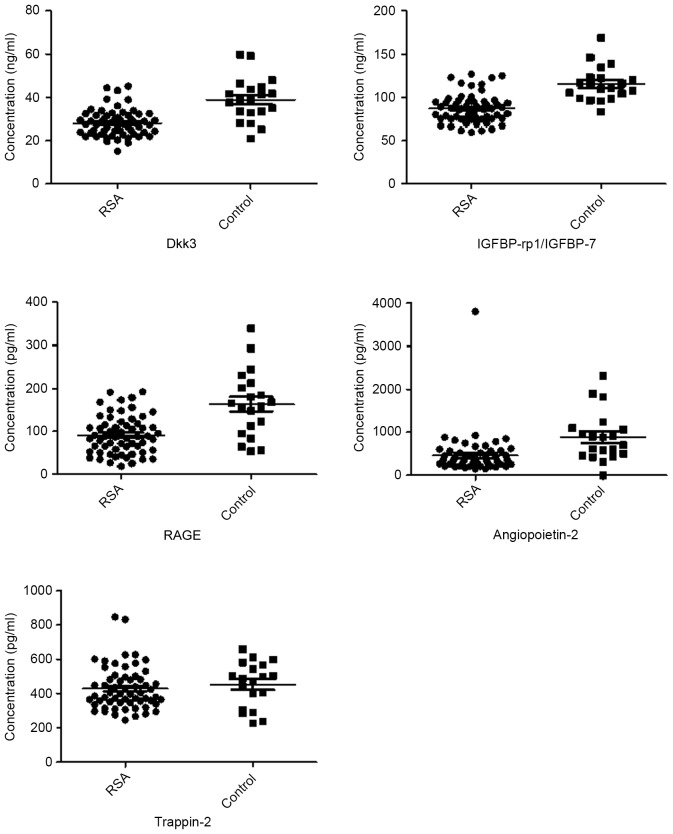
Validation of microarray data by ELISA
A total of five of the 151 proteins were selected for validation assay in 60 RSA and 20 control samples. Serum levels of trappin-2, IGFBP-rp1/IGFBP-7, RAGE, Dkk3 and angiopoietin-2 were selected to be measured by ELISA based on the results from the microarray experiments, previous reports on serum biomarkers in RSA and the availability of commercial test kits. Levels of IGFBP-rp1/IGFBP-7, Dkk3, RAGE and angiopoietin-2 were downregulated in RSA patients compared with healthy controls, which was consistent with the microarray results (P<0.05; Table IV and Fig. 4).
Table IV.
ELISA analysis of cytokine levels in the serum of patients with RSA and healthy controls.
| Patients vs. control | |||||
|---|---|---|---|---|---|
| Cytokine | RSA | Control | F-test | t-test P-value | Fold-change |
| Trappin-2 | 429.17±125.17 | 453.26±132.34 | 0.718 | 0.473 | 0.947 |
| IGFBP-rp1/IGFBP-7 | 86.94±16.49 | 115.63±20.12 | 0.246 | 0.000a | 0.752 |
| RAGE | 91.29±44.28 | 163.64±76.99 | 0.001 | 0.001a | 0.558 |
| Dkk3 | 28.16±6.22 | 38.96±10.05 | 0.005 | 0.000 | 0.723 |
| Angiopoietin-2 | 461.34±484.38 | 887.72±576.22 | 0.312 | 0.002a | 0.520 |
P<0.05, RSA vs. control. Data are expressed as the mean ± standard deviation. IGFBP-rp1/IGFBP-7, insulin-like growth factor-binding protein-related protein 1/insulin-like growth factor-binding protein 7; Dkk3, Dickkopf-related protein 3; RAGE, receptor for advanced glycation end products; RSA, recurrent spontaneous abortion.
Figure 4.
Validation of five differentially expressed proteins obtained from microarray analysis, and validated by ELISA. Concentrations of these factors in serum samples obtained from RSA patients and healthy controls were calculated using the four parameters method. IGFBP-rp1/IGFBP-7, insulin-like growth factor-binding protein-related protein 1/insulin-like growth factor-binding protein 7; Dkk3, Dickkopf-related protein 3; RAGE, receptor for advanced glycation end products; RSA, recurrent spontaneous abortion.
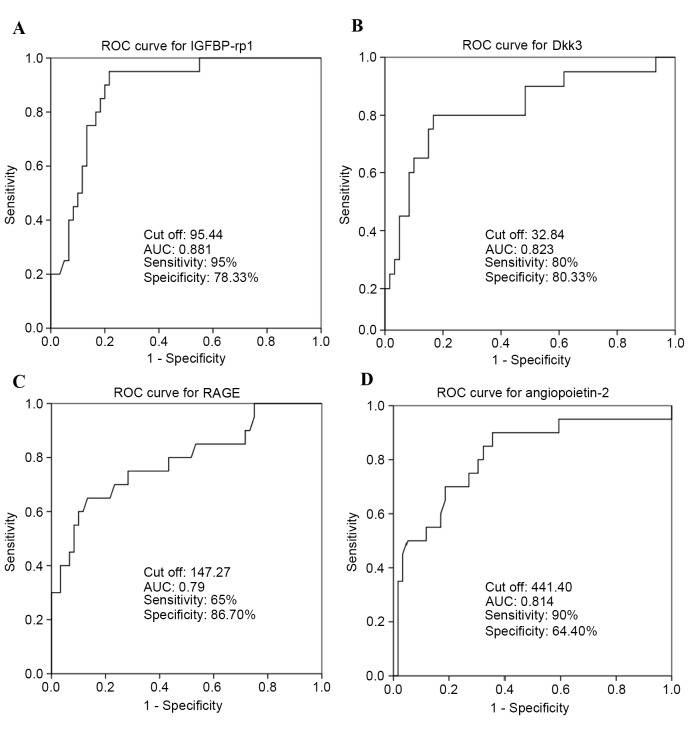
Analysis of sensitivity and specificity of serum biomarkers for RSA
To validate whether IGFBP-rp1/IGFBP-7, Dkk3, RAGE and angiopoietin-2 may be used as biomarkers for predicting RSA, ROC curves were used to analyze sensitivity and specificity. Area-under-ROC-curve values for IGFBP-rp1/IGFBP-7 (Fig. 5A), Dkk3 (Fig. 5B), RAGE (Fig. 5C) and angiopoietin-2 (Fig. 5D) cytokines were 0.881, 0.823, 0.79 and 0.814, respectively. IGFBP-rp1/IGFBP-7 had a sensitivity of 95% and specificity of 78.33%. Dkk3 had a sensitivity of 80% and specificity of 83.33%. RAGE had a sensitivity of 65% and specificity of 86.70%. Angiopoietin-2 had a sensitivity of 90% and specificity of 64.40%. All these were deemed suitable biomarkers for the prediction of RSA.
Figure 5.
ROC curve analysis for the four upregulated serum cytokines as validated by ELISA. The area under the ROC curve (AUC) indicates the mean sensitivity of the biomarkers (A) IGFBP-rp1, (B) Dkk3, (C) RAGE and (D) angiopoietin-2 (Fig. 5D). 0.5≤AUC≤1, the biomarker is strongly differential between patients and controls; AUC≤0.5, no predictive value. ROC, receiver operating characteristics curve; IGFBP-rp1/IGFBP-7, insulin-like growth factor-binding protein-related protein 1/insulin-like growth factor-binding protein 7; Dkk3, Dickkopf-related protein 3; RAGE, receptor for advanced glycation end products; RSA, recurrent spontaneous abortion.
Discussion
Potential biomarkers of RSA have previously been reported. Khonina et al (20) investigated whether mixed lymphocyte reaction blocking factor may be used as an indicator of the efficacy for immunotherapy with paternal lymphocytes in females with RSA. Metwally et al (21) performed a proteomic analysis of obese and overweight women with RSA by 2-D gel electrophoresis, principle component analysis and mass spectrometry, and demonstrated that RSA patients exhibit a significant increase in haptoglobin expression. Ibrahim et al (22) demonstrated that pentraxin-3 indicates the presence of abnormally exaggerated intrauterine inflammation that may cause pregnancy failure in females with unexplained RSA. Kim et al (23) identified RSA-associated factors in human blood samples by 2-D gel electrophoresis, and analyzed spots samples with matrix-assisted laser desorption/ionization-time of flight/mass spectrometry, and reported that in RSA patients, inter-α-trypsin inhibitor heavy chain family member 4 (ITI-H4) expression was low and exhibited a molecular weight of 120 kDa in controls; however, ITI-H4 was expressed at higher levels and at a modified molecular weight of 36 kDa in the RSA patient group. This indicated that ITI-H4 may be used as biomarker of RSA.
The present study used antibody array technology for a primary screening of RSA biomarkers on pooled samples. The array results revealed that the levels of eight cytokines were significantly increased in the RSA patient group compared with controls, and the levels of 143 of the tested 1,000 proteins were significantly reduced in the RSA patient group compared with controls. A total of 5 proteins, trappin-2, IGFBP-rp1/IGFBP-7, Dkk3, RAGE and angiopoietin-2, were selected for ELISA validation assay in a larger cohort of patient and control subjects. ELISA results for these proteins were in accordance with the array results. Sensitivity and specificity analysis by ROC revealed that these four cytokines may be used as biomarkers of RSA.
To the best of our knowledge, the association between IGFBP-rp1/IGFBP-7, Dkk3 and angiopoietin-2, and RSA has not been reported. However, an isoform of the RAGE protein, sRAGE, has been reported to be associated with RSA (24).
IGFBP-rp1/IGFBP-7
IGFs, which have characteristics of tissue growth factors and circulating growth hormones, are potent mitogens and anti-apoptotic agents (25). IGFs include the hormones IGF-I and -II and their corresponding receptors, and the IGFBPs (26). The IGFBP superfamily includes six members (IGFBP-1-6) and 10 associated proteins (IGFBP-rp1-10) (27). IGFBP-7 has been demonstrated to be a tumor suppressor in a variety of cancers. Benatar et al (28) reported that treatment with IGFBP-7 may have therapeutic potential for triple-negative breast cancer. Liu et al (29) demonstrated that IGFBP-7 was downregulated in gastric cancer, and that it may be used as an indicator of poor prognosis in patients with gastric cancer. IGFBP-7 has additionally been proposed as a novel biomarker for assessing the risk of acute kidney injury (30) and heart failure with reduced ejection fraction, and has been demonstrated to have links to the presence and severity of echocardiographic parameters of abnormal diastolic function (31).
Dkk3
The Wnts are an evolutionarily conserved family of secreted glycoproteins characterized by numerous conserved cysteine residues (32). The Dkk proteins are secreted Wnt inhibitors, inducing removal of the Wnt co-receptor low-density lipoprotein receptor-related protein, and consist of four primary members in vertebrates (Dkk1-4) (33,34). Dkk-3 is downregulated in various types of cancer cells. Loss of Dkk3 protein expression is associated with poor prognoses in patients with gastric cancer, indicating that it may be a biomarker for predicting lymph node involvement in these patients (35). Dkk3 has recently been implicated in clear cell renal cell carcinoma, and may present a novel molecular target for its diagnosis and treatment (36). Additionally, Dkk3 may represent a therapeutic target for the treatment of heart failure following myocardial infarction (37).
RAGE
RAGE is a cell-surface receptor that interacts with AGEs, and is a member of the immunoglobulin superfamily (38). RAGE activation via its multiple ligands, including S100 calcium-binding protein (S100A) 4 (39), high mobility group box 1 protein (40) and amyloid-β protein (41), serves important roles in certain diseases. Dahlmann et al (42) demonstrated that the activity of S100A4-RAGE induces RAGE-dependent increases in the migratory and invasive capabilities of colorectal cancer cells. Guo et al (43) identified RAGE as a potential prognostic biomarker in renal cell carcinoma. RAGE/S100A7 signaling has been demonstrated to have a functional role in linking inflammation to aggressive breast cancer development; therefore, RAGE expression is currently regarded as a potential biomarker for triple-negative breast cancer (44). Additionally, overexpression of RAGE may be a useful marker to predict gastric cancer progression (45).
Angiopoietin-2
As a member of the angiopoietin family, angiopoietin-2 has complex and unique roles in regulating angiogenesis, and has additional unconventional functions, including stimulating tumor angiogenesis, invasion and metastasis via Tie2-independent signaling pathways, involving integrin-mediated signaling. Therefore, angiopoietin-2 may have great potential as a therapeutic target, prognostic marker and inhibitor of human cancer (46). Angiopoietin-2 is expressed during vascular remodeling, thus preventing vascular stability (47). A study by Morrissey et al (48) demonstrated that angiopoietin-2 inhibition impeded tumor growth of LuCaP 23.1 prostate cancer xenografts, and suggested that angiopoietin-2 inhibition in combination with other treatments is a potential therapy for metastatic disease patients. Calfee et al (49) reported that lowering plasma angiopoietin-2 with fluid conservative therapy may be beneficial, in part by decreasing endothelial inflammation. Goede et al (50) demonstrated that serum angiopoietin-2 represents a candidate biomarker for the outcome of metastatic colorectal cancer patients treated with bevacizumab-containing therapy. Additionally, angiopoietin-2 has been associated with other diseases, including chronic kidney disease (51) and cerebral malaria (52).
In conclusion, the present study used a microarray platform to detect 1,000 proteins to identify dysregulated serum factors in RSA samples. This method was demonstrated to be effective in investigating dynamic alterations in protein profiles, and to select target proteins for further RSA research. The results indicated that IGFBP-rp1/IGFBP-7, Dkk3, RAGE and angiopoietin-2 expression were downregulated in RSA patients, suggesting that they may be important in the pathological process of RSA. Furthermore, upregulating them may inhibit the development of RSA. Therefore, these biomarkers represent potential predictive and diagnostic markers for RSA due to their high sensitivity and specificity. However, larger-scale studies are required to confirm the diagnostic value of these markers.
Acknowledgements
The present study was supported by Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding (China; grant no. ZYLX201510). The authors would like to thank Mr Xiangfu Ren (Beijing KeZhongZhi Biotechnology Co., Ltd., Beijing, China) for technical assistance and Ms. Hong Shao (Beijing KeZhongZhi Biotechnology Co., Ltd.) for valuable discussions.
Glossary
Abbreviations
- RSA
recurrent spontaneous abortion
- AUC
area under the curve
References
- 1.Ford HB, Schust DJ. Recurrent pregnancy loss: Etiology, diagnosis, and therapy. Rev Obstet Gynecol. 2009;2:76–83. [PMC free article] [PubMed] [Google Scholar]
- 2.Tang AW, Alfirevic Z, Turner MA, Drury J, Quenby S. Prednisolone trial: Study protocol for a randomized controlled trial of prednisolone for women with idiopathic recurrent miscarriage and raised levels of uterine natural killer (uNK) cells in the endometrium. Trials. 2009;10:102. doi: 10.1186/1745-6215-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen DI, Hong ZB. Research on the pathogenesis diagnosis and treatment of female reproductive disorders. J Shanghai Jiaotong Univ. 2012;32:1161–1165. [Google Scholar]
- 4.Li TC, Markris M, Tomsu M, Tuckerman E, Laird S. Recurrent miscarriage: Aetiology, management and prognosis. Hum Reprod Update. 2002;8:463–481. doi: 10.1093/humupd/8.5.463. [DOI] [PubMed] [Google Scholar]
- 5.Regan L. Recurrent miscarriage. BMJ. 1991;302:543–544. doi: 10.1136/bmj.302.6776.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuan Z, Jie D, Hao X, Junhua B, Mengjing G, Liguo P, Yousheng Y, Hong L, Zhenghao H. Associations between androgen receptor CAG & GGN repeat polymorphism & recurrent spontaneous abortions in Chinese women. Indian J Med Res. 2014;139:730–736. [PMC free article] [PubMed] [Google Scholar]
- 7.Shankarkumar U, Pradhan VD, Patwardhan MM, Shankarkumar A, Ghosh K. Autoantibody profile and other immunological parameters in recurrent spontaneous abortion patients. Niger Med J. 2011;52:163–166. doi: 10.4103/0300-1652.86126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang L, Wei Z, Li O, Huang R, Qin J, Chen H, Fan X, Chen ZJ. An increase in vascular endothelial growth factor (VEGF) and VEGF soluble receptor-1 (Sflt-1) are associated with early recurrent spontaneous abortion. PLoS One. 2013;8:e75759. doi: 10.1371/journal.pone.0075759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rull K, Tomberg K, Kõks S, Männik J, Möls M, Sirotkina M, Värv S, Laan M. Increased placental expression and maternal serum levels of apoptosis-inducing TRAIL in recurrent miscarriage. Placenta. 2013;34:141–148. doi: 10.1016/j.placenta.2012.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burska A, Boissinot M, Ponchel F. Cytokines as biomarkers in rheumatoid arthritis. Mediators Inflamm. 2014;2014:545493. doi: 10.1155/2014/545493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsangaris GT, Anagnostopoulos AK, Tounta G, Antsaklis A, Mavrou A, Kolialexi A. Application of proteomics for the identification of biomarkers in amniotic fluid: Are we ready to provide a reliable prediction? EPMA J. 2011;2:149–155. doi: 10.1007/s13167-011-0083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stortoni P, Cecati M, Giannubilo SR, Sartini D, Turi A, Emanuelli M, Tranquilli AL. Placental thrombomodulin expression in recurrent miscarriage. Reprod Biol Endocrinol. 2010;8:1. doi: 10.1186/1477-7827-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao SH, Shuai W, Tong J, Wang L, Chen P, Duan T. Increased Dickkopf-1 expression in patients with unexplained recurrent spontaneous miscarriage. Clin Exp Immunol. 2013;172:437–443. doi: 10.1111/cei.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang RP, Burkholder B, Jones Sloane V, Jiang WD, Mao YQ, Chen QL, Shi Z. Cytokine antibody arrays in biomarker discovery and validation. Curr Proteomics. 2012;9:55–70. doi: 10.2174/157016412799746209. [DOI] [Google Scholar]
- 15.Wilson JJ, Burgess R, Mao YQ, Luo S, Tang H, Jones VS, Weisheng B, Huang RY, Chen X, Huang RP. Antibody arrays in biomarker discovery. Adv Clin Chem. 2015;69:255–324. doi: 10.1016/bs.acc.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Burkholder B, Huang RY, Burgess R, Luo S, Jones VS, Zhang W, Lv ZQ, Gao CY, Wang BL, Zhang YM, Huang RP. Tumor-induced perturbations of cytokines and immune cell networks. Biochim Biophys Acta. 2014;1845:182–201. doi: 10.1016/j.bbcan.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Liao J, Wang J, Liu Y, Li J, Duan L, Chen G, Hu J. Modern researches on blood stasis syndrome 1989–2015: A bibliometric analysis. Medicine (Baltimore) 2016;95:e5533. doi: 10.1097/MD.0000000000005533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park B, Yun KJ, Jung J, You S, Lee JA, Choi J, Kang BK, Alraek T, Birch S, Lee MS. Conceptualization and utilization of blood stasis syndrome among doctors of Korean medicine: Results of a web-based survey. Am J Transl Res. 2014;6:857–868. [PMC free article] [PubMed] [Google Scholar]
- 19.Mahlknecht P, Stemberger S, Sprenger F, Rainer J, Hametner E, Kirchmair R, Grabmer C, Scherfler C, Wenning GK, Seppi K, et al. An antibody microarray analysis of serum cytokines in neurodegenerative Parkinsonian syndromes. Proteome Sci. 2012;10:71. doi: 10.1186/1477-5956-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khonina NA, Broitman EV, Shevela EY, Pasman NM, Chernykh ER. Mixed lymphocyte reaction blocking factors (MLR-Bf) as potential biomarker for indication and efficacy of paternal lymphocyte immunization in recurrent spontaneous abortion. Arch Gynecol Obstet. 2013;288:933–937. doi: 10.1007/s00404-013-2832-x. [DOI] [PubMed] [Google Scholar]
- 21.Metwally M, Preece R, Thomas J, Ledger W, Li TC. A proteomic analysis of the endometrium in obese and overweight women with recurrent miscarriage: Preliminary evidence for an endometrial defect. Reprod Biol Endocrinol. 2014;12:75. doi: 10.1186/1477-7827-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibrahim MI, Harb HM, Ellaithy MI, Elkabarity RH, Abdelgwad MH. First trimester assessment of Pentraxin-3 levels in women with primary unexplained recurrent pregnancy loss. Eur J Obstet Gynecol Reprod Biol. 2012;165:37–41. doi: 10.1016/j.ejogrb.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Kim MS, Gu BH, Song S, Choi BC, Cha DH, Baek KH. ITI-H4, as a biomarker in the serum of recurrent pregnancy loss (RPL) patients. Mol Biosyst. 2011;7:1430–1440. doi: 10.1039/c0mb00219d. [DOI] [PubMed] [Google Scholar]
- 24.Ota K, Yamagishi S, Kim M, Dambaeva S, Gilman-Sachs A, Beaman K, Kwak-Kim J. Elevation of soluble form of receptor for advanced glycation end products (sRAGE) in recurrent pregnancy losses (RPL): Possible participation of RAGE in RPL. Fertil Steril. 2014;102:782–789. doi: 10.1016/j.fertnstert.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Neuhouser ML, Platz EA, Till C, Tangen CM, Goodman PJ, Kristal A, Parnes HL, Tao Y, Figg WD, Lucia MS, et al. Insulin-like growth factors and insulin-like growth factor binding proteins and prostate cancer risk: Results from the prostate cancer prevention trial. Cancer Prev Res (Phila) 2013;6:91–99. doi: 10.1158/1940-6207.CAPR-12-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bower NI, Johnston IA. Transcriptional regulation of the igf signaling pathway by amino acids and insulin-like growth factors during myogenesis in Atlantic salmon. PLoS One. 2010;5:e11100. doi: 10.1371/journal.pone.0011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto T, Hess S, Kajiyama H, Sakairi T, Saleem MA, Mathieson PW, Nojima Y, Kopp JB. Proteomic analysis identifies insulin-like growth factor-binding protein-related protein-1 as a podocyte product. Am J Physiol Renal Physiol. 2010;299:F776–F784. doi: 10.1152/ajprenal.00597.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benatar T, Yang W, Amemiya Y, Evdokimova V, Kahn H, Holloway C, Seth A. IGFBP7 reduces breast tumor growth by induction of senescence and apoptosis pathways. Breast Cancer Res Treat. 2012;133:563–573. doi: 10.1007/s10549-011-1816-4. [DOI] [PubMed] [Google Scholar]
- 29.Liu L, Yang Z, Zhang W, Yan B, Gu Q, Jiao J, Yue X. Decreased expression of IGFBP7 was a poor prognosis predictor for gastric cancer patients. Tumour Biol. 2014;35:8875–8881. doi: 10.1007/s13277-014-2160-1. [DOI] [PubMed] [Google Scholar]
- 30.Kashani K, Al-Khafaji A, Ardiles T, Artigas A, Bagshaw SM, Bell M, Bihorac A, Birkhahn R, Cely CM, Chawla LS, et al. Discovery and validation of cell cycle arrest biomarkers in human acute kidney injury. Crit Care. 2013;17:R25. doi: 10.1186/cc12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandhi PU, Gaggin HK, Sheftel AD, Belcher AM, Weiner RB, Baggish AL, Motiwala SR, Liu PP, Januzzi JL., Jr Prognostic usefulness of insulin-like growth factor-binding protein 7 in heart failure with reduced ejection fraction: A novel biomarker of myocardial diastolic function? Am J Cardiol. 2014;114:1543–1549. doi: 10.1016/j.amjcard.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 33.Dellinger TH, Planutis K, Jandial DD, Eskander RN, Martinez ME, Zi X, Monk BJ, Holcombe RF. Expression of the Wnt antagonist Dickkopf-3 is associated with prognostic clinicopathologic characteristics and impairs proliferation and invasion in endometrial cancer. Gynecol Oncol. 2012;126:259–267. doi: 10.1016/j.ygyno.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 35.Park JM, Kim MK, Chi KC, Kim JH, Lee SH, Lee EJ. Aberrant loss of dickkopf-3 in gastric cancer: Can it predict lymph node metastasis preoperatively? World J Surg. 2015;39:1018–1025. doi: 10.1007/s00268-014-2886-3. [DOI] [PubMed] [Google Scholar]
- 36.Guo CC, Zhang XL, Yang B, Geng J, Peng B, Zheng JH. Decreased expression of Dkk1 and Dkk3 in human clear cell renal cell carcinoma. Mol Med Rep. 2014;9:2367–2373. doi: 10.3892/mmr.2014.2077. [DOI] [PubMed] [Google Scholar]
- 37.Bao MW, Cai Z, Zhang XJ, Li L, Liu X, Wan N, Hu G, Wan F, Zhang R, Zhu X, et al. Dickkopf-3 protects against cardiac dysfunction and ventricular remodelling following myocardial infarction. Basic Res Cardiol. 2015;110:25. doi: 10.1007/s00395-015-0481-x. [DOI] [PubMed] [Google Scholar]
- 38.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 39.Yammani RR, Carlson CS, Bresnick AR, Loeser RF. Increase in production of matrix metalloproteinase 13 by human articular chondrocytes due to stimulation with S100A4: Role of the receptor for advanced glycation end products. Arthritis Rheum. 2006;54:2901–2911. doi: 10.1002/art.22042. [DOI] [PubMed] [Google Scholar]
- 40.Yang H, Antoine DJ, Andersson U, Tracey KJ. The many faces of HMGB1: Molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol. 2013;93:865–873. doi: 10.1189/jlb.1212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer's disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 42.Dahlmann M, Okhrimenko A, Marcinkowski P, Osterland M, Herrmann P, Smith J, Heizmann CW, Schlag PM, Stein U. Rage mediates S100A4-induced cell motility via MAPK/ERK and hypoxia signaling and is a prognostic biomarker for human colorectal cancer metastasis. Oncotarget. 2014;5:3220–3233. doi: 10.18632/oncotarget.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Y, Xia P, Zheng JJ, Sun XB, Pan XD, Zhang X, Wu CZ. Receptors for advanced glycation end products (rage) is associated with microvessel density and is a prognostic biomarker for clear cell renal cell carcinoma. Biomed Pharmacother. 2015;73:147–153. doi: 10.1016/j.biopha.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Nasser NW, Wani NA, Ahirwar DK, Powell CA, Ravi J, Elbaz M, Zhao H, Padilla L, Zhang X, Shilo K, et al. Rage mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor microenvironment. Cancer Res. 2015;75:974–985. doi: 10.1158/0008-5472.CAN-14-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, Li T, Ye G, Shen Z, Hu Y, Mou T, Yu J, Li S, Liu H, Li G. Overexpression of the receptor for advanced glycation endproducts (rage) is associated with poor prognosis in gastric cancer. PLoS One. 2015;10:e0122697. doi: 10.1371/journal.pone.0122697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu B, Cheng SY. Angiopoietin-2: Development of inhibitors for cancer therapy. Curr Oncol Rep. 2009;11:111–116. doi: 10.1007/s11912-009-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bupathi M, Kaseb A, Janku F. Angiopoietin 2 as a therapeutic target in hepatocellular carcinoma treatment: Current perspectives. Onco Targets Ther. 2014;7:1927–1932. doi: 10.2147/OTT.S46457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrissey C, Dowell A, Koreckij TD, Nguyen H, Lakely B, Fanslow WC, True LD, Corey E, Vessella RL. Inhibition of angiopoietin-2 in LuCaP 23. 1 prostate cancer tumors decreases tumor growth and viability. Prostate. 2010;70:1799–1808. doi: 10.1002/pros.21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calfee CS, Gallagher D, Abbott J, Thompson BT, Matthay MA, et al. NHLBI ARDS Network: Plasma angiopoietin-2 in clinical acute lung injury: Prognostic and pathogenetic significance. Crit Care Med. 2012;40:1731–1737. doi: 10.1097/CCM.0b013e3182451c87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goede V, Coutelle O, Neuneier J, Reinacher-Schick A, Schnell R, Koslowsky TC, Weihrauch MR, Cremer B, Kashkar H, Odenthal M, et al. Identification of serum angiopoietin-2 as a biomarker for clinical outcome of colorectal cancer patients treated with bevacizumab-containing therapy. Br J Cancer. 2010;103:1407–1414. doi: 10.1038/sj.bjc.6605925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang FC, Lai TS, Chiang CK, Chen YM, Wu MS, Chu TS, Wu KD, Lin SL. Angiopoietin-2 is associated with albuminuria and microinflammation in chronic kidney disease. PLoS One. 2013;8:e54668. doi: 10.1371/journal.pone.0054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conroy AL, Glover SJ, Hawkes M, Erdman LK, Seydel KB, Taylor TE, Molyneux ME, Kain KC. Angiopoietin-2 levels are associated with retinopathy and predict mortality in Malawian children with cerebral malaria: A retrospective case-control study*. Crit Care Med. 2012;40:952–959. doi: 10.1097/CCM.0b013e3182373157. [DOI] [PMC free article] [PubMed] [Google Scholar]