Abstract
Human cytomegalovirus (HCMV) persists as a subclinical, lifelong infection in the normal human host, but reactivation from latency in immunocompromised subjects results in serious disease. Latency and reactivation are defining characteristics of the herpesviruses and are key to understanding their biology; however, the precise cellular sites in which HCMV is carried and the mechanisms regulating its latency and reactivation during natural infection remain poorly understood. Here we present evidence, based entirely on direct analysis of material isolated from healthy virus carriers, to show that myeloid dendritic cell (DC) progenitors are sites of HCMV latency and that their ex vivo differentiation to a mature DC phenotype is linked with reactivation of infectious virus resulting from differentiation-dependent chromatin remodeling of the viral major immediate-early promoter. Thus, myeloid DC progenitors are a site of HCMV latency during natural persistence, and there is a critical linkage between their differentiation to DC and transcriptional reactivation of latent virus, which is likely to play an important role in the pathogenesis of HCMV infection.
Primary infection of healthy individuals with human cytomegalovirus (HCMV) is often asymptomatic and results in lifelong persistence in the host, a characteristic of all herpes viruses. However, primary infection and reactivation of latent HCMV causes serious disease in immunosuppressed transplant recipients and in advanced HIV infection (1–3). Transfusion-mediated HCMV disease can be prevented by leukocyte depletion (4) of blood, but infectious virus cannot be detected in the blood of healthy carriers, suggesting that HCMV is transmitted as latent virus in the peripheral blood leukocyte population. Accumulating evidence has shown that HCMV is carried latently in mononuclear cells of the myeloid lineage during lifelong latency in naturally infected individuals (5–8). Differentiation of monocytes to macrophages in vitro is reported to induce immediate-early (IE) lytic gene expression from latent virus (9), and groups have intermittently reported that infectious virus can be recovered after differentiation of monocytes to macrophages through explant culture (10) and, more recently, by allogeneic T cell stimulation (11), suggesting that reactivation of latent virus is associated with both the differentiation and activation state of myeloid cells.
The major IE genes of HCMV, driven by the viral major IE promoter/enhancer (MIEP), are the two most abundantly transcribed genes at IE times of virus lytic infection (12), and their proteins play a critical role in control of viral early and late gene expression (for review, see ref. 12). Consistent with the differentiation-dependent induction of IE gene expression observed above, there is a clear correlation between the differentiation state of the cell and the regulation of HCMV IE gene expression in vitro. Thus, in transfection assays, the MIEP is transcriptionally repressed in undifferentiated but transcriptionally active in differentiated, monocytic cell lines (13–15). Similarly, in model systems experimentally infected in vitro HCMV IE gene expression only occurs in differentiated myeloid cells (13, 16), and this observation is correlated with a closed chromatin conformation around the MIEP after infection of nonpermissive monocytic cells but an open chromatin conformation after infection of macrophages (17).
Thus, although experimental infections in vitro indicate that HCMV may be carried in cells of the myeloid lineage and that viral gene expression depends on their state of differentiation, the myeloid lineage represents a heterogeneous population of cells and the precise site of HCMV latency and mechanism of its reactivation in vivo remain undefined. We wished to determine specifically whether myeloid lineage dendritic cells (DC) are a significant site for latency of HCMV during natural persistent infection and, if so, how latency and reactivation in this cell type might be controlled.
DC are specialized antigen-presenting cells which, in common with several other cell types, such as endothelial cells, can be experimentally infected with HCMV in vitro (18, 19). There are at least two types of myeloid lineage-derived DC: the Langerhans DC that are located in the epithelium (and could, therefore, be one of the first cell types to encounter HCMV after infection) and the interstitial DC that reside in deeper-lying tissues (reviewed in ref. 20). Consequently, we took advantage of recent methods for generating defined DC populations ex vivo. Highly purified populations of Langerhans-like DC derived from purified CD34+ cells (21) or interstitial DC derived from monocytes (22) were isolated from HCMV seropositive and seronegative individuals and analyzed for the presence of viral genome. It must be emphasized that all of the data we show refer to the specific analysis of naturally acquired HCMV in cells isolated from healthy, long-term HCMV seropositive individuals. Our study thus represents an ex vivo analysis of factors that regulate the latency and reactivation of naturally acquired HCMV in healthy virus carriers.
Materials and Methods
Purification of CD34+ Hematopoietic Stem Cell Leukapheresis Products. Normal hematopoietic stem cell transplant donors to whom granulocyte colony-stimulating factor had been administered for stem cell mobilization, with >1% of CD34+ cells as a constituent of their peripheral blood mononuclear cells, were asked to donate part of a standard (60 ml) leukapheresis. Informed consent was obtained from all patients, and the protocol was approved by the Cambridge Local Research Ethics Committee. CD34+ stem cells were purified from the leukapheresis material by using a midiMACS positive selection system (CD34+ progenitor isolation kit). Cells were isolated as described by the manufacturer (Miltenyi Biotec, Auburn, CA). The purity of CD34+ cells after selection varied from 80% to 95%. Purified CD34+ cells were frozen at concentrations of 5 × 106 per aliquot.
Purification of Monocytes from Peripheral Blood. The mononuclear cells from 50 ml of blood, donated by healthy volunteers, were isolated on a lymphoprep density gradient. Further isolation of the monocyte fraction was then performed by using a midi-MACS negative selection system (Monocyte isolation kit) as described by the manufacturer (Miltenyi Biotec).
Culture of DC. CD34+ cells were seeded at 2 × 105 per ml in X-vivo 15 (BioWhittaker) supplemented with type β TGF (500 ng/ml), TNF-α (2.5 ng/ml), stem cell factor (20 ng/ml), Flt-3L (100 ng/ml), granulocyte/macrophage colony-stimulating factor (GM-CSF) (100 ng/ml), and 2 mM l-glutamine and cultured for 7 days as described in ref. 21. Alternatively, monocytes were seeded at 5 × 105 per ml in X-vivo 15 supplemented with 10% Human AB Serum (Sigma-Aldrich), 2 mM l-glutamine, IL-4 (1,000 units/ml; PeproTech) and GM-CSF (1,000 units/ml; PeproTech, Rocky Hill, NJ) for 6 days (22). Maturation of immature DCs was promoted by addition of LPS for 48 h (50 ng/ml; Sigma-Aldrich).
Amplification of HCMV DNA by IE-PCR. A 310-bp fragment from the IE region of the HCMV genome (between nucleotides 172468 and 172778) was amplified from cellular DNA purified from seropositive donors' cells by PCR with sense primer (5′-CGT CCT TGA CAC GAT GGA GT-3′) and antisense primer (5′-ATT CTT CGG CCA ACT CTG GA-3′). PCR products were transferred to nitrocellulose and hybridized to an IE-specific [32P]radiolabeled probe (Amersham Pharmacia). A 201-bp probe fragment was generated from the IE region of the HCMV genome (between nucleotides 172535 and 172736) by using sense primer (5′-CCC TGA TAA TCC TGA CGA GG-3′) and antisense primer (5′-CAT AGT CTG CAG GAA CGT CGT-3′). All amplifications by PCR were performed by using Amplitaq Gold (Applied Biosystems) and the addition of 3× MasterAmp PCR Enhancer (Cambio, Cambridge, U.K.). The cycle parameters for all PCR amplifications were a 95°C phase for 5 min to activate the Amplitaq Gold DNA polymerase, followed by 65 cycles of 94°C (40 s), 55°C (40 s), and 72°C (90 s).
Amplification of IE-RNA by RT-PCR. The primers used to amplify viral DNA were used to amplify cellular RNA isolated from seropositive donors' cells by RT-PCR with the same conditions outlined for the IE-PCR for detection of HCMV DNA. The primers spanned the intron between exon 2 and exon 3 and, thus gave, rise to a 196-bp product by RT-PCR. Reverse transcription of mRNA was achieved by using an amphotropic murine leukemia virus reverse transcriptase kit (Roche Diagnostics) as described by the manufacturer.
Amplification of Cellular DNA and RNA by PCR. A 302-bp product was amplified from the cellular β-globin gene by using sense primer (5′-TGT CCA CTC CTG ATG CTG TT-3′) and antisense primer (5′-GGA TTC TAA ACT GTA CCC TG-3′) in a 25-cycle PCR.
A 370-bp product was amplified from cellular histidyl tRNA synthetase mRNA by RT-PCR with a sense primer (5′-TCA TCA GGA CCC AGC TGT GC-3′) and an antisense primer (5′-CTT CAG GGA GAG CGC GTG CG-3′) in a 35-cycle PCR.
Chromatin Immunoprecipitation (ChIP) Assay. ChIPs were carried out essentially as described in refs. 17 and 23.
CD34+ cells, CD34+ cell-derived DCs, monocytes, and monocyte-derived DCs were fixed with 1% formaldehyde and then lysed. DNA associated with histones was immunoprecipitated with control serum (Sigma-Aldrich), antiacetyl histone H4 antiserum (ChIP grade, 1:200 dilution; Upstate Biotechnology, Charlottesville, VA) or anti-heterochromatin protein 1 (HP1) antiserum (1:200 dilution; Serotec) as described in ref. 17.
For detection of the MIEP of HCMV, DNA from disrupted nucleosomes was precipitated and amplified by PCR with sense primer (5′-TGG GAC TTT CCT ACT TGG-3′) and antisense primer (5′-CCA GGC GAT CTG ACG GTT-3′) complementary to positions –272 and +13 relative to the MIEP start site. PCR products were transferred to nitrocellulose and hybridized to an MIEP-specific [32P]radiolabeled probe (Amersham Pharmacia). The probe fragment was generated by PCR of HCMV DNA by using sense primer (5′-ATT ACC ATG GTG ATG CGG TT-3′) and antisense primer (5′-GGC GGA GTT GTT ACG ACA T-3′). All amplifications by PCR were performed by using Amplitaq Gold and the addition of 2× MasterAmp PCR Enhancer. The cycle parameters for amplification by PCR were 95°C for 5 min and then 50 cycles at 94°C (40 s), 50°C (40 s), and 72°C (90 s).
Amplification of the HS4 gene by PCR was performed used sense primer (5′-TGG CAT CTA GCG CAA TGA CTT-3′) and antisense primer (5′-GGG CAA GCC ATC TCA TAG CTG-3′), which have been used in previous analyses of this region (24).
Western Blot Analysis of Protein Expression. Protein samples from CD34+ cells, CD34+-derived DCs, monocytes, and monocyte-derived DCs were separated by SDS/PAGE electrophoresis on 10% polyacrylamide gels. After transfer to nitrocellulose filters, blots were incubated with anti-YY1 (Yin Yang 1) antibody (clone H414 used at 40 ng/ml; Santa Cruz Biotechnology), anti-ERF (Ets-2 repressor factor) antibody (Santa Cruz Biotechnology; used at 40 ng/ml) or anti-HDAC (histone deacetylase) 1 polyclonal antiserum (25). Primary antibodies were detected with an appropriate horseradish peroxidase-conjugated antibody. Protein bands were detected by using enhanced chemiluminescence (Amersham Pharmacia) according to the manufacturer's instructions.
Coculture of CD34+ Cell-Derived DCs with Human Fibroblasts (HFs). Mature CD34+ cell-derived DCs (5 × 106) were cultured on a confluent monolayer of HF. Samples of supernatants were taken and used to inoculate fresh HF to test for infectious virus. Twenty-four hours after infection, the HF were permeabilized (70% ethanol at –20°C for 10 min) and stained for IE protein expression in the nuclei by using a mouse anti-IE72/IE86 antibody (clone E13, Argene, Varilhes, France). Detection was performed by using an FITC-conjugated anti-mouse IgG antibody (Sigma).
Results
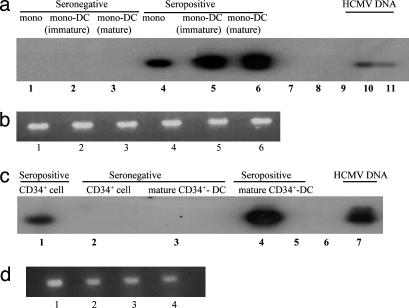
Monocytes isolated from seropositive and seronegative donors were differentiated to monocyte-derived DC, and then the DNA isolated from equivalent numbers of monocyte-derived DC and their monocyte precursors was subjected to PCR for HCMV DNA. As expected (see ref. 7), a 308-bp HCMV IE-specific PCR product was consistently amplified from DNA isolated from the monocytes of seropositive subjects (Fig. 1a, lane 4, and Figs. 6a and 7b, which are published as supporting information on the PNAS web site). However, the same 308-bp PCR product was also detected in DNA from highly purified immature and mature monocyte-derived DC of seropositive individuals [Figs. 1a (lanes 5 and 6), 6a, and 7b] establishing that monocyte-derived DC also carry naturally acquired HCMV. No viral DNA was detected in the monocytes, immature monocyte-derived DC or mature monocyte-derived DC of seronegative individuals [Figs. 1a (lanes 1–3) and 6a].
Fig. 1.
Naturally acquired HCMV genome is carried throughout myeloid differentiation to DC. (a) DNA from monocytes (lane 1), immature monocyte-derived DC (lane 2), and mature monocyte-derived DC (lane 3) from a seronegative donor and monocytes (lane 4), immature monocyte-derived DC (lane 5), and mature monocyte-derived DC (lane 6) from a seropositive donor were amplified in an IE-specific PCR. Negative water controls (lanes 7–9) and positive HCMV DNA controls (lanes 10 and 11) are shown. (b) DNA from monocytes (lane 1), immature monocyte-derived DC (lane 2), and mature monocyte-derived DC (lane 3) from a seronegative donor shown in a and monocytes (lane 4), immature monocyte-derived DC (lane 5), and mature monocyte-derived DC (lane 6) from a seropositive donor shown in a were amplified in a β-globin PCR. (c) DNA from CD34+ cells (lane 2) and mature CD34+ cell-derived DC (lane 3) from a seronegative donor and CD34+ cells (lane 1) and mature CD34+ cell-derived DC (lane 4) from a seropositive donor were amplified in an IE-specific PCR. Negative water controls (lanes 5 and 6) and a positive HCMV DNA control are shown (lane 7). (d) DNA from CD34+ cells (lane 1) and mature CD34+ cell-derived DC (lane 2) from a seronegative donor shown in c and CD34+ cells (lane 3) and mature CD34+ cell-derived DC (lane 4) from a seropositive donor shown in c were amplified by β-globin PCR.
We performed the same analysis on CD34+ myeloid progenitor-derived DC and their precursors. The pure populations of mature CD34+ cell-derived DC were generated from mobilized CD34+ cells isolated from the blood of stem cell donors, and although we have previously shown that CD34+ cells isolated directly from the bone marrow of normal donors carry naturally acquired HCMV DNA (8), it was necessary to confirm that these CD34+ cells mobilized with granulocyte colony-stimulating factor also carried HCMV DNA. Again, viral genome was specifically detected in 1 μg of DNA isolated from 105 mobilized CD34+ cells of seropositive donors [Figs. 1c (lane 1), 6 b and c, and 7b] but not in equivalent CD34+ cells taken from seronegative donors [Figs. 1c (lane 2), and 6 b and c]. When the CD34+ cells were differentiated to a DC phenotype, viral genomes were also detected in mature CD34+ cell-derived DC from seropositive donors [Figs. 1c (lane 4), 6 b and c, and 7b] but not CD34+ cell-derived DC from seronegative donors cultured concurrently [Figs. 1c (lane 3) and 6 b and c].
Interestingly, a comparison of the DNA PCR results from monocytes and mature monocyte-derived DC and from CD34+ cells and mature CD34+ cell-derived DC regularly showed that the differentiated cells contained more HCMV DNA than their undifferentiated counterparts in multiple donors (eight seropositive individuals were analyzed in total). Because genomic DNA from equivalent cell numbers was always assayed (Fig. 1 b and d), this result implies that differentiation results in an induction of viral DNA replication. Further experiments were performed in which DNA samples isolated from healthy, long-term HCMV seropositive individuals were amplified by IE-PCR (Fig. 7b) concurrently with a titration of viral genomes of a known copy number (Fig. 7a). After Southern blot hybridization, a direct comparison with the titration performed concurrently (Fig. 7a) showed ≈100 copies of viral genome were detected in 105 CD34+ cells which, upon differentiation, increased in copy number by at least 10-fold to ≈1,000 copies of viral genome in mature CD34+-derived DC (Fig. 7b). However, to more accurately define the genome copy number per cell, it would be necessary to perform either single-cell PCR or limiting dilution analyses. Nevertheless, such increases in the amount of viral genome, as monocytes or CD34+ cells differentiate to mature DC cells, could represent the activation of viral DNA replication and, if true, this replication would require the induction of a viral IE gene expression that is critically important for the subsequent expression of the viral early and late gene products, which include the viral DNA replication machinery (12).
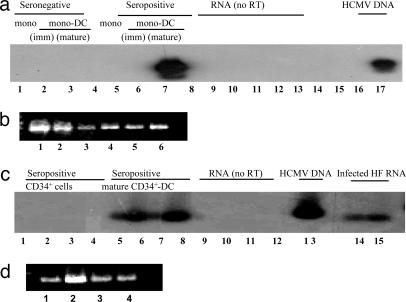
To determine whether DC differentiation does indeed induce lytic gene expression, RT-PCR was performed on 10 μg of total RNA isolated from monocytes, monocyte-derived DC, CD34+ cells and CD34+ cell-derived DC (Fig. 2). No IE gene expression could be detected in monocytes or immature monocyte-derived DC [Fig. 2a (lanes 5 and 6) and Fig. 8 (lane 3), which is published as supporting information on the PNAS web site] even though they both clearly carried naturally acquired viral DNA [Figs. 1a (lanes 4 and 5) and 6a (lane 1)]. However, differentiation of these cells to mature monocyte-derived DC [Figs. 2a (lane 7) and 8 (lane 4)] resulted in induction of IE gene expression from this naturally acquired latent virus. Similarly, the same analysis performed on CD34+ cells and mature CD34+ cell-derived DC also showed that differentiation resulted in induction of IE gene expression from naturally acquired virus. Specifically, no IE gene expression was detected in CD34+ cells from duplicate samples of two seropositive donors (Fig. 2c, lanes 1–4) even though they clearly carried viral genome [Figs. 1c (lane 1) and 6b (lane 5)]. However, differentiation of these cells to mature CD34+ cell-derived DC resulted in an induction of viral IE gene expression (Fig. 2c, lanes 5–8). We did not detect IE gene expression in immature monocyte-derived DC even though they did appear to increase DNA copy number (Fig. 1a, lane 5). We presume that this finding is due to the sensitivity of the IE-RT-PCR and that the additional time culturing immature DC to mature DC leads to increases in steady-state levels of IE RNA that is then detectable by PCR. We confirmed that the lack of IE gene expression in CD34+ cells and monocytes was not due to a nonspecific inability to amplify mRNA from CD34+ cells or monocytes by a histidyl tRNA synthetase RT-PCR in which an amplified product was observed from all samples (Fig. 2 b and d). Thus, terminal differentiation of monocytes or CD34+ cells to their mature DC phenotype, in all four seropositive individuals analyzed, resulted in an induction of HCMV lytic IE gene expression and an apparent increase in viral DNA.
Fig. 2.
Differentiation to mature DC induces reactivation of IE gene expression from naturally acquired latent HCMV. (a) RNA from monocytes (lane 1), immature DC (lane 2), and mature monocyte-derived DC (lane 3) from a seronegative donor and monocytes (lane 5), immature DC (lane 6), and mature monocyte-derived DC (lane 7) from a seropositive donor were amplified in an IE-specific RT-PCR. Negative water controls (lanes 4 and 14–16) and a positive HCMV DNA control (lane 17) are shown. RNA samples 1–3 and 5–7 but with no prior RT are shown (in lanes 8–10 and 11–13, respectively) (b) RNA from monocytes (lane 1), immature monocyte-derived DC (lane 2), and mature monocyte-derived DC (lane 3) from the seronegative donor shown in A and monocytes (lane 4), immature monocyte-derived DC (lane 5), and mature monocyte-derived DC (lane 6) from the seropositive donor also shown in A were amplified in a histidyl tRNA synthetase RT-PCR. (c) Duplicate RNA samples from CD34+ cells (lanes 1–4) and mature CD34+ cell-derived DC (lanes 5–8) from two seropositive donors were amplified in an IE-specific RT-PCR. RNA samples 1, 3, 5, and 7 but with no prior RT were amplified by an IE-specific RT-PCR (lanes 9–12, respectively). HCMV DNA (lane 13) and RNA from infected HF (lanes 14 and 15) are shown. (d) RNA from the CD34+ cells (lanes 1 and 2) and mature CD34+ cell-derived DC (lane 3 and 4) from the two seropositive donors shown in c were amplified in a histidyl tRNA synthetase RT-PCR.
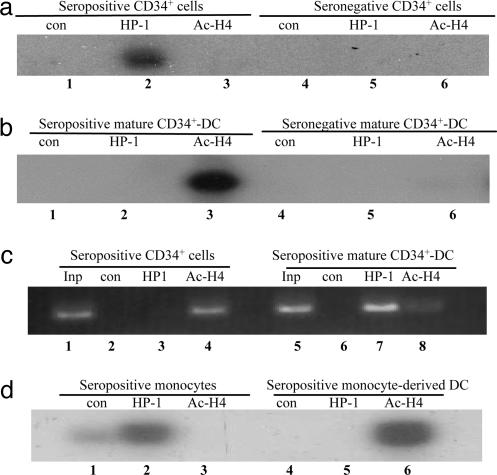
Our previous observations showing that after experimental infection of monocytes and monocyte-derived macrophages in vitro the viral MIEP becomes chromatinized in vitro (17), led us to ask whether chromatin remodeling of the viral MIEP plays a role in authentic reactivation of HCMV from latency in vivo. Consequently, ChIP assays were used to analyze the acetylation state of histones associated with the naturally acquired viral MIEP in undifferentiated and differentiated CD34+ cells. To do this experiment, chromatin-bound DNA from 5 × 106 mobilized CD34+ cells or 5 × 106 mature CD34+ cell-derived DC, cultured under conditions that have been observed to promote reactivation of HCMV IE gene expression, was immunoprecipitated with a control antibody or antibodies specific for acetylated histone H4 (indicative of open, transcriptionally active chromatin; refs. 26 and 27) or HP1 (indicative of closed, transcriptionally repressed chromatin; refs. 27 and 28) as described in ref. 17. A PCR specific for the viral MIEP was then carried out to determine whether the MIEP was associated with acetylated histones or HP1 protein. Fig. 3 shows that the MIEP of latent naturally acquired HCMV in CD34+ cells is clearly associated with HP1 (Fig. 3a, lane 2) and not acetylated histones (Fig. 3a, lane 3), which is consistent with the lack of IE gene expression seen in these cells (Fig. 2c, lanes 1 and 2). Differentiation of the CD34+ cells to mature DC results in a loss of HP1 association with the viral MIEP (Fig. 3b, lane 2), where it is replaced with an association with acetylated histones (Fig. 3b, lane 3), which is consistent with the detection of IE gene expression in these cells observed previously (Fig. 2c, lanes 5 and 6). We included a control PCR on the samples targeting the HS4 region of the cellular β-globin locus control region to preclude the possibility that any negative immunoprecipitation was due to a nonspecific inability to immunoprecipitate chromatin. The HS4 region of the cellular β-globin promoter is acetylated in CD34+ cells and becomes deacetylated/methylated upon differentiation (29). Consistent with this analysis, we observed that the HS4 region was associated with acetylated histones in CD34+ cells (Fig. 3c, lane 4), but in mature CD34+ cell-derived DC, it became associated with HP1 (Fig. 3c, lane 7) although some histones remained acetylated (Fig. 3c, lane 8). Thus, we can be confident that the changes in chromatinization of the MIEP were a result of differentiation and not due to the failure of the antibodies to bind chromatin in some samples.
Fig. 3.
Chromatin remodeling of the latent viral MIEP occurs upon differentiation of monocytes or CD34+ cells to mature DC with reactivation of IE gene expression. DNA associated with histones was immunoprecipitated and used in PCR assays with primers complementary to the MIEP (a. b, and d) or HS4 region of the β-globin gene (c). (a) Seropositive CD34+ cells (lanes 1–3) and seronegative CD34+ cells (lanes 4–6) were incubated with control serum (lanes 1 and 4), anti-HP1 (lanes 2 and 5) or antiacetylated histone H4 (lanes 3 and 6) antibodies. (b) Seropositive mature CD34+ cell-derived DC (lanes 1–3) and seronegative mature CD34+ cell-derived DC (lanes 4–6) were incubated with control serum (lanes 1 and 4), anti-HP1 (lanes 2 and 5) or antiacetylated histone H4 (lanes 3 and 6) antibodies. (c) Seropositive CD34+ cells (lanes 2–4) and seropositive mature CD34+ cell-derived DC (lanes 5–8) were incubated with control serum (lanes 2 and 6), anti-HP1 (lanes 3 and 7) or antiacetylated histone H4 (lanes 4 and 8) antibodies. Ten percent of input controls with no ChIP are shown (lanes 1 and 5) (d) Seropositive monocytes (lanes 1–3) and seropositive mature monocyte-derived DC (lanes 4–6) were incubated with control serum (lanes 1 and 4), anti-HP1 (lanes 2 and 5), or antiacetylated histone H4 (lanes 3 and 6) antibodies.
We next asked whether differentiation of monocytes to monocyte-derived DC with concomitant reactivation of viral IE expression (see Fig. 2), was also associated with chromatin remodeling of the naturally acquired viral MIEP as observed in CD34+ cells. ChIP assays of monocytes or mature monocyte-derived DC showed that the viral MIEP is not associated with acetylated histones (Fig. 3d, lane 3) but with HP1 (Fig. 3d, lane 2) in undifferentiated monocytes. However, in differentiated mature monocyte-derived DC, the histones associated with the MIEP of naturally acquired virus are predominantly acetylated (Fig. 3d, lane 6) and the MIEP is no longer associated with the HP1 protein (Fig. 3d, lane 5). These changes in the architecture of the chromatin bound to the naturally acquired viral MIEP associated with differentiation of CD34+ cells or monocytes to mature DC are entirely consistent with reactivation of virus from latency occurring through chromatin-mediated transcriptional activation of the viral MIEP.
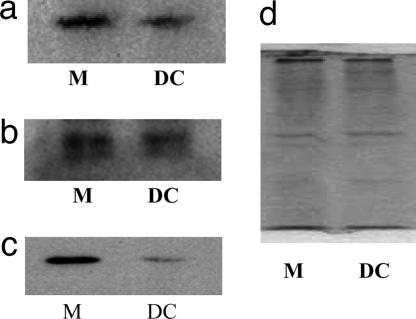
The differentiation-dependent remodeling of the viral MIEP in monocytes or CD34+ cells and their mature DC derivatives clearly raises the question of what factors regulate such chromatin remodeling of this viral promoter. Indeed, a number of defined cellular transcription factors that can bind to specific sequences in the MIEP and also repress MIEP activity in in vitro assays have been identified (30–34) and, intriguingly, two such factors identified, YY1 (30) and ERF (32), may require HDACs as corepressors to exert their function (35, 47). Because differentiation-dependent changes in the levels of these factors appear to be important during the switch in phenotype from nonpermissiveness to permissiveness for HCMV infection in model cell lines (17), we analyzed the expression of these proteins during DC differentiation. Western blot analysis (Fig. 4) shows that the significant changes in YY1 (Fig. 4a) and ERF (Fig. 4b) expression did not occur upon differentiation of monocytes (lane 1) to monocyte-derived DC (lane 2). In contrast, a significant decrease in the levels of HDAC1 (Fig. 4c) upon differentiation of monocytes (lane 1) to mature monocyte-derived DC (lane 2) was observed. This finding is consistent with a previous analysis of embryonal carcinoma cells (17), which are normally nonpermissive for experimental HCMV infection because of a block in viral IE promoter activity but which can be differentiated to a permissive phenotype with retinoic acid (36) and entirely consistent with increased acetylation of histones bound to the MIEP after differentiation (Fig. 3b).
Fig. 4.
HDAC1 expression decreases upon differentiation of monocytes to mature monocyte-derived DC. A total of 105 monocytes (M) or mature monocyte-derived DC (DC) were analyzed by Western blot analysis for expression of the cellular transcription factors YY1 (a), ERF (b), and HDAC1 (c), which are known to be associated with the regulation of the viral MIEP. HDAC1 levels decreased upon differentiation but there was no significant change in expression of YY1 or ERF. A coomassie stain (d) shows that equivalent levels of protein from monocytes (M) and mature DC were loaded.
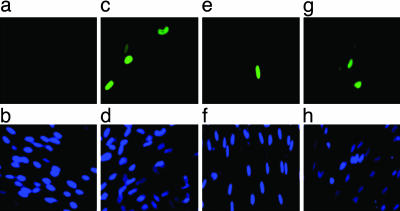
Finally, having observed that differentiation of CD34+ cells to mature DC results in the remodeling of the viral MIEP and induction of lytic gene expression from naturally acquired HCMV, we asked whether the differentiation of CD34+ cells to mature CD34+ cell-derived DC resulted in the reactivation and release of infectious virus. These analyses were performed specifically on the same mature CD34+ cell-derived DC in which IE gene expression was detected (Fig. 2c). Cell-free supernatants taken from mature CD34+ cell-derived DC cocultured with primary HF for 12–15 days were assayed for infectious virus by addition to fresh fibroblasts. Twenty-four hours later, the fibroblasts were stained for the presence of viral major IE antigens. The analysis of these mature CD34+ cell-derived DC samples from three HCMV seropositive and one HCMV seronegative donors is shown (Fig. 5). Infectious virus was clearly detectable in the supernatants of mature CD34+ cell-derived DC from all seropositive individuals (Fig. 5 c, e, and g). Two of these individuals were the same donors in which the induction of lytic gene expression had been observed (Fig. 2c) and one in which chromatin remodeling was observed (Fig. 3 a and b). In contrast, no infectious virus was detected in supernatants from the mature CD34+ cell-derived DC supernatant from the seronegative subject (Fig. 5a).
Fig. 5.
Reactivation of naturally acquired HCMV from mature CD34+ cell-derived DC. Mature CD34+ cell-derived DC were cocultured with primary HFs for 13 days, and then the supernatant was transferred to new HF monolayers that were subsequently stained for IE gene expression. Supernatants collected from seronegative mature CD34+ cell-derived DC cocultures (a) and three seropositive mature CD34+-derived DC cocultures (c, e, and g) were tested. The nuclei of infected HF were counterstained with Hoechst 33342 (b, d, f, and h).
In total, we have analyzed four mature CD34+ cell-derived DC samples from seropositive donors for such reactivation of infectious virus. In three of four cases, virus reactivation was routinely observed in triplicate fibroblast cocultures. Interestingly, in the single case where HCMV reactivation was not observed, no IE gene expression could be detected in the mature CD34+-derived DC by RT-PCR (data not shown). One possibility is that the “latent viral HCMV load,” which is the frequency of CD34+ cells carrying HCMV genomes, may be an important determinant of whether viral reactivation can be observed in vitro. For instance, the seropositive individual from which reactivation could not be detected had markedly lower levels of viral DNA in the mature CD34+-derived DC (Fig. 6c) in comparison with the cells of donors observed to reactivate HCMV (Figs. 1c and 6b). Interestingly, it has been shown that reactivation of HSV1 from latently infected ganglia may depend on the viral copy number of the latently infected neuron (37).
Discussion
Analyses of the mechanisms that control HCMV latency and reactivation have been limited by the relatively low frequency of cells carrying viral genomes in vivo. Indeed, the fraction of cells carrying HCMV genomes in healthy seropositive subjects has been estimated to be only 0.01% of the total granulocyte colony-stimulating factor mobilized (bone marrow-derived) mononuclear cell population (6). Consequently, studies of HCMV latency have relied heavily on the use of experimental infection of nonpermissive myeloid precursors to generate populations of cells that are infected at high frequencies and then used as models of latency in vitro (38–41). After such experimental infection of CD34+ stem cells in vitro, latent HCMV can be detected in a subpopulation of CD33+ precursor cells that are believed to represent progenitors of myeloid DC (40). The monocyte-derived macrophages from which HCMV was reported to be reactivated, derived from T cell-stimulated monocytes (11), also expressed some markers of myeloid DC populations (11). However, the precise identity of the cells carrying latent HCMV in vivo has, thus far, not been defined. The ability to generate pure populations of DC ex vivo allowed us to analyze the role of this specific cell population in HCMV latency, in detail.
Our results clearly show that HCMV DNA remains latent in myeloid DC progenitors of naturally infected carriers, and carriage of HCMV is maintained upon specific differentiation of these cells to a mature DC phenotype. Further, upon such differentiation and maturation, the induction of viral lytic gene expression from latent virus and an increase in viral genome copy number was observed. Our results show that this induction of lytic gene expression and apparent replication of viral DNA was concomitant with the reactivation of infectious virus from the DC of some healthy seropositive individuals, consistent with DC being a biologically significant site of viral reactivation in vivo.
The induction of IE lytic gene expression from latent viruses represents the critical event required for the switch from latency to reactivation. Analyses in vitro have shown that transcriptional activation of viral IE gene expression from the viral MIEP requires the action of cellular transcription factors (13–15, 30–32) and chromatinization of the MIEP (17). These studies led us to analyze whether chromatin remodeling of the viral MIEP played any role in reactivation of HCMV from latency upon DC differentiation. Our analyses clearly showed that latent HCMV in DC progenitors, in vivo, is in a closed, transcriptionally silent chromatin conformation as indicated by its predominant association with the silencing protein HP1. In contrast, differentiation of these progenitors to mature DC resulted in specific chromatin remodeling of the MIEP whereby its association with HP1 was replaced with an association with acetylated histones, consistent with open, transcriptionally active chromatin and the reactivation of viral lytic IE gene expression. It is highly likely that this remodeling of chromatin around the viral MIEP precedes induction of lytic gene expression and virus reactivation; it seems much less likely that such remodeling could result from viral genomes that have reactivated, and subsequently infected, surrounding DC.
The chromatin remodeling of the MIEP upon ex vivo differentiation of DC progenitors to mature DC was also linked with changes in expression of specific cellular proteins. Although YY1 and ERF, two cellular proteins that have been suggested to repress the viral MIEP in vitro, showed no differential expression, differentiation of DC progenitors to mature DC resulted in the down-regulation of HDAC1 protein. HDAC1 is a known corepressor of a number of transcriptional repressors and has been suggested to be intimately involved with transcriptional repression mediated by both YY1 (35) and ERF (47). Consequently, these observations are entirely consistent with the differentiation-dependent increase in association of the viral MIEP with acetylated histones. Thus, the latency and reactivation of HCMV in vivo appears intimately associated with changes in the host cell transcriptional milieu as myeloid progenitors differentiate to mature DC.
Sporadic episodes of controlled reactivation of HCMV in vivo, consequent on periodic differentiation of myeloid cells to mature DC, might provide a mechanism for the HCMV persistence and intermittent virus excretion seen in healthy seropositive individuals. There is good evidence that in the healthy host, HCMV infection is controlled by the adaptive immune response, particularly by CD8+ cytotoxic T lymphocytes (CTL), and we speculate that two observations concerning this immune control might be related to the persistence of HCMV in DC. First, we and others have reported strikingly high frequencies of HCMV-specific CD8+ CTL in normal virus carriers, apparently higher than in most other persistent virus infections (42, 43). Reactivation of HCMV within DC might be predicted to be a particularly efficient method of continually restimulating virus-specific CTL, and at least, in part, explain these high frequencies. Second, the large number of mechanisms HCMV has evolved to inhibit CTL surveillance, reflected in the multiple “immune evasion” genes it encodes, might be taken to reflect an especially close relationship between the virus and antigen-presenting cells such as DC and macrophages (44–46).
In conclusion, our results show that naturally acquired HCMV genomes are carried in highly purified populations of DC, and differentiation of myeloid cells to mature DC results in the induction of lytic gene expression from this HCMV. Furthermore, our analyses show that the MIEP of naturally acquired HCMV is subject to chromatinization, and chromatin remodeling is a key determinant of latency and reactivation of naturally acquired HCMV in vivo. These data represent analyses of naturally acquired HCMV genomes during latency and the mechanism of their reactivation in vivo. In showing how the virus selectively utilizes the differentiation pathway of key antigen presenting cells to persist in its human host, they also point toward a better understanding of how it may cause disease.
Supplementary Material
Acknowledgments
We thank Mike Scott for coordinating the collection of leukophoresis samples from granulocyte colony-stimulating factor mobilized patients and providing stem cell factor. This work was supported by the U.K. Medical Research Council and the Wellcome Trust. M.B.R. was funded by an Medical Research Council studentship. P.J.L. is a Wellcome Clinical Senior Fellow.
Author contributions: M.B.R., J.G.P.S., and J.H.S. designed research; M.B.R. performed research; P.A.M. contributed new reagents/analytic tools; M.B.R., P.J.L., J.G.P.S., and J.H.S. analyzed data; and M.B.R., P.J.L., J.G.P.S., and J.H.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; DC, dendritic cells; HCMV, human cytomegalovirus; HDAC, histone deacetylase; HF, human fibroblast; HP1, heterochromatin protein 1; IE, immediate-early; MIEP, major IE promoter.
References
- 1.Adler, S. P. (1983) Rev. Infect. Dis. 5, 977–993. [DOI] [PubMed] [Google Scholar]
- 2.Drew, W. L. (1988) J. Infect. Dis. 158, 449–456. [DOI] [PubMed] [Google Scholar]
- 3.Rubin, R. H. (1990) Rev. Infect. Dis. 12, Suppl. 7, S754–S766. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert, G. L., Hayes, K., Hudson, I. L. & James, J. (1989) Lancet 1, 1228–1231. [DOI] [PubMed] [Google Scholar]
- 5.Kondo, K., Xu, J. & Mocarski, E. S. (1996) Proc. Natl. Acad. Sci. USA 93, 11137–11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slobedman, B. & Mocarski, E. S. (1999) J. Virol. 73, 4806–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor-Wiedeman, J., Sissons, J. G., Borysiewicz, L. K. & Sinclair, J. H. (1991) J. Gen. Virol. 72, 2059–2064. [DOI] [PubMed] [Google Scholar]
- 8.Mendelson, M., Monard, S., Sissons, P. & Sinclair, J. (1996) J. Gen. Virol. 77, 3099–3102. [DOI] [PubMed] [Google Scholar]
- 9.Taylor-Wiedeman, J., Sissons, P. & Sinclair, J. (1994) J. Virol. 68, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diosi, P., Moldovan, E. & Tomescu, N. (1969) Br. Med. J. 4, 660–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soderberg-Naucler, C., Fish, K. N. & Nelson, J. A. (1997) Cell 91, 119–126. [DOI] [PubMed] [Google Scholar]
- 12.Stenberg, R. M. (1996) Intervirology 39, 343–349. [DOI] [PubMed] [Google Scholar]
- 13.Sinclair, J. H., Baillie, J., Bryant, L. A., Taylor-Wiedeman, J. A. & Sissons, J. G. (1992) J. Gen. Virol. 73, 433–435. [DOI] [PubMed] [Google Scholar]
- 14.Ghazal, P., Lubon, H. & Hennighausen, L. (1988) J. Virol. 62, 1076–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meier, J. L. & Stinski, M. F. (1997) J. Virol. 71, 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lathey, J. L. & Spector, S. A. (1991) J. Virol. 65, 6371–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy, J. C., Fischle, W., Verdin, E. & Sinclair, J. H. (2002) EMBO J. 21, 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riegler, S., Hebart, H., Einsele, H., Brossart, P., Jahn, G. & Sinzger, C. (2000) J. Gen. Virol. 81, 393–399. [DOI] [PubMed] [Google Scholar]
- 19.Hertel, L., Lacaille, V. G., Strobl, H., Mellins, E. D. & Mocarski, E. S. (2003) J. Virol. 77, 7563–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banchereau, J., Briere, F., Caux, C., Davoust, J., Lebecque, S., Liu, Y. J., Pulendran, B. & Palucka, K. (2000) Annu. Rev. Immunol. 18, 767–811. [DOI] [PubMed] [Google Scholar]
- 21.Strobl, H., Bello-Fernandez, C., Riedl, E., Pickl, W. F., Majdic, O., Lyman, S. D. & Knapp, W. (1997) Blood 90, 1425–1434. [PubMed] [Google Scholar]
- 22.Sallusto, F. & Lanzavecchia, A. (1994) J. Exp. Med. 179, 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, R. X., Postigo, A. A. & Dean, D. C. (1998) Cell 92, 463–473. [DOI] [PubMed] [Google Scholar]
- 24.Bottardi, S., Aumont, A., Grosveld, F. & Milot, E. (2003) Blood 102, 3989–3997. [DOI] [PubMed] [Google Scholar]
- 25.Richon, V. M., Emiliani, S., Verdin, E., Webb, Y., Breslow, R., Rifkind, R. A. & Marks, P. A. (1998) Proc. Natl. Acad. Sci. USA 95, 3003–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 27.Lusser, A. (2002) Curr. Opin. Plant Biol. 5, 437–443. [DOI] [PubMed] [Google Scholar]
- 28.Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C. & Kouzarides, T. (2001) Nature 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 29.McMorrow, T., van den Wijngaard, A., Wollenschlaeger, A., van de Corput, M., Monkhorst, K., Trimborn, T., Fraser, P., van Lohuizen, M., Jenuwein, T., Djabali, M., et al. (2000) EMBO J. 19, 4986–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, R., Baillie, J., Sissons, J. G. & Sinclair, J. H. (1994) Nucleic Acids Res. 22, 2453–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zweidler-Mckay, P. A., Grimes, H. L., Flubacher, M. M. & Tsichlis, P. N. (1996) Mol. Cell. Biol. 16, 4024–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bain, M., Mendelson, M. & Sinclair, J. (2003) J. Gen. Virol. 84, 41–49. [DOI] [PubMed] [Google Scholar]
- 33.Huang, L. & Stinski, M. F. (1995) J. Virol. 69, 7612–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, T. H., Oka, T., Asai, T., Okada, T., Merrills, B. W., Gertson, P. N., Whitson, R. H. & Itakura, K. (1996) Nucleic Acids Res. 24, 1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas, M. J. & Seto, E. (1999) Gene 236, 197–208. [DOI] [PubMed] [Google Scholar]
- 36.Gonczol, E., Andrews, P. W. & Plotkin, S. A. (1985) J. Gen. Virol. 66, 509–515. [DOI] [PubMed] [Google Scholar]
- 37.Sawtell, N. M. (1998) J. Virol. 72, 6888–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo, K., Kaneshima, H. & Mocarski, E. S. (1994) Proc. Natl. Acad. Sci. USA 91, 11879–11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Movassagh, M., Gozlan, J., Senechal, B., Baillou, C., Petit, J. C. & Lemoine, F. M. (1996) Blood 88, 1277–1283. [PubMed] [Google Scholar]
- 40.Hahn, G., Jores, R. & Mocarski, E. S. (1998) Proc. Natl. Acad. Sci. USA 95, 3937–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodrum, F. D., Jordan, C. T., High, K. & Shenk, T. (2002) Proc. Natl. Acad. Sci. USA 99, 16255–16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wills, M. R., Carmichael, A. J., Mynard, K., Jin, X., Weekes, M. P., Plachter, B. & Sissons, J. G. (1996) J. Virol. 70, 7569–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillespie, G. M., Wills, M. R., Appay, V., O'Callaghan, C., Murphy, M., Smith, N., Sissons, P., Rowland-Jones, S., Bell, J. I. & Moss, P. A. (2000) J. Virol. 74, 8140–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arrode, G. & Davrinche, C. (2003) Curr. Top. Microbiol. Immunol. 276, 277–294. [DOI] [PubMed] [Google Scholar]
- 45.Johnson, D. C. & Hegde, N. R. (2002) Curr. Top. Microbiol. Immunol. 269, 101–115. [DOI] [PubMed] [Google Scholar]
- 46.Lehner, P. J., Karttunen, J. T., Wilkinson, G. W. G. & Cresswell, P. (1997) Proc. Natl. Acad. Sci. USA 94, 6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright, E., Bain, M., Teague, L., Murphy, J. & Sinclair, J. (2005) J. Gen. Virol. 86, 535–544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.