Abstract
Lactobacillus acidophilus NCFM is a probiotic bacterium that has been produced commercially since 1972. The complete genome is 1,993,564 nt and devoid of plasmids. The average GC content is 34.71% with 1,864 predicted ORFs, of which 72.5% were functionally classified. Nine phage-related integrases were predicted, but no complete prophages were found. However, three unique regions designated as potential autonomous units (PAUs) were identified. These units resemble a unique structure and bear characteristics of both plasmids and phages. Analysis of the three PAUs revealed the presence of two R/M systems and a prophage maintenance system killer protein. A spacers interspersed direct repeat locus containing 32 nearly perfect 29-bp repeats was discovered and may provide a unique molecular signature for this organism. In silico analyses predicted 17 transposase genes and a chromosomal locus for lactacin B, a class II bacteriocin. Several mucus- and fibronectin-binding proteins, implicated in adhesion to human intestinal cells, were also identified. Gene clusters for transport of a diverse group of carbohydrates, including fructooligosaccharides and raffinose, were present and often accompanied by transcriptional regulators of the lacI family. For protein degradation and peptide utilization, the organism encoded 20 putative peptidases, homologs for PrtP and PrtM, and two complete oligopeptide transport systems. Nine two-component regulatory systems were predicted, some associated with determinants implicated in bacteriocin production and acid tolerance. Collectively, these features within the genome sequence of L. acidophilus are likely to contribute to the organisms' gastric survival and promote interactions with the intestinal mucosa and microbiota.
Keywords: adhesion, stress response, proteolytic system, sugar metabolism, insilico analysis
Lactobacilli are important inhabitants of the gastrointestinal (GI) tract and some species are considered to have probiotic properties, offering a number of benefits to health and well being (1). The most well known members of this group are classified as the “acidophilus complex,” composed of six species of closely related lactobacilli that have historically been isolated from the GI tract of humans and animals (2–4). Of these, Lactobacillus acidophilus remains to be the most widely recognized and commercially distributed probiotic culture. The organism was first isolated by Moro in 1900 from infant feces and is characterized as a homofermentative, short Gram-positive rod (2–10 μm) that grows optimally from 37°C to 42°C (34).
The genome sequences of a number of different species isolated from the human GI tract have been published recently and include Bifidobacterium longum (5), Lactobacillus johnsonii (6), Bacteroides fragilis (GenBank accession no. NC_006347), and Lactobacillus plantarum (7). Others are nearing completion (8) and, collectively, these genomes will provide a solid platform for comparative genomic analysis of organisms that survive passage through and inhabit the gastrointestinal tract of humans.
The objective of this study was to determine and analyze the genome sequence of L. acidophilus NCFM, a probiotic culture that has been widely investigated for its physiological, biochemical, genetic, and fermentative properties (9). Consistent with two other closely related species of the “acidophilus” complex, L. johnsonii and Lactobacillus gasseri, the organism lacked biosynthetic capacity for most vitamins and amino acids, but encoded considerable transporter and fermentative capacities, expected for organism's residing within the nutrient rich conditions of upper GI tract. Bioinformatic analysis and comparisons to other probiotic genomes revealed a number common and unique features that are likely to be important to the organisms intestinal residence and roles.
Materials and Methods
Genome Sequencing and Assembly. Genomic DNA from NCFM was used to construct a genomic library with the Stratagene Lambda Fix II/XhoI Partial Fill-in Vector kit and a small insert (1.6–2.2 kb) pUC18 plasmid library. DNA sequencing was performed by using standard primers on an ABI377 automated sequencer. All procedures were performed according to standard procedures described elsewhere. DNA traces were assembled by using the phred/phrap/consed software package (www.phrap.org). The draft genome was subcontracted to Fidelity Systems (Gaithersburg, MD) for direct genome sequencing to fill the remaining 380 gaps. Detailed descriptions of genome sequencing, closure, and assembly can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Bioinformatic Analyzes. The complete genome sequence was subjected to an automated annotation process, performed by gamola (10). The gene model was determined by using glimmer (11). Sequence similarity analyses were performed with the gapped blastp algorithm (12), by using the nonredundant database provided by National Center for Biotechnology Information (ftp://ftp.ncbi.nih.gov/blast/db). A functional classification was applied by using the clusters of orthologous proteins (COG) database (applied threshold 1e-10) (13). Protein motifs were determined by hmmer (http://hmmer.wustl.edu) using pfam hmm libraries, with global and local alignment models (http://pfam.wustl.edu) (applied threshold, 0.001). tRNAs were identified by using trnascan-se with both the relaxed and the stringent parameter sets (14). Nucleotide repeats were identified and visualized by using the kodon software package (Applied Maths, Austin) and reputer (15). Genome atlas visualizations were obtained by using genewiz (16). Necessary sequence analyses were performed by in-house-developed software solutions. Pathway reconstructions employing the ORFeome of L. acidophilus NCFM were performed by using the in-house developed software-suite pathwayvoyager (unpublished data) in conjunction with the KEGG (Kyoto Encyclopedia of Genes and Genomes) on-line database (www.genome.ad.jp/kegg/kegg2.html) (17). Automated computer annotations were manually verified. The predicted gene model was reviewed and start positions were altered based on protein sequence alignments and potential ribosomal binding sites.
The complete DNA sequence and the corresponding annotation of L. acidophilus NCFM is available from GenBank (accession no. CP000033).
Results and Discussion
General Genome Features. The complete genome of L. acidophilus NCFM consisted of 1,993,564 nucleotides with an average GC content of 34.71%. In silico analyses revealed the presence of 1,864 ORFs resulting in a coding percentage of 87.9% (Fig. 1, circle 3). One or more protein families (pfam) were attributed to 75% of these ORFs, and 89% showed similarities to at least one COG. As a result of the manual annotation curation, only 11.7% of the ORFs remained unknown and 15.8% showed similarities to unclassified genes of other organisms. Of the predicted ORFs, 72.5% were assigned to a defined function.
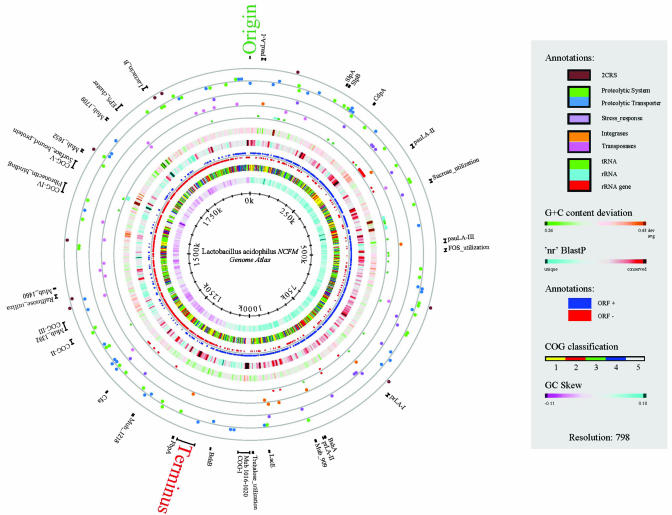
Fig. 1.
Genome atlas of L. acidophilus NCFM. The atlas represents a circular view of the complete genome sequence of L. acidophilus NCFM. The key describes the single circles in the top-down outermost-innermost direction. The circle was created by using GENEWIZ (16) and in-house-developed software. Innermost circle 1 shows GC-skew. Circle 2 shows COG classification. Predicted ORFs were analyzed by using the COG database and grouped into the four major categories: 1, information storage and processing; 2, cellular processes and signaling; 3, metabolism; 4, poorly characterized; and 5, ORFs with uncharacterized COGs or no COG assignment. Circle 3 shows ORF orientation. ORFs in sense orientation (ORF+) are shown in blue; ORFs oriented in antisense direction (ORF–) are shown in red. Circle 4 shows BLAST similarities. Deduced amino acid sequences compared against the nonredundant (nr) database by using gapped BLASTP (12). Regions in blue represent unique proteins in NCFM, whereas highly conserved features are shown in red. The degree of color saturation corresponds to the level of similarity. Circle 5 shows G+C content deviation. Deviations from the average GC-content are shown in either green (low GC spike) or orange (high GC spike). A box filter was applied to visualize contiguous regions of low or high deviations. Circle 6 shows ribosomal machinery. tRNAs, rRNAs, and ribosomal proteins are shown as green, cyan, or red lines, respectively. Clusters are represented as colored boxes to maintain readability. Circle 7 shows mobile elements. Predicted transposases are shown as light purple, and phage-related integrases are shown as orange dots. Circle 8 shows eptide and amino acid utilization. Proteases and peptidases are shown in green, and nonsugar related transporters are shown in light blue dots. Outermost circle 9 shows two-component regulators (2CRS). Each 2CRS is represented as brown dots, consisting of a response regulator and a histidine kinase. In circles 7–9, each full dot represents one predicted ORF, and clusters of ORFs are represented by stacked dots. Selected features representing single ORFs and ORF clusters are shown outside of circle 9 with bars indicating their absolute size. Origin and terminus of DNA replication are identified in green and red, respectively. Other features are: SlpA and -B (S-layer proteins), CdpA (Cell division protein; ref. 50), sugar utilization (sucrose, FOS, trehalose, raffinose), LacE (PTS-sugar transporter), BshA and -B (bile salt hydrolases), Mub-909 to Mub-1709 (mucus-binding proteins, numbers correspond to the La-number scheme), FbpA (fibronectin binding protein), Cfa (cyclopropane fatty acid synthase), Fibronectin_binding (fibronectin-binding protein cluster), EPS_cluster (exopolysaccharides), Lactacin_B (bacteriocin), pauLA-I to pauLA-III (potential autonomous units), and prLA-I and prLA-II (phage remnants).
The origin and terminus of replication was predicted by GC-skew analysis and the ORF orientation shift (Fig. 1, circles 1 and 3). Both are placed fairly symmetrical in the genome. More detailed information can be found in General Genome Features in Supporting Text and Fig. 4, which are published as supporting information on the PNAS web site.
Analysis of the GC-content distribution showed localized peak deviations from the average GC content of the genome (Fig. 1, circle 5). Without exceptions, GC-content spikes were found to harbor the four rRNA loci (average GC content of 50.88%), whereas the two neighboring low GC-regions at 1.75 Mbp (average GC content of 28.5%) revealed a gene cluster predicted to encode production of exo-polysaccharides (EPS, see below and Table 1, which is published as supporting on the PNAS web site) and a large uncharacterized region, unique to L. acidophilus NCFM.
Identified within the genome were 61 tRNAs, representing all 21 amino acids, with redundant tRNAs for all amino acids except cysteine and tryptophan. Only eight tRNAs were located on the lagging strand, mostly clustered around an rRNA locus. Ribosomal proteins were mainly assembled around one locus at 260 kb. Four ribosomal RNA loci were identified throughout the genome. Three of them were clustered within the first 500 kb and oriented in the same sense-direction, whereas the fourth rRNA locus, located at ≈1.6 Mbp, is oriented in the opposite direction. Thus, all rRNA loci were in phase with the direction of DNA replication (Fig. 1, circle 6).
The COG database classifies paralogous proteins of at least three lineages into functionally related groups. The graphical representation of the COG distribution (Fig. 1, circle 2) shows that the majority of predicted proteins (64.4%) could be classified into the three functional classes and only 19% were assigned to the “poorly characterized” group. However, 6.6% of COGs could not be assigned into any classification, designated here as COG category 5 (Fig. 1). Of those, five genome regions stand out (Fig. 1, COG-I, II, III, IV, and V) as all of the genes present were predicted to be involved in cell-adherence and initial host–cell recognition (i.e., La1016–La1020, La1377, La1392: mucus+binding proteins; La1606–La1612: fibronectin-binding proteins; and La1633–La1636: surface-bound proteins). Further analyses of other organisms may suggest the need for an additional COG group for extracellular structures (functional category W).
The NCFM genomic DNA sequence was analyzed for repetitive DNA by a “repeat and match” analysis. One intergenic region of 2.4 kb between La1550 (DNA polymerase I, polA) and La1551 (putative phosphoribosylamine-glycine ligase, purD) featured characteristic of a spacers interspersed direct repeats (SPIDR) locus. Within this region, the SPIDR locus was ≈1.5 kb and contained 32 nearly perfect repeats of 29 bp separated by unique 32-bp spacers (Fig. 5, which is published as supporting information on the PNAS web site). The SPIDR locus constitutes a previously undescribed family of repeat sequences that are present in Bacteria and Archaea but not in Eukarya (18). The repeat loci typically consist of repetitive stretches of nucleotides with a length of 25–37 bp alternated by nonrepetetive DNA spacers of approximately equal size as the repeats (Fig. 2). To date, SPIDR loci have been identified in >40 microorganisms (18), but only within the Streptococcus species of the lactic acid bacteria (LAB). Despite their discovery >15 years ago in Escherichia coli (19), no physiological function has been attributed to these regions.
Fig. 2.
The SPIDR regions and the embedded sequence repeats were visualized by using REPUTER (15). The genome is represented by the green and purple horizontal lines. Repeats are shown by colored boxes on the genome, and the vertical lines indicate similar repeats. Sequence repeat lengths, in bp, are displayed by color coding as indicated by the colored boxes. The sequence 5′-AGGATCACCTCCACTTTCGTGGAGAAAAT-3′ was repeated 32 times in this 1,953-bp region.
Prophages and PAUs. Prophages are a common feature among prokaryotic genomes and several phages have been reported in LAB (20, 21). In silico analysis of the genome of L. acidophilus NCFM did not reveal any complete prophages. However, several isolated prophage remnants (Fig. 1, prLA-I and prLA-II) and single ORFs with similarities to phage-genes were identified (Table 2, which is published as supporting information on the PNAS web site). Detailed information about the identified prophage remnants can be found in Prophages in Supporting Text.
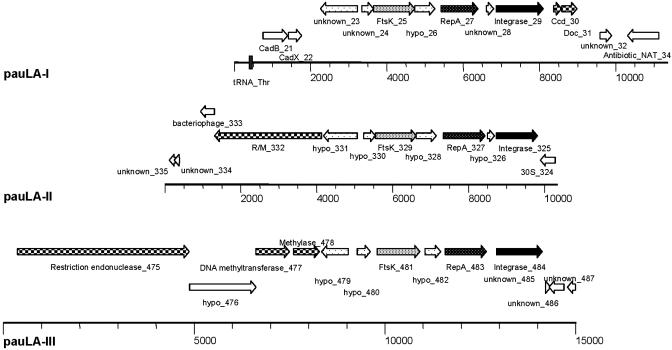
Clustered within the first 500 ORFs of the genome were three PAUs, designated as pauLA-I (La23 to La29), pauLA-II (La331 to La325), and pauLA-III (La479 to La484) (Fig. 1). Each PAU features a core region of seven ORFs. Synteny and ORF sizes are highly conserved, with the exception of pauLA-III, where one small unclassified ORF was absent (Fig. 3). In silico analyses predicted a core consisting of an integrase, IntG, (La29, La325, and La484), a replication protein, RepA, (La27, La327, and La483), and a DNA segregation ATPase, FtsK, involved in DNA partitioning (La25, La329, and La481). However, amino acid alignments between the PAUs revealed significant variations in the degree of similarity between the respective proteins. PauLA-III showed significant sequence differences for all core proteins, whereas pauLA-I and pauLA-II were more closely related. This finding suggests that pauLA-III either evolved in a different organism and was acquired later or was the most ancient integration event into the chromosome, followed by a duplication that formed pauLA-I. The high degree of similarity of RepA, FtsK, and the two hypothetical proteins flanking FtsK between pauLA-I and pauLA-II (88%, 89%, 79%, and 65%, respectively) suggested a more recent duplication of pauLA-I, resulting in pauLA-II. Adjacent to the core, pauLA-II and pauLA-III feature one (La332) and two (La477 and La478) putative DNA methyltransferases, respectively. La475 revealed striking similarities (e-value, 0) to type II restriction endonucleases. Downstream of pauLa-I, ORF La31 showed similarities to COG3654, describing a prophage maintenance system killer protein, Doc, previously reported for E. coli phage P1 (18). Doc acts as a molecular poison, capable of killing the host cell. A second protein, Phd, coexpressed with Doc, counteracts the killer protein. Absence of Phd causes cell death, usually induced upon phage or plasmid curing (22). A second ORF, La30, separated from La31 by only one codon, was also predicted, revealing striking similarities to the systems described for P1 and phage F. Both ORFs are very small (213 and 399 bp, respectively), likely organized in an operon, and both exhibit low sequence similarities to other known proteins. Both are located near the putative origin of replication, and arranged such that the antidote (Phd) is transcribed before the lethal protein (Doc). Translational coupling suggests dominant expression of Phd expression over Doc. Closely related Doc-like proteins were also found in Brevibacterium linens, Bifidobacterium longum, and L. gasseri ATCC33323 (GenBank accession no. ZP_00046979). L. johnsonii NCC533 (6), closely related to L. gasseri, does not share this feature. Although the genetic organization of phd and doc in L. gasseri is very similar to L. acidophilus, they are not linked to any prophage elements or PAUs and are flanked by two putative transcriptional regulators. In L. acidophilus, it cannot be discerned whether or not the core-flanking regions were introduced by the PAUs.
Fig. 3.
PAUs. The genome is represented by a black line, and base-pair intervals are shown in bp. The three PAUs were aligned at the 5′ end of the integrase, which is shown as a black arrow, and are drawn in scale. The potential replication protein RepA is represented by white-dotted arrows, and the DNA segregation ATPase FtsK is represented by densely black-dotted arrows. ORFs with no predicted functions that are assumed to be part of a PAU are shown as sparsely black-dotted arrows. Checker-boarded ORFs indicate proteins potentially involved in unit stabilization. Adjacent ORFs are shown as open arrows. tRNAs are drawn as dark gray boxes on the genome line.
Putative Transposase Genes. Insertion sequences (IS) are defined as small (<2.5 kb) segments of DNA with a simple organization, capable of inserting at multiple sites in a target molecule and generally only encode genetic information required for transposition (23). IS elements have been documented in many species of LAB. Seventeen putative transposase genes, representing seven different families, were identified in NCFM (Fig. 1, circle 7, and Table 3, which is published as supporting information on the PNAS web site). Three of these families appear in multiple copies ranging from two to six replicas and are highly conserved indicating recent integration and multiplication events. Detailed information about classification of the identified transposase genes is in Putative Transposase Genes in Supporting Text. L. acidophilus NCFM harbors a significantly larger diversity of mobile elements than other probiotic LAB that have been sequenced thus far. In L. johnsonii and L. plantarum, 14 and 13 IS elements were identified, respectively. Although this is a similar number when compared to L. acidophilus NCFM, these mobile elements were grouped into only three families in L. johnsonii and two in L. plantarum. In contrast, 13 of the 17 IS elements of L. acidophilus were grouped into three major clusters and four single copy genes were grouped on separate branches of a phylogenetic tree, representing distinct families (data not shown).
Bacteriocins. Bacteriocins are small antimicrobial peptides produced widely by LAB. Their spectrum of activity is typically quite narrow and limited to only closely related species. Lactacin B is a class II bacteriocin produced by L. acidophilus NCFM (24). The genome sequence of L. acidophilus NCFM identified a 9.5-kb region containing 12 putative genes (La1791–La1803) implicated in both production and processing of lactacin B (Fig. 6 and Table 4, which are published as supporting information on the PNAS web site). The genetic organization of this locus, typical of other class II bacteriocins, exhibited sensory and regulatory machinery, a number of small putative structural genes, and an export system (25). Detailed information about the operon structure can be found in Bacteriocins in Supporting Text.
Biosynthetic Capabilities. In silico analyses of the genome of L. acidophilus NCFM indicated the potential to synthesize three amino acids (cysteine, serine, and aspartate) de novo. From these three amino acids, a series of seven other derivatives could be generated. No de novo or conversion pathways could be predicted for the remaining 10 amino acids.
Like other LAB (6, 7), L. acidophilus shows only a partial citrate cycle. Also, NCFM appears to be capable of assimilating ammonia by conversion into l-glutamate via l-glutamine as intermediates (glnA, La1501; asnH, La158), or by synthesizing l-asparagine via l-aspartate (asnA, La1896). l-aspartate may also be produced directly from ammonia, as several genes with some similarities to an aspartate-ammonia lyase were found. The proposed model for nitrogen assimilation is supported by the presence of amtB (La473), an ammonia permease. Assimilation of sulfur in the form of sulfate or H2S appears unlikely.
Similar to other lactobacilli, L. acidophilus appears unable to synthesize most cofactors and vitamins like riboflavine, vitamin B6, nicotinate, nicotinamide, biotin, and folate. Nevertheless, some partial pathways could be reconstructed, allowing conversions of some of these molecules. The complete pathway for de novo synthesis of purines was present, whereas only a partial pathway for pyrimidine synthesis could be reconstructed.
Based on the in silico analyses, L. acidophilus NCFM is likely to be auxotrophic for 14 amino acids, most vitamins and cofactors with the possible exceptions of pantothenate, CoA, and CoQ, and for UTP and dTTP. This high degree of auxotrophy is found in most other lactobacilli and reflects their demanding nutritional requirements when grown on synthetic media (26). Detailed information about amino acid and cofactor metabolism can be found in Biosynthetic Capabilities in Supporting Text.
Proteolytic System. The considerable auxotrophy for amino acids predicted for L. acidophilus was consistent with the presence of a large number of peptidases/proteases and related transport systems for amino acids and peptides. In silico analysis of predicted transporters revealed the presence of nine ATP-binding cassette (ABC)-type transporters, translocating both amino acids and oligopeptides. Two separate di- and oligopeptide transporting systems (oppA, -B, and -C) were predicted. Scattered throughout the genome, six additional genes coding for distinct periplasmatic components (OppA) were identified, and likely broaden the opp-transporter specificities (La1216, La1347, La1400, La1665, La1958, and La1961). The remaining transporters were predicted to recognize glutamate, glutamine, branched, and polar amino acids. Also, a separate ABC-type transporter for both spermidine and putrescine was identified (potA, La709; potB, La711; potC, La712; and potD, La713). Furthermore, 22 permeases for amino acids were found, often with no predicted specificity. Only one permease-type transporter for di- and tri-peptides (dtpT, La1848) was located (Fig. 1, circle 9, proteolytic transporter). This vast number of amino acid and peptide transporters was complimented by an array of 20 peptidases and proteases (Fig. 1, circle 9, proteases/peptidases and Table 5, which is published as supporting information on the PNAS web site). Detailed information about the peptidases is presented in Proteolytic System in Supporting Text.
Interestingly, a significant number of peptidases were located directly adjacent to transport systems or regulators, often forming operon like structures. A prolyl amino peptidase (PAP) (La92), was followed by an ABC transporter consisting of a permease (La93) and an ATPase component (La94). La95 was predicted to encode for an AraC type transcriptional regulator and is divergently oriented to the upstream located ABC-transporter and PAP. Analysis of the intergenic region between La94 and La95 revealed the presence of two promoter-like structures with partially overlapping -10-regions (data not shown). Similarly, the di- and oligopeptide transport system (Opp) (La197 through La203) was flanked by the dipeptidases PepG (La195) and PepE (La204). Only a few peptidases were found to be accompanied by permease-type transporters, like PepN (La1849), which is preceded by DtpT (La1848), a di- and tripeptide permease.
Cell-envelope proteases (PrtP) are critical for gowth of LAB in milk (23), because they hydrolyze casein into >100 smaller peptide fragments. PrtP is synthesized as an inactive precursor molecule and requires a membrane-bound lipoprotein (PrtM) for its autocatalytic maturation process (27). In silico analyses revealed the presence of both PrtP (La1512) and PrtM (La1588) in NCFM, sharing significant similarities to L. gasseri, L. johnsonii, Lactobacillus rhamnosus, L. plantarum WCFS1, Lactobacillus paracasei, and Lactococcus lactis subsp. cremoris (PrtM, >65% similarity; PrtP, >46% similarity). The presence of both genes in NCFM suggested that the organism can digest large proteins extracellularly and generate small peptides and essential amino acids that are internalized by Opp transporters and amino acid permeases.
Sugar Metabolism. L. acidophilus has the ability to use a variety of carbohydrates, including mono-, di-, and polysaccharides, as shown by its API50 sugar fermentation pattern (Biomerieux, Durham, NC) (data not shown). In particular, complex dietary carbohydrates that escape digestion in the upper GI-tract, such as raffinose and fructooligosaccharides (28, 29) (Fig. 1), can be used. The NCFM genome encodes a large variety of genes related to carbohydrate utilization, including 20 phosphoenol-pyruvate sugar-transferase systems (PTS) and five ABC families of transporters (Fig. 7 and Table 6, which are published as supporting information on the PNAS web site). Putative PTS transporters were identified for trehalose (La1012), fructose (La1777), sucrose (La401), glucose and mannose (La452-La456), melibiose (La1705), gentiobiose and cellobiose (1369), salicin (La876-La879), arbutin (La884), and N-acetyl glucosamine (La146). Putative ABC transporters were identified for FOS (La502-La504, La506), raffinose (La1439-La1442), and maltose (La1854-La1857). A putative lactose-galactose permease was also identified (La1463). Most of these transporters share a genetic locus with a glycosidase and a transcriptional regulator, allowing localized transcriptional control.
In silico analyses of the genome revealed the presence of genes representing the complete glycolysis pathway. Additionally, members of the general carbohydrate utilization regulation network were identified, namely HPr (La639, ptsH), EI (La640, ptsI), CcpA (La431, ccpA), and HPrK/P (La676, ptsK), indicating an active carbon catabolite repression network based on sugar availability. More information can be found in Sugar Metabolism in Supporting Text.
Stress Response. The genome of L. acidophilus encodes a number of stress-related proteins, including several proteases involved in the stress response and most of the highly conserved SOS regulon genes (Fig. 1, circle 8).
Mechanisms of acid resistance used by Gram-positive bacteria include proton pumps, amino acid decarboxylation, electrogenic transport systems, chaperones involved in repair/degradation of damaged proteins, incremental expression of regulators that promote local or global responses, and structural alterations in the cell envelope (30). The F1F0-ATPase system has been well characterized in L. acidophilus (31) and is encoded by the atp operon. Also encoded were a number of amino acid decarboxylases, such as an ornithine decarboxylase (La996), which has already been shown to play a role in acid tolerance (32). L. acidophilus is among the least oxygen-tolerant lactobacilli (33), although it has been reported to produce superoxide dismutase (SOD) (34). In contrast, the NCFM genome did not reveal a putative SOD homolog. Consequently, genes involved in the disulfide-reducing pathway and elimination of reactive oxygen species are of obvious importance. L. acidophilus NCFM encodes a thioredoxin system (La422, La439, La679, La1581, La1898, and La1901) and the glutathione reductase (La1107). Additionally, putative NADH-oxidase (La1418 and La1421) and NADH-peroxidase (La887) genes have been detected. RecA (La666), FlpA (La1969), and FlpB (La544) have been reported elsewhere to be involved in repairing DNA damage (for reviews, see ref. 35).
Only one deduced protein (La818) in the genomic sequence showed high similarity to CspA, a cold shock protein of L. delbrueckii subsp. bulgaricus whose transcription increased after a temperature downshift from 42°C to 25°C (36). Lastly, a gene (relA, La932) was identified that encodes an enzyme putatively involved in osmotolerance by synthesis and hydrolysis of (p)p-pGpp (37). More information on heat shock proteins and the atp operon is provided in Stress Response in Supporting Text.
Regulation. The interaction of alternative sigma factors with RNA polymerase is one of the most efficient methods known for adaptive regulation. Only one major (RpoD, La1196) and one putative minor alternative sigma factor (RpoE, La351) were found in the genome sequence of L. acidophilus. Orthologs of RpoD and RpoE were also identified by psi-blast analyses in the genomes of L. gasseri, L. johnsonii, and L. plantarum. Detailed information on in silico analyses of RpoD and RpoE can be found in Regulation in Supporting Text.
Ninety-six transcriptional regulators were identified (comprising >5% of NCFM total genes) based on the presence of conserved functional domains. Most of the unambiguously identified regulators are repressors: HipB (eight genes), TetR (six genes), RpiR (six genes), PhnF (six genes), MarR (five genes), and NagC (four genes). NCFM also has six predicted PurR-type repressors involved in sugar metabolism that include the sucrose operon transcriptional repressor (ScrR), MsmR and MsmR2 repressors (29). Only 15 genes are transcriptional activators, with LysR being the most represented family (9 genes) (Table 7, which is published as supporting information on the PNAS web site).
Nine two-component regulatory systems (comprising ≈1% of NFCM total genes) were identified (Fig. 1, circle 10 and Table 8, which is published as supporting information on the PNAS web site), each composed of a histidine kinase and the corresponding response regulator. Two are sensor–responder pairs that appear to be potentially associated with lactacin-B production (La1798 and La1799) and one pair (La1524 and La1525) with similarities to the acid tolerance 2CRS of Listeria monocytogenes (38). Additionally, we identified four response regulators that contained the LytTR DNA-binding motif (La248, La403, La1542, and La1775) (39), but were not associated with a histidine kinase. Also notable was the presence of typical eukaryotic regulators: a putative serine-threonine protein kinase (La1317) and two serine–threonine protein phosphatases (La489 and La1318).
The EPS Cluster. The EPS cluster consisted of 14 genes including the highly conserved proteins EpsA–EpsF (La1732–La1737), EpsJ (La1725 and La1726), and EpsI (La1724) and five variable proteins (La1727–La1731) representing glycosyl transferases and polysaccharide polymerases. Together, this set shows high synteny to reported EPS clusters in streptococci (40) and recently reported in L. gasseri and L. johnsonii (6). Scanning electron microscopy of NCFM did not detect an external polysaccharide layer (41), and it remains unclear whether the EPS cluster is functional or whether any EPS produced is excreted rather than anchored. Three ORFs in the NCFM EPS cluster encode for two UDP-galactopyranose mutases and a membrane protein involved with the export of O-antigen and teichoic acid. Other teichoic acid associated ORFs include a tandem set of teichoic acid biosynthesis and transport proteins (La524 and La525), another predicted biosynthetic protein (La519), two more polysaccharide transporters specific to O-antigen and teichoic acid (La1614 and La1917), along with a cell wall teichoic acid glycosylation protein (La621) (Table 9, which is published as supporting information on the PNAS web site). An exaggerated inflammatory response from intestinal epithelial cells to Gram-negative bacteria can be tempered by teichoic acids from lactobacilli (42), suggesting an intimate involvement of teichoic acids and the immune system. The uncharacterized low GC regions and the EPS cluster are centered on two divergently oriented transposases (La1722, La1721, and La1720). The exceptionally low GC content and the presence of mobile elements could indicate the acquisition of this region by horizontal gene transfer.
Adherence. Adherence by lactobacilli to tissues of the human intestinal mucosa is purported to be mediated by cell-surface proteins and polysaccharides (43, 44). The presence of these bacteria on mucosal surfaces is thought to modulate both immune responses (45, 46) and promote mucosal integrity and maintenance of the normal microflora (47). NCFM adheres to human fetal intestinal cells and Caco-2 cells, although the mechanisms underlying attachment remains to be determined (48). Analysis of the genome revealed six ORFs with a Grampositive cell-wall anchor motif (PFAM PF00746), of which one (La1740) contained SIRK-type signal sequences, suggesting secretion and anchoring to the cell surface.
Five predicted proteins in NCFM share similarity (E < 1–33) with the mucus-binding protein of L. reuteri, a 358-kDa protein reported to specifically bind mucin glycoproteins (44). The most striking of these, La1392, is composed of 4326 amino acid residues and represents the largest ORF in the NCFM genome. As is common with proteins of this family, it contains four imperfect C-terminal repeats of 18 aa. Multiple copies of Mub homologs are present in L. gasseri (seven), L. johnsonii (four), and L. plantarum (two). The abundance of these proteins in probiotic lactobacilli suggests an important role in adhesion and/or colonization of intestinal mucosal surfaces.
A predicted fibronectin binding protein, FbpA (La1148), suggests that L. acidophilus has the ability to bind fibronectin, a cell-surface dimeric glycoprotein. Homologs of FbpA can be found in L. gasseri (draft genome), L. johnsonii (Lj1182), and L. plantarum (Lp1793). A correlation between fibronectin binding and adherence of lactobacilli to intestinal cells in vitro was reported (49) and a subsequent in vivo relationship suggested. The distribution of selected proteins potentially involved in adherence is shown in Fig. 1. Additional ORFs suspected to be involved in adhesion are described in Adherence in Supporting Text.
Conclusions
In silico analyses of the complete genome sequence of L. acidophilus NCFM revealed significant similarities to other prokaryotes and in particular to other probiotic LAB. Detailed in silico comparative analyzes, in particular to the published genomes of L. plantarum, L johnsonii, and L. gasseri, are required. A series of strain-specific genes and gene clusters were identified that are suspected to encode some of the unique features exhibited by NCFM. The presence of a SPIDR region could facilitate the development of strain and potentially species specific identification methods. The programmed ability to use a diverse number of fermentable sugars and produce a variety of cell surface proteins implicated in adherence to epithelial cells and mucus provides mechanistic support for the intestinal roles of probiotic bacteria. Ongoing research, now sustained by a full genome foundation, is actively investigating genetic targets potentially linked to survival, adherence, bacteriocin production, and metabolism. L. acidophilus is one of a few prokaryotes with no intact prophages. However, its large number of phage-related proteins and the newly discovered potential autonomous units suggests a history of inactivation or elimination of integrated prophages. The predicted presence of a phage killer maintenance system appears to be unusual in lactobacilli and lactococci, because only L. gasseri was found thus far to harbor a similar gene set. Consequently, this system might be used for development of highly stable genomic integration systems. Reconstruction of complete and partial metabolic pathways revealed a significant degree of auxotrophy, reflecting the organisms adaptation to the nutrient-rich conditions of the upper human GI-tract.
Supplementary Material
Acknowledgments
We thank E. Durmaz for help and technical assistance and T. Cox, A. Davis, V. Willoughby, and D. Doherty for assistance and sequencing efforts. This work was supported in part by the North Carolina Agricultural Research Service, Danisco, Inc., Dairy Management, Inc., the Southeast Dairy Foods Research Center, the North Carolina Dairy Foundation, the California Dairy Research Foundation, and the Environmental Biotechnology Institute. R.B. and T.D. were partially supported by National Science Foundation/Integrative Graduate Education and Research Trainee Functional Genomics Fellowships; W.M.R. was partially supported by a Graduate Assistance in Areas of National Need fellowship in biotechnology; and B.L.B. and A.D. were partially supported by National Institutes of Health Biotechnology Fellowships. M.C. and S.L. contributed to manual annotation; A.H. and R.C. provided DNA sequence and assembly data. Contract sequencing for genome finishing and polishing was carried out by Fidelity Systems (Gaithersburg, MD).
Abbreviations: GI, gastrointestinal; COG, cluster of orthologous groups of proteins; EPS, exo-polysaccharides; SPIDR, spacers interspersed direct repeats; LAB, lactic acid bacteria; PAU; potential autonomous unit; ABC, ATP-binding cassette.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. CP000033).
See accompanying Biography on page 3903.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
References
- 1.Reid, G., Sanders, M. E., Gaskins, H. R., Gibson, G. R., Mercenier, A., Rastall, R., Roberfroid, M., Rowland, I., Cherbut, C. & Klaenhammer, T. R. (2003) J. Clin. Gastroenterol. 37, 105–118. [DOI] [PubMed] [Google Scholar]
- 2.Klaenhammer, T. R. & Russell, W. M. (2000) in Encyclopedia of Food Microbiology (Academic, Amsterdam), Vol. 2, pp. 1151–1157. [Google Scholar]
- 3.Johnson, J. L., Phelps, C. F., Cummins, C. S., London, J. & Gasser, F. (1980) Int. J. Syst. Bacteriol. 30, 53–68. [Google Scholar]
- 4.Heilig, H. G., Zoetendal, E. G., Vaughan, E. E., Marteau, P., Akkermans, A. D. & De Vos, W. M. (2002) Appl. Environ. Microbiol. 68, 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M. C., Desiere, F., Bork, P., Delley, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pridmore, R. D., Berger, B., Desiere, F., Vilanova, D., Barretto, C., Pittet, A. C., Zwahlen, M. C., Rouvet, M., Altermann, E., Barrangou, R., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 2512–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleerebezem, M., Boekhorst, J., van Kranenburg, R., Molenaar, D., Kuipers, O. P., Leer, R., Tarchini, R., Peters, S. A., Sandbrink, H. M., Fiers, M. W., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1990–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klaenhammer, T., Altermann, E., Arigoni, F., Bolotin, A., Breidt, F., Broadbent, J., Cano, R., Chaillou, S., Deutscher, J., Gasson, M., et al. (2002) Antonie Leeuwenhoek 82, 29–58. [DOI] [PubMed] [Google Scholar]
- 9.Sanders, M. E. & Klaenhammer, T. R. (2001) J. Dairy Sci. 84, 319–331. [DOI] [PubMed] [Google Scholar]
- 10.Altermann, E. & Klaenhammer, T. R. (2003) OMICS 7, 161–169. [DOI] [PubMed] [Google Scholar]
- 11.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurtz, S. & Schleiermacher, C. (1999) Bioinformatics 15, 426–427. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, L. J., Friis, C. & Ussery, D. W. (1999) Res. Microbiol. 150, 773–777. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. (2004) Nucleic Acids Res. 32, D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen, R., van Embden, J. D., Gaastra, W. & Schouls, L. M. (2002) OMICS 6, 23–33. [DOI] [PubMed] [Google Scholar]
- 19.Ishino, Y., Shinagawa, H., Makino, K., Amemura, M. & Nakata, A. (1987) J. Bacteriol. 169, 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desiere, F., Lucchini, S., Canchaya, C., Ventura, M. & Brussow, H. (2002) Antonie Leeuwenhoek 82, 73–91. [PubMed] [Google Scholar]
- 21.Foschino, R., Picozzi, C. & Galli, A. (2001) J. Appl. Microbiol. 91, 394–403. [DOI] [PubMed] [Google Scholar]
- 22.Lehnherr, H., Maguin, E., Jafri, S. & Yarmolinsky, M. B. (1993) J. Mol. Biol. 233, 414–428. [DOI] [PubMed] [Google Scholar]
- 23.Romero, D. A. & Klaenhammer, T. R. (1993) J. Dairy Sci. 76, 1–19. [DOI] [PubMed] [Google Scholar]
- 24.Klaenhammer, T. R. (1993) FEMS Microbiol. Rev. 12, 39–86. [DOI] [PubMed] [Google Scholar]
- 25.Eijsink, V. G., Axelsson, L., Diep, D. B., Havarstein, L. S., Holo, H. & Nes, I. F. (2002) Antonie Leeuwenhoek 81, 639–654. [DOI] [PubMed] [Google Scholar]
- 26.Morishita, T., Deguchi, Y., Yajima, M., Sakurai, T. & Yura, T. (1981) J. Bacteriol. 148, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haandrikman, A. J., Meesters, R., Laan, H., Konings, W. N., Kok, J. & Venema, G. (1991) Appl. Environ. Microbiol. 57, 1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson, G. R. & Roberfroid, M. B. (1995) J. Nutr. 125, 1401–1412. [DOI] [PubMed] [Google Scholar]
- 29.Barrangou, R., Altermann, E., Hutkins, R., Cano, R. & Klaenhammer, T. R. (2003) Proc. Natl. Acad. Sci. USA 100, 8957–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cotter, P. D. & Hill, C. (2003) Microbiol. Mol. Biol. Rev. 67, 429–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kullen, M. J. & Klaenhammer, T. R. (1999) Mol. Microbiol. 33, 1152–1161. [DOI] [PubMed] [Google Scholar]
- 32.Azcarate-Peril, M., Altermann, E., Hoover-Fitzula, R., Cano, R. & Klaenhammer, T. R. (2004) Appl. Environ. Microbiol. 70, 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Archibald, F. S. & Fridovich, I. (1981) J. Bacteriol. 146, 928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez, S. N., Apella, M. C., Romero, N., Pesce de Ruiz Holgado, A. A. & Oliver, G. (1989) Chem. Pharm. Bull. (Tokyo) 37, 3026–3028. [DOI] [PubMed] [Google Scholar]
- 35.Girgis, H. S., Smith, J., Luchansky, J. B. & Klaenhammer, T. R. (2003) in Microbial Stress Adaptation and Food Safety, eds. Yousef, A. E. & Juneja, V. K. (CRC Press, Boca Raton, FL).
- 36.Serror, P., Dervyn, R., Ehrlich, S. D. & Maguin, E. (2003) FEMS Microbiol. Lett. 226, 323–330. [DOI] [PubMed] [Google Scholar]
- 37.Okada, Y., Makino, S., Tobe, T., Okada, N. & Yamazaki, S. (2002) Appl. Environ. Microbiol. 68, 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cotter, P. D., Emerson, N., Gahan, C. G. & Hill, C. (1999) J. Bacteriol. 181, 6840–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolskaya, A. N. & Galperin, M. Y. (2002) Nucleic Acids Res. 30, 2453–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stingele, F., Neeser, J. R. & Mollet, B. (1996) J. Bacteriol. 178, 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hood, S. K. & Zottola, E. A. (1987) J. Food Sci. 52, 791–805. [Google Scholar]
- 42.Vidal, K., Donnet-Hughes, A. & Granato, D. (2002) Infect. Immun. 70, 2057–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Granato, D., Perotti, F., Masserey, I., Rouvet, M., Golliard, M., Servin, A. & Brassart, D. (1999) Appl. Environ. Microbiol. 65, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos, S. & Jonsson, H. (2002) Microbiology 148, 433–442. [DOI] [PubMed] [Google Scholar]
- 45.Schiffrin, E. J., Rochat, F., Link-Amster, H., Aeschlimann, J. M. & Donnet-Hughes, A. (1995) J. Dairy Sci. 78, 491–497. [DOI] [PubMed] [Google Scholar]
- 46.Valeur, N., Engel, P., Carbajal, N., Connolly, E. & Ladefoged, K. (2004) Appl. Environ. Microbiol. 70, 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tannock, G. W. (1999) Antonie Leeuwenhoek 76, 265–278. [PubMed] [Google Scholar]
- 48.Greene, J. D. & Klaenhammer, T. R. (1994) Appl. Environ. Microbiol. 60, 4487–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapczynski, D. R., Meinersmann, R. J. & Lee, M. D. (2000) Curr. Microbiol. 41, 136–141. [DOI] [PubMed] [Google Scholar]
- 50.Altermann, E., Buck, L. B., Cano, R. & Klaenhammer, T. R. (2004) Gene 342, 189–197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.