Abstract
Endothelial dysfunction and apoptosis have key roles in the initiation and progression of atherosclerosis (AS). AS has been demonstrated to be associated with a high-fat diet, which may increase endothelial permeability and apoptosis; however, the exact mechanisms underlying the development of AS remain poorly understood. MicroRNAs (miRNAs) are vital for the regulation of cardiovascular disease, and dysregulated miRNAs have been implicated in AS. The present study investigated whether miRNA (miR)-126 regulates high-fat diet-induced endothelial permeability and apoptosis by targeting transforming growth factor β (TGFβ), a secreted protein that controls cellular proliferation and apoptosis. In the present study, apolipoprotein E (apoE)−/− mice were fed a high-fat diet in order to establish a model of AS. Mice were subcutaneously injected with a miR-126 mimic, a miR-126 antagomir or control miRNA. Reverse transcription-quantitative polymerase chain reaction was used to assess miR-126 expression, and a fluorometric assay was used to evaluate caspase-3 activity. The effects of miR-126 on the endothelial permeability of the aortic intima were also explored. Western blotting and immunohistochemical analysis were used to investigate the effects of miR-126 on B-cell lymphoma-2 (Bcl-2) and transforming growth factor (TGF) β protein expression levels. Furthermore, a luciferase assay was performed to verify whether TGFβ may be a direct target gene of miR-126. In apolipoprotein E-knockout mice, a high-fat diet reduced miR-126 expression and induced apoptosis as determined by the upregulation of caspase-3 activity. A miR-126 antagomir increased endothelial permeability and apoptosis in mice fed a high-fat diet. By contrast, an miR-126 mimic attenuated endothelial permeability and apoptosis. The reduction in miR-126 was associated with a reduction in protein expression levels of Bcl-2 and an increase of TGFβ in mice fed a high-fat diet. In addition, the present study demonstrated that miR-126 reduced TGFβ expression following binding to the 3′-untranslated region of TGFβ mRNA. The current study demonstrated a role for miR-126 in AS and identified TGFβ as a direct target of miR-126. Furthermore, the present study demonstrated that miR-126 contributed to endothelial permeability and apoptosis, and suggested that the downregulation of TGFβ may be involved in the molecular mechanisms underlying the actions of miR-126. miR-126 may therefore have potential as a novel therapeutic target for the treatment of AS.
Keywords: microRNA-126, permeability, apoptosis, transforming growth factor β, high-fat diet
Introduction
Atherosclerosis (AS) is characterised by autoimmune and immunological mechanisms leading to plaque formation (1–5). Endothelial dysfunction and apoptosis are often seen as the initiating factors (3–6). A high-fat diet promotes the development of AS by increasing endothelial permeability and apoptosis (7–10). The discovery of microRNAs (miRNAs) has provided a novel perspective for AS research. miRNAs are small, single-stranded, non-coding RNA molecules (~22 nucleotides) that are encoded within the genome and derived from endogenous small hairpin precursors (3,4,11). miRNAs are involved in the post-transcriptional regulation of gene expression via binding to the 3′-untranslated regions (3′-UTRs) of specific mRNAs, which subsequently inhibits the transcription or translation of the target mRNAs (12,13). It has been reported that miRNAs have key roles in various physiological cellular activities, including development, growth, proliferation, metabolism, differentiation and apoptosis. The occurrence and development of numerous cardiovascular diseases, including AS, hypertension and cardiac fibrosis, is associated with aberrant expression of miRNAs (14–17).
miRNA (miR)-126 is currently the only known miRNA expressed specifically in endothelial cells and hematopoietic stem cells, and is closely associated with AS, coronary heart disease and other cardiovascular diseases (18–22). Our recent study demonstrated that miR-126 exerts anti-apoptotic effects in human umbilical vein endothelial cells (HUVECs) by inhibiting the expression of tumour necrosis factor receptor-associated factor 7 and the production of reactive oxygen species (23).
Transforming growth factor β (TGFβ) is a cytokine that has key roles in numerous biological processes, including inflammatory responses and apoptosis (24–26). Studies have indicated that TGFβ is closely associated with endothelial cells and the pathogenesis of AS (27–31). The effect of miR-126 on high-fat diet-induced endothelial permeability and apoptosis remains unclear. The results of the present study suggested that miR-126 inhibited endothelial permeability and apoptosis by inhibiting TGFβ expression via binding to the 3′-UTR of TGFβ mRNA.
Materials and methods
Cell culture and reagents
Primary HUVECs were purchased from Lonza, Inc. (Allendale, NJ, USA). Sulfo-NHS-LC-biotin was purchased from Pierce; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). miRNA oligonucleotides (miR-126 mimic, 5′-CAGUACUUUUGUGUACAA-3′; miR-126 antagomir, 5′-CGCAUUAUUACUCACGGUACGA-3′; and negative control miRNA, 5′-UUCUCCGAACGUGUCACGUTT-3′) and TransMessenger Transfection reagent were obtained from Qiagen China Co., Ltd. (Shanghai, China). Rhodamine600 (XRITC)-avidin was obtained from Pierce; Thermo Fisher Scientific, Inc. HUVECs were cultured in endothelial cell growth medium supplemented with 10% fetal bovine serum and 100 U/ml penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.), and maintained at 37°C in a 5% CO2 atmosphere. The cells at passage 3–4 were used for experiments. All experiments were repeated at least three times.
Animal experimental procedures
All of the experiments performed in the present study were approved by the Ethics Committee of The First Affiliated Hospital of Anhui Medical University (Hefei, China). A total of 60 male apolipoprotein E (apoE)−/− mice (age, 4 weeks; weight, 22±5 g) were obtained from the Institute of Basic Medical Sciences of Peking Union Medical College (Beijing, China). Mice were housed individually in screen-bottomed plastic cages in a temperature-controlled room (25°C) under a 12-h light/dark cycle with free access to food and water. Mice were divided into two groups: Standard diet (n=16) and standard diet plus 5% lard oil and 2% cholesterol (n=44); diets were maintained for 16 weeks. At 12 weeks, 8 control and 8 high-fat diet mice were euthanized for detection of miR-126 expression levels, and the remaining control and high-fat diet mice were randomly divided into the following three groups: Control miRNA oligonucleotides (n=8; 4 control mice and 4 high-fat diet mice); miR-126 antagomir oligonucleotides (n=18; 9 control mice and 9 high-fat diet mice); and miR-126 mimic oligonucleotides (n=18; 9 control mice and 9 high-fat diet mice). Control miRNA, or miR-126 antagomir or mimic was injected subcutaneously at a dose of 10 mg/kg twice in the first week, followed by once a week for 4 weeks. During the study period, 1 mouse died in the control miRNA oligonucleotide group, 2 died in the miR-126 antagomir oligonucleotide group and 2 died in the miR-126 mimic oligonucleotide group. At the end of the experiment, the aorta was removed from mice as previously described (8). Part of the aorta was embedded in optimal cutting temperature compound and snap frozen. The remaining aorta was stored at −80°C.
miR-126 expression assay
Total RNA was extracted from the aorta using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). miR-126 expression was determined using a miRNA Plate Assay kit (cat no. MA-0101; Signosis, Inc., Santa Clara, CA, USA) and an oligo mix specific for miR-126 (cat no. MO-0040; Signosis, Inc.), according to the manufacturer's protocol. The RNA content was normalized to U6 small nuclear (sn)RNA. miR-126 expression was also determined using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) with the following primers: miR-126, forward 5′-UCGUACCGUGAGUAAUAAUGCG-3′, reverse 5′-CAUUAUUACUCACGGUACGAUU-3′; and U6 snRNA, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′. Amplification conditions were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec, and at 60°C for 60 sec. The reaction mixture contained the following: 2X Power SYBR Green Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.), cDNA template, 10 pmol/µl of each primer and sterile H2O. Relative gene expression was quantified according to the comparative Cq method (32) using GraphPad Prism software version 5.01 (GraphPad Software, Inc., La Jolla, CA, USA).
Permeability assay
The permeability assay was performed as previously described (33). Briefly, frozen aorta sections (10-µm thick) were stained at 37°C with Sulfo-NHS-LC-biotin for 30 min, and blocked with 5% non-fat milk at 4°C overnight and subsequently submerged into blocking buffer containing XRITC-avidin (1:500) at room temperature for 1 h. The slides were washed three times with PBS and dried. Images were captured using an Olympus Provis AX70 fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Caspase-3 activity measurement
Caspase-3 activity was detected in aortic samples using a Caspase-3 Fluorometric assay kit (Enzo Life Sciences, Inc., Farmingdale, NY, USA), as previously described (34). Briefly, aortic samples were isolated from anesthetized mice and stored in liquid nitrogen. Samples were homogenized with lysis buffer containing 50 mM Tris-HCl (pH 6.8), 10% glycerol and 2% SDS for 3 min on ice, maintained on ice for 10 min, centrifuged at 21,130 × g for 10 min at 4°C, and supernatants were collected and cryopreserved at −70°C until further use. Protein concentration was determined using a bicinchoninic acid (BCA) assay with the Micro BCA™ Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of extracted protein samples (50 µg) were incubated at 37°C overnight with N-acetyl-Asp-Glu-Val-Asp p-nitroanilide (Ac-DEVD-pNA). The quantity of pNA that was released was estimated at 405 nm using a microplate ELISA reader. Caspase-3 relative activity was calculated as follows: Caspase-3 activity=(mean experimental absorbance/mean control absorbance)x100%.
Western blot analysis
Aortic samples were homogenized, lysed for 3 min on ice in 1X SDS lysis buffer containing 50 mM Tris-HCl (pH 6.8), 10% glycerol and 2% SDS and boiled for 10 min, followed by centrifugation at 16,000 × g for 10 min at room temperature. Protein concentration was determined using a Micro BCA™ Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of extracted protein samples (20 µg) were separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (GE Healthcare Life Sciences, Chalfont, UK). Membranes were blocked using 5% (w/v) bovine serum albumin (BSA; Amresco, LLC, Solon, OH, USA) for 2 h at room temperature, followed by incubation with the following primary antibodies: Anti-TGFβ (cat no. 3711; 1:1,000), anti-B-cell lymphoma (Bcl)-2 (cat no. 2872; 1:1,000), purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA); and anti-GAPDH (cat no. TA505454; 1:5,000; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd., Beijing, China) diluted in TBS containing 0.05% Tween-20 (TBST) at 4°C overnight. Following 3 washes with TBST, horseradish peroxidase (HRP)-conjugated secondary antibodies (cat nos. ZB-2305 and ZB-2301; 1:5,000; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) were added to membranes and incubated at room temperature for 2 h. Protein bands were visualized by enhanced chemiluminescence using ECL reagents (Pierce; Thermo Fisher Scientific, Inc.). GAPDH was used as the loading control. Blots were semi-quantified by densitometric analysis using the Image-Pro Plus software version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA). Experiments were repeated 3 times.
Immunohistochemistry
Immunohistochemistry was performed as previously described (8). Sections (10 µm) of the frozen aorta tissue samples were blocked using PBS containing 0.05% Tween-20 and 1% BSA at room temperature for 30 min, and incubated at 4°C overnight with an anti-TGFβ (cat no. 3711; 1:1,000) primary antibody, purchased from Cell Signaling Technology, Inc. Subsequently, sections were incubated with a HRP-conjugated goat secondary antibody (cat no. ZB-2301; 1:5,000; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) at room temperature for 60 min. A Metal Enhanced 3,3′-diaminobenzidine Substrate kit (Pierce; Thermo Fisher Scientific, Inc.) was used for 3 min to develop the colour, and sections were counterstained with hematoxylin (10%) at room temperature for 30 sec. The integral absorbance was examined under a light microscope and results were quantified using the Image-Pro Plus software version 6.0 (Media Cybernetics, Inc.).
Luciferase assay
The region of the TGFβ 3′-UTR containing the potential binding site of miR-126 (source: NCBI GenBank; https://www.ncbi.nlm.nih.gov/nuccore/NM_011577.2) was predicted using TargetScan version 7.1 (http://www.targetscan.org/vert_71/). The sequence was inserted into the 3′ region of the luciferase gene in a luciferase vector (wt-Luc-TGFβ; Shanghai GeneChem Co., Ltd., Shanghai, China), and a mutated version of this sequence was inserted into the vector (mu-Luc-TGFβ). Plasmid DNA (300 ng) and miR-126 mimic, antagomir or control oligonucleotide (80 nmol/l) were co-transfected using TransMessenger Transfection reagent into HUVECs seeded into 6-well plates at a density of 1–2×105 cells/well (confluence, 60–70%) for 24 h at 37°C. The pRL-TK vector (Promega Corporation, Madison, WI, USA) expressing Renilla luciferase served as a control. The luciferase assay was performed using a Dual-Luciferase® Reporter assay system (Promega Corporation) 24 h following transfection, according to the manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± standard deviation of 3 independent experiments. The statistical significance of the differences between groups was assessed using one-way analysis of variance followed by a Neuman-Keuls post hoc test for multiple comparisons. Comparisons between two groups were based on least significant differences. P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using SPSS software version 21.0 (IBM Corp., Armonk, NY, USA).
Results
Reduction of miR-126 expression and increase in apoptosis of arterial endothelial cells in high-fat diet fed mice
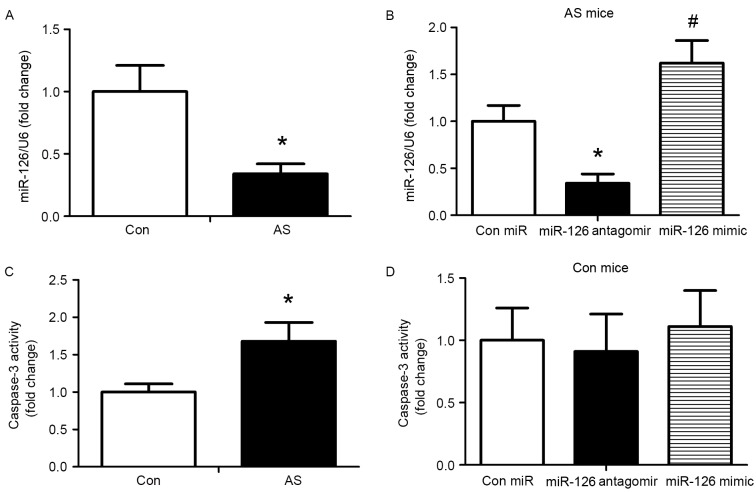
To evaluate the impact of miR-126 on an established AS model, apoE−/− mice were fed a high-fat diet for 12 weeks, followed by subcutaneous injection of 10 mg/kg control miRNA, miR-126 antagomir or miR-126 mimic twice for one week, and once a week for 4 weeks. Subsequently, the expression of miR-126 in the aorta was determined using a miRNA Plate assay kit. miR-126 levels were reduced in AS mice compared with control mice (P<0.05; Fig. 1A). Following 4 weeks of treatment, the miR-126 mimic significantly increased miR-126 expression and the miR-126 antagomir significantly reduced the expression of miR-126 compared with AS mice injected with control miRNA (P<0.05; Fig. 1B). In addition, caspase-3 activity was measured following 4 weeks of treatment with the miRNAs. Compared with the control group, caspase-3 activity was greater in AS mice (P<0.05; Fig. 1C). Treatment with miR-126 antagomir and mimic exhibited no effect on caspase-3 activity in mice fed a standard diet (Fig. 1D). Thus, the standard diet group was not analysed in subsequent experiments.
Figure 1.
miR-126 expression and apoptosis in a mouse model of AS. AS was induced in mice by a high-fat diet; mice on a standard diet served as controls. (A) miR-126 expression levels in control and AS mice. (B) miR-126 expression was detected in AS mice treated with control miR, miR-126 antagomir or miR-126 mimic. Caspase-3 activity was measured in (C) control and AS mice and (D) control mice treated with control miR, miR-126 antagomir or miR-126 mimic. *P<0.05 vs. control; #P<0.05 vs. miR-126 antagomir. miR, microRNA; AS, atherosclerosis; Con, control.
miR-126 expression alleviates endothelial permeability in AS model mice
To investigate the effect of miR-126 on endothelial permeability, NHS-LC-biotin and XRITC-avidin were used to detect the endothelial permeability of aortic intima. Results presented in Fig. 2 demonstrated that the aortic intima from AS mice exhibited greater staining with NHS-LC-biotin compared with control mice, in which only the endothelial surface was stained. When AS mice were treated with an miR-126 antagomir (Fig. 2Ac), an increased quantity of NHS-LC-biotin leaked into the aortic intima compared with AS mice treated with control miRNA (Fig. 2Ab). By contrast, miR-126 mimic (Fig. 2Bc) treatment reduced endothelial permeability compared with control miRNA-treated AS mice (Fig. 2Bb).
Figure 2.
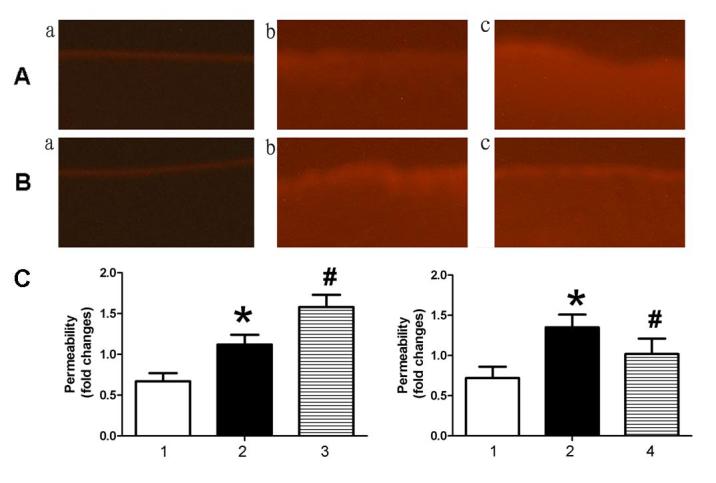
miR-126 expression levels affect endothelial permeability in the aorta. Mice were treated with (A) miR-126 antagomir or (B) miR-126 mimic. (a) Control mice treated with control miR, (b) AS mice treated with control miR and (c) AS mice treated with miR-126 antagomir (A)/mimic (B). (C) Following incubation of aortic intima sections with NHS-LC-biotin for 30 min, Rhodamine600-avidin was used to detect surface-bound biotin. Magnification, ×200. 1, control mice treated with control miR; 2, AS mice treated with control miR; 3, AS mice treated with miR-126 antagomir; and 4, AS mice treated with miR-126 mimic. *P<0.05 vs. control mice treated with control miR; #P<0.05 vs. AS mice treated with control miR. miR, microRNA; AS, atherosclerosis.
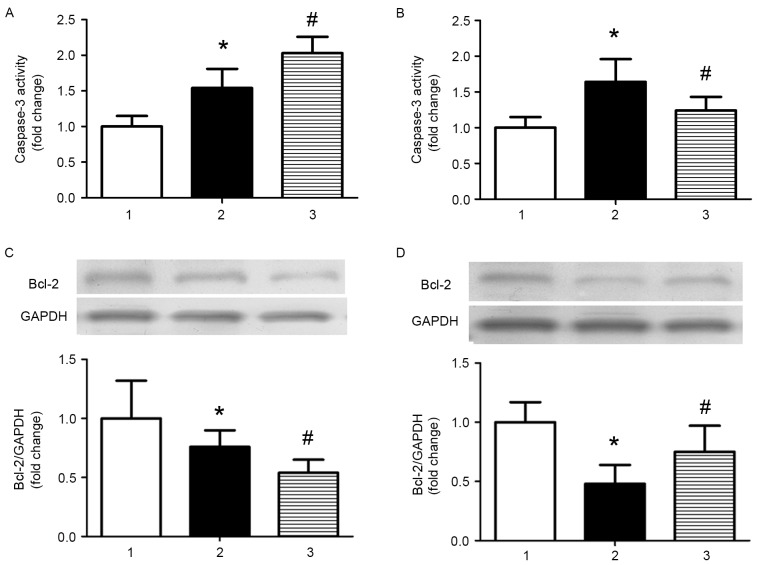
miR-126 inhibits apoptosis in AS model mice
As apoptosis is involved in the development of AS, the effect of miR-126 on apoptosis was investigated in the current study. Caspase-3 activity was significantly increased in AS model mice treated with control miRNA compared with control mice treated with control miRNA, and miR-126 antagomir treatment further increased caspase-3 activity (P<0.05; Fig. 3A). Treatment with an miR-126 mimic partially reversed the increased activity of caspase-3 induced by a high-fat diet (P<0.05; Fig. 3B). The opposite results were observed when the expression levels of the anti-apoptotic protein B-cell lymphoma 2 (Bcl-2) were analysed (Fig. 3C and D).
Figure 3.
miR-126 expression affects caspase-3 activity and Bcl-2 protein expression levels in the aorta. The effect of (A) miR-126 antagomir and (B) miR-126 mimic on caspase-3 activity was measured using a Caspase-3 Fluorometric assay kit. Western blotting was performed to investigate the effect of (C) miR-126 antagomir and (D) miR-126 mimic on Bcl-2 protein expression levels in the aorta of AS mice. 1, control mice treated with control miR; 2, AS mice treated with control miR; and 3, AS mice treated with miR-126 antagomir (A and C)/mimic (B and D). *P<0.05 vs. control mice treated with control miR; #P<0.05 vs. AS mice treated with control miR. miR, microRNA; AS, atherosclerosis; Bcl-2, B-cell lymphoma 2.
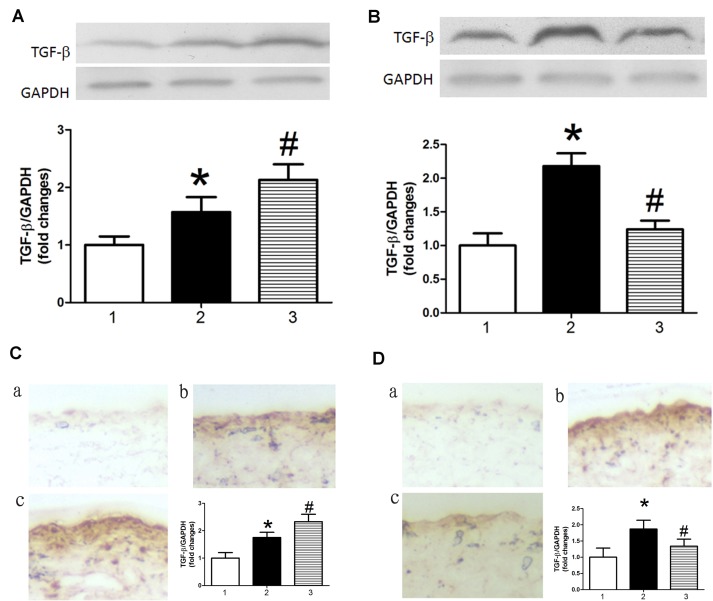
miR-126 modulates TGFβ expression in AS model mice
Previous studies have demonstrated that TGFβ promotes apoptosis in endothelial cells (22–24). Therefore, the present study investigated the effect of miR-126 on the expression of TGFβ. TGFβ protein expression levels were increased in AS model mice treated with control miRNA compared with control mice treated with control miRNA, as determined by western blot analysis (Fig. 4A and B) and immunohistochemistry (Fig. 4C and D). This result indicated that TGFβ may be a target gene of miR-126. Results in Fig. 4 demonstrated that miR-126 antagomir treatment significantly increased levels of TGFβ in AS model mice (Fig. 4A and C), whereas an miR-126 mimic had the opposite effect (Fig. 4B and D).
Figure 4.
miR-126 expression affects TGFβ protein expression levels in the aorta. Western blotting was performed to investigate the effect of (A) miR-126 antagomir and (B) miR-126 mimic on TGFβ protein expression levels. Immunohistochemistry was performed to investigate the effect of (C) miR-126 antagomir and (D) miR-126 mimic on TGFβ protein expression. (a) Control mice treated with control miR, (b) AS mice treated with control miR and (c) AS mice treated with miR-126 antagomir (C)/mimic (D). Magnification ×200. 1, control mice treated with control miR; 2, AS mice treated with control miR; and 3, AS mice treated with miR-126 antagomir (A and C)/mimic (B and D). *P<0.05 vs. control mice treated with control miR; #P<0.05 vs. AS mice treated with control miR. miR, microRNA; TGFβ, transforming growth factor β; AS, atherosclerosis.
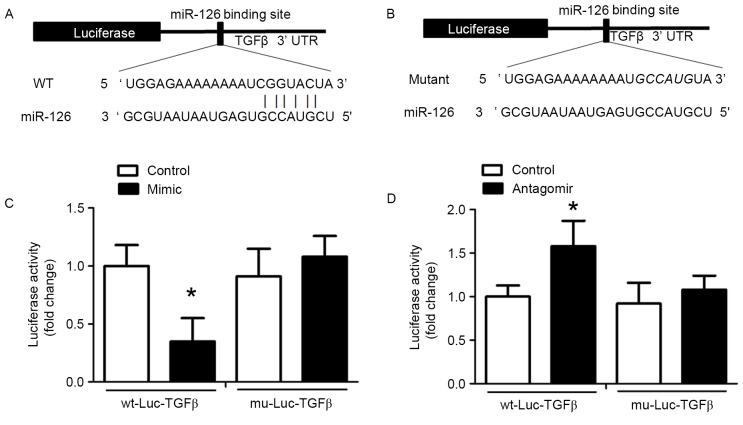
TGFβ is a target of miR-126
miRNAs specifically bind to the 3′-UTR of target mRNAs and induce transcript degradation or translational repression. An miR-126 binding site in the TGFβ 3′-UTR was identified and the alignment between miR-126 and the wild-type and mutant TGFβ 3′-UTR are presented in Fig. 5A and B, respectively. This indicated that miR-126 may cause translation inhibition of TGFβ. To validate this result of bioinformatics analysis, wt-Luc-TGFβ and mu-Luc-TGFβ were inserted into the luciferase vector, which was co-transfected into HUVECs with control miRNA, miR-126 antagomir or miR-126 mimic. Treatment with miR-126 mimic reduced luciferase activity in HUVECs co-transfected with wt-Luc-TGFβ compared with HUVECs co-transfected with wt-Luc-TGFβ and control miRNA (Fig. 5C). In HUVECs co-transfected with mu-Luc-TGFβ and an miR-126 mimic, no inhibition was observed (Fig. 5C). miR-126 antagomir significantly promoted luciferase activity in the wt-Luc-TGFβ-transfected HUVECs, but had no effect on mu-Luc-TGFβ-transfected HUVECs (Fig. 5D).
Figure 5.
miR-126 binds to and inhibits TGFβ expression in HUVECs. A Dual-Luciferase assay was performed. (A) A segment of the TGFβ 3′UTR containing the predicted miR-126 binding site was inserted downstream of the luciferase-coding sequence. Sequence alignment demonstrates TGFβ 3′UTR complementarity with the 5′ end of miR-126. (B) Sequence alignment of miR-126 and the mutated TGFβ 3′UTR exhibited no complementarity. (C) HUVECs were co-transfected with the wt-Luc-TGF-β or mu-Luc-TGFβ plasmid and miR-126 mimic or control miR. (D) HUVECs were co-transfected with the wt-Luc-TGFβ or mu-Luc-TGFβ plasmid and miR-126 antagomir or a control miR. *P<0.05 vs. control. miR, microRNA; TGFβ, transforming growth factor β; HUVECs, human umbilical vein endothelial cells; UTR, untranslated region; wt, wild-type; mu, mutant; Luc, luciferase.
Discussion
The present study investigated the effect of alterations in miR-126 expression on endothelial cell permeability and apoptosis in a mouse model of AS. The present results demonstrated that TGFβ was a direct target gene for miR-126, thus suggesting that miR-126 may bind to TGFβ and suppresses its expression. These findings indicated a novel mechanism by which miR-126 may have a vital role in endothelial permeability and apoptosis in AS model mice.
It has been reported that a high-fat diet is one of the factors that promotes AS development (7,31,35). Jakob et al (36) demonstrated that the expression of miR-126 in patients with chronic heart failure was significantly reduced compared with healthy controls. Furthermore, cardiac function was markedly improved by transfection with miR-126. The present study demonstrated that arterial wall permeability and apoptosis was increased in AS, and inhibition of miR-126 promoted this pathological process. Overexpression of miR-126, using an miR-126 mimic, attenuated these effects. Thus, normal levels of miR-126 may be necessary for the integrity of the arterial wall. However, miR-126 inhibited apoptosis as demonstrated by reduced caspase-3 activity and increased protein expression levels of Bcl-2 in mice fed high-fat diets. miR-126 had no effect on basal apoptosis in mice fed standard chow. This indicated that miR-126 may only inhibit apoptosis in mice on a high-fat diet, which inhibits miR-126 expression. However, this requires further investigation.
Typically, miRNAs influence gene expression by inducing post-transcriptional inhibition, mRNA degradation or translation suppression (12,13). It has been reported that TGFβ has key roles in inflammatory responses and apoptosis (24–26), and is closely associated with the pathogenesis of AS (27,28). The present study demonstrated that miR-126 directly binds to TGFβ mRNA and inhibits its expression. The results of the current study demonstrated that a reduction in miR-126 expression caused by a high-fat diet increased TGFβ protein expression levels and caused an increase in endothelial permeability and apoptosis.
In conclusion, the present study identified TGFβ as a direct target gene of miR-126, and demonstrated that reduced miR-126 expression increased TGFβ protein expression. These findings suggested that TGFβ downregulation may be implicated in the increased endothelial leakage and apoptosis that is observed in AS. Based on the results of the current study, miR-126 may have potential as a novel target for the treatment of cardiovascular diseases.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant nos. 81570419, 81470568 and 81270372), the National Natural Science Fund for Distinguished Young Scholars of China (grant no. 81302150) and Grants for Cultivating Program of National Natural Science Foundation for Young Scholars of the First Affiliated Hospital of Anhui Medical University (grant no. 2012KJ10).
References
- 1.Staff AC, Johnsen GM, Dechend R, Redman CW. Preeclampsia and uteroplacental acute atherosis: Immune and inflammatory factors. J Reprod Immunol. 2014;101:120–126. doi: 10.1016/j.jri.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Ooi CY, Sutcliffe MP, Davenport AP, Maguire JJ. Changes in biomechanical properties of the coronary artery wall contribute to maintained contractile responses to endothelin-1 in atherosclerosis. Life Sci. 2014;118:424–429. doi: 10.1016/j.lfs.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 3.Schober A, Nazari-Jahantigh M, Weber C. MicroRNA-mediated mechanisms of the cellular stress response in atherosclerosis. Nat Rev Cardiol. 2015;12:361–174. doi: 10.1038/nrcardio.2015.38. [DOI] [PubMed] [Google Scholar]
- 4.Svoboda P. A toolbox for miRNA analysis. FEBS Lett. 2015;589:1694–1701. doi: 10.1016/j.febslet.2015.04.054. [DOI] [PubMed] [Google Scholar]
- 5.Xu S, Liu Z, Liu P. Targeting hydrogen sulfide as a promising therapeutic strategy for atherosclerosis. Int J Cardiol. 2014;172:313–317. doi: 10.1016/j.ijcard.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd MM, Grima MA, Rayner BS, Hadfield KA, Davies MJ, Hawkins CL. Comparative reactivity of the myeloperoxidase-derived oxidants hypochlorous acid and hypothiocyanous acid with humancoronary artery endothelial cells. Free Radic Biol Med. 2013;65:1352–1362. doi: 10.1016/j.freeradbiomed.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Nahrendorf M, Swirski FK. Lifestyle effects on hematopoiesis and atherosclerosis. Circ Res. 2015;116:884–894. doi: 10.1161/CIRCRESAHA.116.303550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu HQ, Li Q, Dong LY, Zhou Q, Wang H, Wang Y. MicroRNA-29b promotes high-fat diet-stimulated endothelial permeability and apoptosis in apoE knock-out mice by down-regulating MT-1 expression. Int J Cardiol. 2014;176:764–770. doi: 10.1016/j.ijcard.2014.07.095. [DOI] [PubMed] [Google Scholar]
- 9.Kraakman MJ, Kammoun HL, Allen TL, Deswaerte V, Henstridge DC, Estevez E, Matthews VB, Neill B, White DA, Murphy AJ, et al. Blocking IL-6 trans-signaling prevents high-fat diet-induced adipose tissue macrophage recruitment but does not improve insulin resistance. Cell Metab. 2015;21:403–416. doi: 10.1016/j.cmet.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 10.van Bussel BC, Henry RM, Ferreira I, van Greevenbroek MM, van der Kallen CJ, Twisk JW, Feskens EJ, Schalkwijk CG, Stehouwer CD. A healthy diet is associated with less endothelial dysfunction and less low-grade inflammation over a 7-year period in adults at risk of cardiovascular disease. J Nutr. 2015;145:532–540. doi: 10.3945/jn.114.201236. [DOI] [PubMed] [Google Scholar]
- 11.Romaine SP, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart. 2015;101:921–928. doi: 10.1136/heartjnl-2013-305402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreou I, Sun X, Stone PH, Edelman ER, Feinberg MW. miRNAs in atherosclerotic plaque initiation, progression, and rupture. Trends Mol Med. 2015;21:307–318. doi: 10.1016/j.molmed.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Zhang E, Lin C. MicroRNAs in tumor angiogenesis. Life Sci. 2015;136:28–35. doi: 10.1016/j.lfs.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 14.Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116:751–762. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 15.Loyer X, Mallat Z, Boulanger CM, Tedgui A. MicroRNAs as therapeutic targets in atherosclerosis. Expert Opin Ther Targets. 2015;19:489–496. doi: 10.1517/14728222.2014.989835. [DOI] [PubMed] [Google Scholar]
- 16.Nazari-Jahantigh M, Egea V, Schober A, Weber C. MicroRNA-specific regulatory mechanisms in atherosclerosis. J Mol Cell Cardiol. 2015;89:35–41. doi: 10.1016/j.yjmcc.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 17.Orenes-Piñero E, Montoro-García S, Patel JV, Valdés M, Marín F, Lip GY. Role of microRNAs in cardiac remodelling: New insights and future perspectives. Int J Cardiol. 2013;167:1651–1659. doi: 10.1016/j.ijcard.2012.09.120. [DOI] [PubMed] [Google Scholar]
- 18.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. microRNA-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Synetos A, Toutouzas K, Stathogiannis K, Latsios G, Tsiamis E, Tousoulis D, Stefanadis C. microRNAs in arterial hypertension. Curr Top Med Chem. 2013;13:1527–1532. doi: 10.2174/15680266113139990101. [DOI] [PubMed] [Google Scholar]
- 20.Hulsmans M, Holvoet P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res. 2013;100:7–18. doi: 10.1093/cvr/cvt161. [DOI] [PubMed] [Google Scholar]
- 21.Sun X, Zhang M, Sanagawa A, Mori C, Ito S, Iwaki S, Satoh H, Fujii S. Circulating microRNA-126 in patients with coronary artery disease: Correlation with LDL cholesterol. Thromb J. 2012;10:16. doi: 10.1186/1477-9560-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long G, Wang F, Duan Q, Chen F, Yang S, Gong W, Wang Y, Chen C, Wang DW. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci. 2012;8:811–818. doi: 10.7150/ijbs.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Wang F, Wu Y, Zuo L, Zhang S, Zhou Q, Wei W, Wang Y, Zhu H. MicroRNA-126 attenuates palmitate-induced apoptosis by targeting TRAF7 in HUVECs. Mol Cell Biochem. 2015;399:123–130. doi: 10.1007/s11010-015-2460-9. [DOI] [PubMed] [Google Scholar]
- 24.Franken R, den Hartog AW, de Waard V, Engele L, Radonic T, Lutter R, Timmermans J, Scholte AJ, van den Berg MP, Zwinderman AH, et al. Circulating transforming growth factor-β as a prognostic biomarker in Marfan syndrome. Int J Cardiol. 2013;168:2441–2446. doi: 10.1016/j.ijcard.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Yan F, Wang Y, Wu X, Peshavariya HM, Dusting GJ, Zhang M, Jiang F. Nox4 and redox signaling mediate TGF-β-induced endothelial cell apoptosis and phenotypic switch. Cell Death Dis. 2014;5:e1010. doi: 10.1038/cddis.2013.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frei K, Gramatzki D, Tritschler I, Schroeder JJ, Espinoza L, Rushing EJ, Weller M. Transforming growth factor-β pathway activity in glioblastoma. Oncotarget. 2015;6:5963–5977. doi: 10.18632/oncotarget.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rath D, Chatterjee M, Müller I, Müller K, Böckmann C, Droppa M, Stimpfle F, Karathanos A, Borst O, Seizer P, et al. Platelet expression of transforming growth factor beta 1 is enhanced and associated with cardiovascular prognosis in patients with acute coronary syndrome. Atherosclerosis. 2014;237:754–759. doi: 10.1016/j.atherosclerosis.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Hwang JS, Eun SY, Ham SA, Yoo T, Lee WJ, Paek KS, Do JT, Lim DS, Seo HG. PPARδ modulates oxLDL-induced apoptosis of vascular smooth muscle cells through a TGF-β/FAK signaling axis. Int J Biochem Cell Biol. 2015;62:54–61. doi: 10.1016/j.biocel.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Tian H, Liu J, Chen J, Gatza ML, Blobe GC. Fibulin-3 is a novel TGF-β pathway inhibitor in the breast cancer microenvironment. Oncogene. 2015;34:5635–5647. doi: 10.1038/onc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcantoni E, Dovizio M, Gaora OP, Di Francesco L, Bendaya I, Schiavone S, Trenti A, Guillem-Llobat P, Zambon A, Nardelli GB, et al. Dysregulation of gene expression in human fetal endothelial cells from gestational diabetes in response to TGF-β1. Prostaglandins Other Lipid Mediat. 2015;120:103–114. doi: 10.1016/j.prostaglandins.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TG-Fβ triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ Res. 2015;116:1753–1764. doi: 10.1161/CIRCRESAHA.116.305178. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Zhu HQ, Zhou Q, Jiang ZK, Gui SY, Wang Y. Association of aorta intima permeability with myosin light chain kinase expression in hypercholesterolemic rabbits. Mol Cell Biochem. 2011;347:209–215. doi: 10.1007/s11010-010-0630-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res. 2011;92:75–84. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- 35.Hu ZP, Fang XL, Fang N, Wang XB, Qian HY, Cao Z, Cheng Y, Wang BN, Wang Y. Melatonin ameliorates vascular endothelial dysfunction, inflammation, and atherosclerosis by suppressing the TLR4/NF-κB system in high-fat-fed rabbits. J Pineal Res. 2013;55:388–398. doi: 10.1111/jpi.12085. [DOI] [PubMed] [Google Scholar]
- 36.Jakob P, Doerries C, Briand S, Mocharla P, Kränkel N, Besler C, Mueller M, Manes C, Templin C, Baltes C, et al. Loss of angiomiR-126 and 130a in angiogenic early outgrowth cells from patients with chronic heart failure: Role for impaired in vivo neovascularization and cardiac repair capacity. Circulation. 2012;126:2962–2975. doi: 10.1161/CIRCULATIONAHA.112.093906. [DOI] [PubMed] [Google Scholar]