Abstract
Allopurinol, a commonly prescribed medication for gout and hyperuricemia, is a frequent cause of severe cutaneous adverse reactions (SCAR), which include the drug hypersensitivity syndrome, Stevens–Johnson syndrome, and toxic epidermal necrolysis. The adverse events are unpredictable and carry significant morbidity and mortality. To identify genetic markers for allopurinol–SCAR, we carried out a case-control association study. We enrolled 51 patients with allopurinol–SCAR and 228 control individuals (135 allopurinol-tolerant subjects and 93 healthy subjects from the general population), and genotyped for 823 SNPs in genes related to drug metabolism and immune response. The initial screen revealed strong association between allopurinol–SCAR and SNPs in the MHC region, including BAT3 (encoding HLA-B associated transcript 3), MSH5 (mutS homolog 5), and MICB (MHC class I polypeptide-related sequence B) (P < 10–7). We then determined the alleles of HLA loci A, B, C, and DRB1. The HLA-B*5801 allele was present in all (100%) 51 patients with allopurinol–SCAR, but only in 20 (15%) of 135 tolerant patients [odds ratio 580.3 (95% confidence interval, 34.4–9780.9); corrected P value = 4.7 × 10–24] and in 19 (20%) of 93 of healthy subjects [393.51 (23.23–6665.26); corrected P value = 8.1 × 10–18]. HLA alleles A*3303, Cw*0302, and DRB1*0301 were in linkage disequilibrium and formed an extended haplotype with HLA-B*5801. Our results indicated that allopurinol–SCAR is strongly associated with a genetic predisposition in Han Chinese. In particular, HLA-B*5801 allele is an important genetic risk factor for this life-threatening condition.
Keywords: adverse drug reactions, genetic polymorphism, Stevens–Johnson syndrome, toxic epidermal necrolysis, pharmacogenetics
Allopurinol is widely used for hyperuricemia-related diseases, such as gout, Lesch–Nyhan syndrome, and recurrent urate kidney stones (1). Although alternative uric-acid lowering agents, such as probenecid and sulfinpyrazone, are available at the present time, allopurinol is still the most frequently used antihyperuricemic agent because of its convenient once-daily regimen and its advantages to treat both urate overproduction and urate underexcretion (2). However, allopurinol is also one of the most frequent causes of adverse drug reactions, accounting for 5% of all cases of severe cutaneous adverse reactions (SCARs) (3).
Allopurinol–SCAR includes drug hypersensitivity syndrome (HSS), Stevens–Johnson syndrome (SJS), and toxic epidermal necrolysis (TEN). Even though rare, the mortality rate can be as high as 26% (4, 5). SJS/TEN is characterized by a rapidly developing blistering exanthema of macules and target-like lesions accompanied by mucosal involvement and skin detachment. HSS has systemic manifestations with multiorgan involvement, in addition to the exanthema (5). Currently, there are no tests that can be used to predict who will develop SCAR from allopurinol.
The manifestations of these serious, life-threatening reactions are likely to be immune-mediated, because rechallenging with the same drug typically shortens the incubation period and results in more severe manifestations. There is also evidence supporting that the pathogenesis of SCAR involves MHC-restricted presentation of drug or drug metabolites for T cell activation (6). Furthermore, cell-mediated immunity to allopurinol and its metabolite, oxypurinol, has been demonstrated in immunohistochemistry studies and in vitro lymphocyte stimulation (7).
Susceptibility to such idiosyncratic reactions is thought to be genetically determined, and familial predisposition to allopurinol hypersensitivity has been reported (8). However, the responsible genetic factors for allopurinol–SCAR have yet to be identified. As genetic factors influencing immune response and drug metabolism may be involved in the pathogenesis of the severe cutaneous adverse reactions, we hypothesize that the genetic polymorphisms of these genes might confer the susceptibility of adverse events. We aimed in this study to identify genetic markers for allopurinol–SCAR.
Methods
Patients and Control Subjects Recruitment. From 1996 to 2004, 93 individuals who fulfilled the diagnostic criteria of SJS, TEN, or HSS induced by allopurinol were identified in Chang Gung Memorial Hospital (CGMH) Health System, National Taiwan University Hospital, Taipei Medical University Hospital, and Taichung Veterans General Hospital in Taiwan. Among them, 51 patients agreed to participate in the study. All patients were assessed by two dermatologists, who reviewed photographs, pathological slides, and medical records. The diagnostic criteria of SJS/TEN were based on the clinical morphology defined by Roujeau (9). We define SJS as skin detachment of <10% of body-surface area, overlap SJS/TEN as skin detachment of 10–30%, and TEN as >30%. The criteria for HSS were cutaneous rash (e.g., diffuse macuopapular, exfoliative dermatitis), plus two of the following symptoms: eosinophilia, atypical circulating lymphocytes, leukocytosis, acute hepatocellular injury, or worsening renal function (5). In all enrolled cases, allopurinol was regarded as the offending drug if the onset of SCAR symptoms occurred within first 3 months of allopurinol exposure and the symptoms resolved upon withdrawal of the drug. Patients with an absence of symptoms after reexposure to allopurinol and patients with milder skin eruption that did not meet the criteria of HSS, SJS, and TEN were excluded.
Two control groups were used in this study. The first control group was the 135 consecutive patients who received allopurinol for at least 6 months without evidence of adverse reactions. These allopurinol-tolerant patients were recruited from the Rheumatology Clinic of the same regional hospitals where allopurinol–SCAR patients were recruited. The second control group consisted of 93 healthy subjects randomly selected from a biobank under a nationwide population study, in which 3,312 Han Chinese descendants were recruited based on the geographic distribution across Taiwan. There was no selfreport of adverse events by any of these 93 subjects. All participants were unrelated Han Chinese residing in Taiwan. There was no difference in the ethnic background among the SCAR patients, tolerant controls, and healthy subjects. The Han Chinese forms the largest ethnic group in Taiwan, making up ≈98% of the population. None of the participants were aboriginal Taiwanese, which account for the remaining 2% of the Taiwan's population.
The study was approved by the institutional review board, and informed consent was obtained from all of the participants.
SNP Genotyping. A total of 823 SNPs were selected from the National Center for Biotechnology Information's SNP database for genotyping (dbSNP build 118) (10). These included 197 SNPs from 4 Mb of the MHC region on chromosome 6p21.3 (11) and 626 SNPs selected from genes encoding immune-related molecules and drug metabolizing enzymes (for SNP ID, gene symbol, and chromosome position, see Table 4, which is published as supporting information on the PNAS web site). The 197 SNPs selected in the MHC region included all 201 SNPs reported by Walsh et al. (11), except for the four SNPs that failed to be genotyped (rs1264314, rs1264440, rs1755038, and rs589428). The other 626 SNPs included genes of apoptosis-related molecules (death-associated proteins, caspases), immune mediators (complement components, immune cell receptors, cytokines), and drug transporters (ATP-binding cassettes). We specifically included genes involved in allopurinol metabolism, which included xanthine oxidase (43 SNPs), aldehyde oxidase (24 SNPs), purine nucleoside phosphorylase, and hypoxanthine phosphoribosyl transferase.
Genomic DNA was isolated by using PUREGENE DNA purification system (Gentra Systems). SNP genotyping was performed by using high-throughput MALDI-TOF mass spectrometry. Briefly, primers and probes were designed by using SpectroDESIGNER software (Sequenom, San Diego). Multiplex PCRs were performed, and unincorporated dNTPs were dephosphorylated by using shrimp alkaline phosphatase (Hoffman-LaRoche, Basel) followed by primer extension. The purified primer extension reaction was spotted onto a 384-element silicon chip (SpectroCHIP, Sequenom) and analyzed in the Bruker Biflex III MALDI-TOF SpectroREADER mass spectrometer (Sequenom), and the resulting spectra processed with SpectroTYPER (Sequenom).
HLA Genotyping. HLA alleles A, B, C, and DRB1, were determined by sequence-specific oligonucleotide reverse lineblot (Dynal, Bromborough, U.K.) (12). Potential ambiguities were resolved by sequencing-based typing (13). The primers used for HLA-B allele typing were: Bin1-TA-M13F, 5′-TGTAAAACGACGGCCAGTGGCGGGGGCGCAGGACCTGA-3′ (forward); Bin1-CGM13F, 5′-TGTAAAACGACGGCCAGTCGGGGGCGCAGGACCCGG-3′ (forward); and Bin3-M13R, 5′-CAGGAAACAGCTATGACCGGAGGCCATCCCCGGCGACCTAT-3′. Primers for HLA-A allele typing were: Ain1-A-M13F, 5′-TGTAAAACGACGGCCAGTGGGGCGCARGACCCGGGA-3′ (forward); Ain1-G-M13F, 5′-TGTAAAACGACGGCCAGTGGGCGCAGGACCGGGG-3′ (forward); Ain1-T-M13F, 5′-TGTAAAACGACGGCCAGTGGGRCGCAGGACCCGGGT-3′ (forward); and Ain3–62-M13R, 5′-CAGGAAACAGCTATGACCGTCCCAATTGTCTCCCCTCCTT-3′ (reverse). Primers for HLA-C allele typing were: 5Cln1–61, 5′-AGCGAGGKGCCCGCCCGGCGA-3′ (forward); and 3BCln3–12, 5′GGAGATGGGGAAGGCTCCCCACT-3′ (reverse). The DRB1 alleles are separated into eight groups, DR1, DR2, DR3/11/6, DR4, DR7, DR8/12, DR9, and DR10, for sequencing-based typing as described (14). The PCR was set up in a volume of 16 μl, containing 50–100 ng of genomic DNA, 1× PCR buffer [16 mM (NH4)2SO4/67 mM Tris·HCl, pH 8.8/0.01% Tween 20], 2.5 mM MgCl2, 200 μM each of dNTP, 5% DMSO, 0.02 mg/ml cresol red, and 0. 5 units of EuroTaq polymerase (Euroclone, Spa, Milan). Cycling was carried out by using the described sequencing-based typing protocol (13, 14).
Statistical Analysis. Categorical data were compared between groups with use of Fisher's exact tests, and continuous data (presented as median, with range given in parentheses) were compared with use of t tests. All P values were two-tailed; a P value of <0.05 was considered to indicate statistical significance. Allelic association screen was carried out by the Cochran–Armitage Trend test for each SNP (15). Odds ratios were calculated with Haldane's modification, which adds 0.5 to all cells to accommodate possible zero counts (16). The corrected P (Pc) values were adjusted by using Bonferroni's correction for multiple comparisons to account for the observed alleles (17 for HLA-A, 40 for HLA-B, 19 for HLA-C, and 30 for HLA-DRB1). Therefore, the Pc values were multiplied by a factor of 387,600.
Results
Characteristics of Patients and Controls. The clinical manifestations of the 51 patients with allopurinol–SCAR, which included SJS (13 cases), SJS/TEN (5 cases), TEN (3 cases), and HSS (30 cases), are summarized in Table 1. The onset of symptoms for all patients was within the first 3 months of allopurinol exposure, and three patients had a second attack within 2 days of reexposure (Table 1). Twenty-one patients received multiple drugs in addition to the allopurinol, but their medical records revealed no adverse drug reactions when these concomitant medications were taken without allopurinol. All patients received allopurinol because of hyperuricemia with or without gouty arthritis (Table 2).
Table 1. Clinical characteristics of patients with allopurinol-induced SCAR.
| Patient no. | Age/sex | Phenotype | Dose, mg/d/duration of drug exposure, days | Exanthema %/blister or detachment,† % | Mucous membrane erosions, number:site | Systemic manifestations |
|---|---|---|---|---|---|---|
| 1 | 80/F | SJS | 100/33 | 31-50/2 | (+) 2: Oral, eyes | None |
| 2 | 47/F | HSS | 100/23 | 31-50/0 | (+) 2: Oral, eyes | Fever, LFI, worsening RF |
| 3 | 60/F | SJS | 100/14 | 31-50/5 | (+) 2: Oral, eyes | Fever, worsening RF, leukocytosis, atypical lymphocytosis§ |
| 4 | 25/M | SJS | 200/24 | 51-70/5 | (+) 1: Oral | Fever, LFI, leukocytosis, eosinophilia |
| 5 | 50/F | HSS | 300/33 | 51-70/0 | (+) 1: Oral | Fever, LFI, eosinophilia |
| 6 | 74/F | HSS | 100/22 | 31-50/0 | (+) 2: Oral, eyes | Fever, worsening RF, eosinophilia, atypical lymphocytosis |
| 7 | 78/M | HSS | NA/21 | 71-90/0 | (-) | Worsening RF, eosinophilia |
| 8 | 37/F | SJS/TEN | 100/38 | 51-70/10 | (+) 3: Oral, eyes, genital | Fever, LFI, leukocytosis, eosinophilia |
| 9 | 18/F | HSS | 200/21 | 51-70/0 | (-) | LFI, worsening RF, leukocytosis, eosinophilia |
| 10 | 63/F | SJS | 200/37 | 51-70/5 | (+) 2: Oral, genital | LFI, worsening RF, eosinophilia |
| 11 | 55/F | HSS | 100/35 | 51-70/0 | (-) | Fever, LFI, leukocytosis |
| 12 | 70/F | HSS | NA/56 | 31-50/0 | (+) 2: Oral, eyes | Fever, LFI, leukocytosis, eosinophilia |
| 13 | 52/F | SJS | 100/35 | 51-70/5 | (+) 3: Oral, eyes, genital | Fever, LFI, eosinophilia |
| 14 | 29/F | SJS | NA/28 | 51-70/5 | (+) 2: Oral, eyes | None |
| 15 | 73/M | SJS | 200/30 | 51-70/1 | (+) 3: Oral, eyes, genital | Fever |
| 16 | 78/M | SJS | 100/1‡ | 51-70/1 | (+) 1: Oral | Worsening RF, eosinophilia |
| 17 | 70/M | SJS/TEN | NA/27 | 51-70/10 | (+) 2: Oral, eyes | Fever |
| 18 | 69/F | HSS | 100/53 | 31-50/0 | (+) 1: Oral | LFI, worsening RF, eosinophilia |
| 19 | 91/F | TEN | 100/26 | 71-90/30 | (+) 2: Oral, eyes | Atypical lymphocytosis |
| 20§ | 60/M | SJS/TEN | 200/21 | 51-70/15 | (+) 2: Oral, eyes | Fever, LFI, worsening RF, eosinophilia |
| 21 | 77/F | SJS | NA/14 | 51-70/5 | (+) 2: Oral, genital | Worsening RF |
| 22 | 62/F | HSS | 100/38 | 51-70/0 | (+) 2: Oral, eyes | Fever, LFI, worsening RF |
| 23 | 41/M | HSS | 200/32 | 51-70/0 | (+) 1: Oral | LFI, leukocytosis |
| 24 | 72/F | HSS | 100/45 | >90/0 | (+) 2: Oral, eyes | Fever, LFI, worsening RF |
| 25 | 51/M | SJS | 200/2‡ | 51-70/5 | (+) 3: Oral, eyes, genital | None |
| 26 | 35/M | HSS | NA/30 | 31-50/0 | (-) | Fever, leukocytosis, eosinophilia |
| 27 | 65/F | HSS | 300/49 | 51-70/0 | (-) | Leukocytosis, eosinophilia |
| 28§ | 54/F | SJS | 100/14 | 51-70/3 | (+) 2: Oral, eyes | Fever, worsening RF, eosinophilia |
| 29 | 51/M | SJS | NA/21 | 51-70/5 | (+) 1: Oral | Fever, LFI, atypical lymphocytosis |
| 30 | 52/F | HSS | NA/45 | 51-70/0 | (+) 1: Oral | Fever, LFI, leukocytosis, severe myositis |
| 31 | 50/M | HSS | 50/15 | 31-50/0 | (-) | LFI, worsening RF |
| 32 | 80/M | SJS | NA/12 | 31-50/5 | (+) 2: Oral, genital | None |
| 33 | 80/M | HSS | 100/21 | 31-50/0 | (-) | LFI, worsening RF, leukocytosis, eosinophilia |
| 34 | 70/M | HSS | 50/18 | 31-50/0 | (-) | LFI, leukocytosis, eosinophilia, atypical lymphocytosis |
| 35 | 52/M | HSS | 100/7 | 71-90/0 | (-) | Leukocytosis, eosinophilia |
| 36 | 63/M | HSS | 100/52 | 31-50/0 | (-) | LFI, eosinophilia |
| 37 | 67/F | HSS | 100/21 | 51-70/0 | (-) | Fever, worsening RF, eosinophilia |
| 38 | 55/M | HSS | 100/14 | 51-70/0 | (-) | Fever, worsening RF, eosinophilia |
| 39§ | 66/F | TEN | 100/23 | 71-90/30 | (+) 3: Oral, eyes, genital | Leukopenia |
| 40 | 62/M | HSS | 300/30 | 71-90/0 | (-) | LFI, worsening RF, eosinophilia |
| 41 | 73/F | HSS | 100/39 | 71-90/0 | (+) 2: Oral, eyes | Fever, worsening RF, eosinophilia |
| 42 | 84/M | HSS | 100/30 | 51-70/0 | (-) | Worsening RF, eosinophilia |
| 43 | 24/M | HSS | NA/28 | 51-70/0 | (-) | Fever, worsening RF, eosinophilia |
| 44§ | 78/F | HSS | 100/30 | 71-90/0 | (+) 1: Oral | Fever, worsening RF, eosinophilia, leukocytosis |
| 45 | 69/F | HSS | 100/26 | 51-70/0 | (-) | Worsening RF, leukocytosis, eosinophilia |
| 46 | 73/M | SJS/TEN | 100/1‡ | 51-70/10 | (+) 3: Oral, eyes, genital | LFI, worsening RF |
| 47§ | 73/F | HSS | 100/30 | 71-90/0 | (+) 1: Oral | Fever, LFI, worsening RF, leukocytosis, eosinophilia |
| 48 | 70/F | SJS/TEN | 200/12 | 51-70/15 | (+) 2: Oral, eyes | Fever, leukocytosis |
| 49 | 70/M | HSS | 100/20 | 51-70/0 | (+) 3: Oral, eyes, genital | LFI, worsening RF, eosinophilia, atypical lymphocytosis |
| 50 | 71/M | TEN | 100/26 | 71-90/30 | (+) 1: Oral | Fever, LFI, worsening RF |
| 51 | 71/M | HSS | 100/7 | 51-70/0 | (+) 1: Oral | Fever, LFI, worsening RF, eosinophilia |
See Methods for the diagnostic criteria. NA, data not available; RF, renal function; LFI, liver function impairment; M, male; F, female.
Exanthema, erythematous or purpuric macules/papules; blister or detachment, extent of blisters or epidermal detachment; both exanthema and blister/detachment were expressed as % of total body surface area.
Second attack on reexposure.
Cases deceased.
Table 2. Demographic variables, dosage and duration of allopurinol, and clinical characteristics in allopurinol—SCAR patients and tolerant patients.
| Allopurinol—SCAR (n = 51) | Allopurinol tolerant (n = 135) | P value* | |
|---|---|---|---|
| Characteristic | |||
| Median age (range), yr | 66 (18-91) | 56 (21-84) | 0.0821† |
| Sex, n (%) | 6.6 × 10-11 | ||
| Male | 24 (47.1) | 125 (92.6) | |
| Female | 27 (52.9) | 10 (7.4) | |
| Allopurinol exposure, median (range) | |||
| Dosage, mg/day | 100 (50-300)‡ | 150 (100-400) | 0.0573† |
| Duration | 26 (≈1-56) days | 22 (6-107) months | <0.0001† |
| Use of thiazide diuretics, n (%) | 2 (5.4) | 4 (2.9) | 0.67 |
| Underlying diseases, n (%) | |||
| Hyperuricemia | 51 (100) | 135 (100) | — |
| Gouty arthritis | 32 (62.7) | 124 (91.9) | 6.5 × 10-6 |
| Chronic renal insufficiency§ | 28 (54.9) | 28 (20.7) | 1.2 × 10-5 |
| Hypertension | 25 (49) | 79 (58.5) | 0.2521 |
| Diabetes | 15 (29.4) | 24 (17.8) | 0.1057 |
| Liver disease¶ | 7 (13.7) | 22 (16.3) | 0.8217 |
| Coronary artery disease | 8 (15.7) | 9 (6.7) | 0.0836 |
| Congestive heart failure | 4 (7.8) | 5 (3.7) | 0.2608 |
| Systemic lupus erythematosus | 1 (2) | 1 (1) | 0.4743 |
| Asthma | 4 (7.8) | 5 (3.7) | 0.2608 |
Unless otherwise noted, Fisher's two-tailed exact tests were used for comparisons between groups.
t test was used.
Data available on 41 patients.
Chronic renal insufficiency was defined as serum creatinine level ≥ 1.7 mg/dl for >6 months.
Liver diseases included chronic hepatitis or liver cirrhosis.
A total of 135 hyperuricemia/gouty arthritis patients who had been on allopurinol for at least 6 months (median, 22 months; range, 6–107 months) with no evidence of adverse events were used as the allopurinol-tolerant control. The sex distribution of the tolerant group is comparable to the general prevalence of gout with a male/female ratio ranging from 7:1 to 9:1 (17), but differed from the allopurinol–SCAR group. The difference can be explained by the fact that there were fewer patients with gouty arthritis in the SCAR group than those in the tolerant group. Chronic renal insufficiency, hypertension, diabetes, liver disease, coronary artery disease, congestive heart failure, systemic lupus erythematous, and asthma were present in both allopurinol–SCAR and tolerant patients. The frequencies of diseases were not significantly different in the two groups except for chronic renal insufficiency, which was seen more frequently in the allopurinol–SCAR group (P < 0.0001; Table 2). The presence of chronic renal insufficiency increased the risk for allopurinol–SCAR [odds ratio, 4.7; 95% confidence interval (CI), 2.3–9.3]. The frequencies of patients taking thiazide diuretics were not significantly different in the two groups, although concomitant use of the thiazide has been reported to be a risk factor for allopurinol–SCAR (18). In addition to the tolerant control group, 93 subjects who were randomly selected from the general population of Taiwan were served as an additional control.
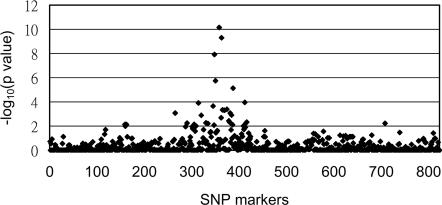
Association Screen for Candidate Gene SNPs. We initially screened 30 patients with allopurinol–SCAR and 60 tolerant subjects for SNP association. Of the total of 823 SNPs screened (197 SNPs in the MHC region and 626 SNPs of immune-related genes and drug metabolizing genes), 29 SNPs in the MHC region were found to have significant association (P < 0.01) with the allopurinol–SCAR patients when compared to the tolerant group (Fig. 1). In particular, three SNPs in the MHC class III region have P values <10–7 (–log10 P value > 7, Fig. 1). These SNPs were rs3117583 of BAT3 (encoding for HLA-B associated transcript 3), rs1150793 of MSH5 (encoding mutS homolog), and rs2855804 of MICB (encoding MHC class I polypeptide-related sequence).
Fig. 1.
Screening of candidate SNPs for association with allopurinol-induced SCAR. On the x axis, 823 SNPs are ordered by their chromosome positions; 197 SNPs in the MHC region are those numbered from 260 to 456. On the y axis, the –log10 P values were calculated by comparison of the genotype frequencies between the allopurinol–SCAR patients and tolerant group.
Outside the MHC region, only three SNPs showed potential associations (P < 0.01). Two SNPs (rs2268791 and rs1594) were located in the CASP8 and FADD-like apoptosis regulator (CFLAR) gene on chromosome 2, and one SNP (rs2304224) belongs to KIR2DL1 (a killer cell Ig-like receptor) gene on chromosome 19. No other significant associations were found in the other SNPs, including the SNPs of allopurinol drug metabolizing enzymes (see Table 4 for the P value of each SNP).
To replicate the initial screening results, we genotyped additional 21 patients with allopurinol–SCAR, 75 allopurinoltolerant controls, and 93 subjects from the general population for all of the SNPs that had P values <0.01. We confirmed the initial observation of the positive association of allopurinol–SCAR with the MHC and CFLAR loci, but not with the KIR2D1 locus.
HLA Allele Frequency. Because the most significant association was seen with the SNPs in the MHC region, we genotyped the individual HLA alleles. As shown in Table 3, alleles HLA-A*3303, B*5801, Cw*0302, and DRB1*0301 occurred at significantly increased frequencies among the allopurinol–SCAR patients compared to the two control groups. In particular, the HLA-B*5801 was present in all 51 (100%) patients with allopurinol–SCAR, but in only 15% (20 of 135) of the allopurinoltolerant group (odds ratio, 580.3; 95% CI, 34.4–9780.9; Pc = 4.7 × 10–24), and 20% (19 of 93) of the general population (odds ratio 393.5; 95% CI, 23.2–6665.26; Pc = 8.1 × 10–18). This association was only seen with allopurinol–SCAR and not with the patients' underlying diseases, such as gout, renal insufficiency, or autoimmune disease, etc. (data not shown).
Table 3.
Frequencies of individual or combined loci of HLA-B* 5801 extended haplotype in patients with allopurinol-induced SCAR, allopurinol tolerant control, and general population control
| Genotype | Allopurinol-SCAR (n = 51) | Tolerant control (n = 135) | Odds ratio | Pc value* | General population control (n = 93) | Odds ratio | Pc value* |
|---|---|---|---|---|---|---|---|
| B* 5801 | 51 (100) | 20 (15) | 580.3 | 4.7 × 10-24 | 19 (20) | 393.5 | 8.1 × 10-18 |
| Cw* 0302 | 48 (94) | 19 (14) | 97.7 | 1.4 × 10-19 | 19 (20) | 62.3 | 2.5 × 10-13 |
| A* 3303 | 34 (67) | 24 (18) | 9.3 | 2.2 × 10-4 | 20 (22) | 7.3 | 4.7 × 10-2 |
| DRB1* 0301 | 33 (65) | 17 (13) | 12.7 | 2.8 × 10-6 | 14 (15) | 10.3 | 8.5 × 10-4 |
| B* 5801, Cw* 0302 | 48 (94) | 19 (14) | 97.7 | 1.4 × 10-19 | 19 (20) | 62.3 | 2.6 × 10-13 |
| B* 5801, Cw* 0302, A* 3303 | 34 (67) | 17 (13) | 13.9 | 5.4 × 10-7 | 16 (17) | 9.6 | 1.7 × 10-3 |
| B* 5801, Cw* 0302, DRB1* 0301 | 30 (59) | 11 (8) | 16.1 | 7.4 × 10-7 | 10 (11) | 11.9 | 7.8 × 10-4 |
| B* 5801, Cw* 0302, A* 3303, DRB1* 0301 | 21 (41) | 9 (7) | 9.8 | 0.039 | 9 (10) | 6.5 | >0.05 |
Numbers in parentheses indicate percentage.
The Pc values were adjusted by using Bonferroni's correction for multiple comparisons to account for the observed alleles.
Guided by five patients (nos. 5, 14, 18, 19, and 35) who were homozygous for the HLA-B*5801 alleles, we analyzed the allele distribution of the combined HLA loci and defined the extended haplotype. The HLA-B*5801 extended haplotype was formed by conserved alleles at closely linked loci as HLA-A*3303-Cw*0302-B*5801-DRB1*0301. This extended haplotype was present in 21 (41%) of the 51 patients with allopurinol–SCAR (Table 3), 7% of the tolerant patients, and 10% of the healthy subjects. This extended haplotype has been reported in Taiwanese in a study of nasopharyngeal carcinoma (19).
Discussion
To our knowledge, this is the largest pharmacogenetic study of allopurinol–SCAR. In this study, we identified a strong association of the allele HLA-B*5801 with the susceptibility of allopurinol-induced HSS, SJS, and TEN in Han Chinese. In fact, the association is 100% in that the HLA-B allele B*5801 was present in all 51 patients with allopurinol-induced SCAR, with an odds ratio exceeding that reported for the association between HLA-B27 and ankylosing spondylitis (20, 21). Although other ethnic allopurinol–SCAR patients were not available for our study, the fact that HLA-B*5801 allele is also present in other populations (7% in Africa, ≈2–7% in Caucasian, and 8% in Asian Indian) (22) suggests that this association may also exist in other ethnic groups. However, as pharmacogenetic results can vary by the study populations, it remains to be seen to what extent this study applies to other populations.
A relation between allopurinol–SCAR and decreased creatinine clearance has been documented, and adjusting the dose of allopurinol based on the creatinine clearance has been proposed (23). Our data showed that patients with chronic renal insufficiency had an increased risk for allopurinol–SCAR (4.7-fold). The drug-accumulation hypothesis has been proposed to explain the association of renal insufficiency with allopurinol hypersensitivity (5). However, adjusting allopurinol dosage has no significant effect on reducing allopurinol hypersensitivity (24). Moreover, lowering dosage may lead to inefficient reduction of serum uric acid levels for effective control of gout (25). Our data suggest that genotyping the B*5801 allele may provide a better guidance of avoiding allopurinol-induced SCAR in patients with renal insufficiency than dosage adjusting.
Associations of particular HLA alleles with adverse events have been reported in patients taking various drugs. For example, HLA-A29, B12, and DR7 were found more frequently in sulfonamide-induced SJS; A2 and B12 were found more frequently in NSAIDs-induced SJS (26, 27); HLA-B59 was found more frequently in SJS with ocular involvement (28); and HLA-AW33 and -B17/BW58 were found more frequently in allopurinol drug eruption (29). However, none of the associations have a sufficiently high predictive value that can be clinically useful for screening before drugs are prescribed. The weak associations in the previous studies could be due to differences in patient ascertainment and/or lack of a precise genotyping method for HLA allele subtypes at that time.
We have recently reported a strong association of a specific HLA-B allele (B*1502) in carbamazepine induced SJS (30). Hypersensitivity to abacavir, a drug for AIDS, was reported to be associated with another HLA-B allele (B*5701) and a haplotypic Hsp70-Hom variant in the MHC region (31, 32). This study, together with the previous reports, suggests that HLA-B alleles and/or other genetic polymorphisms in the MHC region might play a major role in the pathogenesis of immune-mediated SCAR. A specific HLA-B molecule may function as antigenic presentation of certain drug or its metabolite for HLA restricted T cell activation. Further studies along these lines may increase our understanding of the pathogenesis of these potentially life-threatening clinical conditions.
Although all allopurinol–SCAR patients in our study carried at least one HLA-B*5801 allele, there were tolerant patients who also carried the allele. This finding suggests that HLA-B*5801 is necessary but not sufficient for allopurinol–SCAR. There could be other cofactors, such as renal insufficiency or virus infection, which have been implicated to be risk factors for the development of allopurinol–SCAR (5, 33). Our findings provided the evidence that renal insufficiency is indeed a risk factor for allopurinol–SCAR. Furthermore, other genes may also be involved in the mechanism of pathogenesis, such as costimulatory molecules involved in the interaction between antigen-presenting cells and T cell interaction. The present study revealed that CFLAR, an apoptosis regulator, could also be involved in allopurinol–SCAR (34–36).
In summary, we have shown that allopurinol-induced severe cutaneous adverse reactions are associated with a strong genetic predisposition. Genetic polymorphisms in the MHC region, particularly the HLA-B*5801 allele, is highly associated with individuals who are at risk for allopurinol-induced HSS, SJS, or TEN.
Supplementary Material
Acknowledgments
This research project was supported by grants from the National Science and Technology Program for Genomic Medicine, National Science Council, Taiwan (National Clinical Core and National Genotyping Core), and the Genomics and Proteomics Program, Academia Sinica.
Author contributions: S.-I.H., W.-H.C., P.-C.L., K.-H.W., J.-Y.W., and Y.-T.C. designed research; S.-I.H., W.-H.C., L.-B.L., C.-C.C., M.L., H.-P.H., Y.-L.L., J.-L.L., L.-C.Y., H.-S.H., M.-J.C., M.-S.W., C.-Y.C., and K.-H.W. performed research; S.-I.H., W.-H.C., C.-C.C., M.L., P.-C.L., C.-Y.C., C.-H.C., C.S.F., and Y.-T.C. analyzed data; and S.-I.H., W.-H.C., J.-Y.W., and Y.-T.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SCAR, severe cutaneous adverse reaction; HSS, hypersensitivity syndrome; SJS, Stevens–Johnson syndrome; TEN, toxic epidermal necrolysis; Pc, corrected P.
References
- 1.Wortmann, R. L. (2002) Curr. Opin. Rheumatol. 14, 281–286. [DOI] [PubMed] [Google Scholar]
- 2.Terkeltaub, R. A. (2003) N. Engl. J. Med. 349, 1647–1655. [DOI] [PubMed] [Google Scholar]
- 3.Roujeau, J. C., Kelly, J. P., Naldi, L., Rzany, B., Stern, R. S., Anderson, T., Auquier, A., Bastuji-Garin, S., Correia, O., Locati, F., et al. (1995) N. Engl. J. Med. 333, 1600–1607. [DOI] [PubMed] [Google Scholar]
- 4.Aubock, J. & Fritsch, P. (1985) Br. Med. J. 290, 1969–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arellano, F. & Sacristan, J. A. (1993) Ann. Pharmacother. 27, 337–343. [DOI] [PubMed] [Google Scholar]
- 6.Zanni, M. P., von Greyerz, S., Schnyder, B., Brander, K. A., Frutig, K., Hari, Y., Valitutti, S. & Pichler, W. J. (1998) J. Clin. Invest. 102, 1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braden, G. L., Warzynski, M. J., Golightly, M. & Ballow, M. (1994) Clin. Immunol. Immunopathol. 70, 145–151. [DOI] [PubMed] [Google Scholar]
- 8.Melsom, R. D. (1999) Rheumatology (Oxford) 38, 1301. [DOI] [PubMed] [Google Scholar]
- 9.Roujeau, J. C. (1994) J. Invest. Dermatol. 102, 28S–30S. [DOI] [PubMed] [Google Scholar]
- 10.Smigielski, E. M., Sirotkin, K., Ward, M. & Sherry, S. T. (2000) Nucleic Acids Res. 28, 352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh, E. C., Mather, K. A., Schaffner, S. F., Farwell, L., Daly, M. J., Patterson, N., Cullen, M., Carrington, M., Bugawan, T. L., Erlich, H., et al. (2003) Am. J. Hum. Genet. 73, 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begovich, A. B. & Erlich, H. A. (1995) J. Am. Med. Assoc. 273, 586–591. [PubMed] [Google Scholar]
- 13.Tilanus, M. G. J., Hansen, J. A. & Hurley, C. K., eds. (2002) Genomic Analysis of the Human MHC: DNA-Based Typing for HLA Alleles and Linked Polymorphisms (International Histocompatibility Working Group, Seattle), technical manual.
- 14.Chu, C. C., Lin, M., Nakajima, F., Lee, H. L., Chang, S. L., Juji, T. & Tokunaga, K. (2001) Tissue Antigens 58, 9–18. [DOI] [PubMed] [Google Scholar]
- 15.Schaid, D. J. & Jacobsen, S. J. (1999) Am. J. Epidemiol. 149, 706–711. [DOI] [PubMed] [Google Scholar]
- 16.Haldane, J. B. (1956) Ann. Hum. Genet. 20, 309–311. [DOI] [PubMed] [Google Scholar]
- 17.Kramer, H. M. & Curhan, G. (2002) Am. J. Kidney Dis. 40, 37–42. [DOI] [PubMed] [Google Scholar]
- 18.Young, J. L., Jr., Boswell, R. B. & Nies, A. S. (1974) Arch. Intern. Med. 134, 553–558. [DOI] [PubMed] [Google Scholar]
- 19.Hildesheim, A., Apple, R. J., Chen, C. J., Wang, S. S., Cheng, Y. J., Klitz, W., Mack, S. J., Chen, I. H., Hsu, M. M., Yang, C. S., et al. (2002) J. Natl. Cancer Inst. 94, 1780–1789. [DOI] [PubMed] [Google Scholar]
- 20.Schlosstein, L., Terasaki, P. I., Bluestone, R. & Pearson, C. M. (1973) N. Engl. J. Med. 288, 704–706. [DOI] [PubMed] [Google Scholar]
- 21.Brewerton, D. A., Hart, F. D., Nicholls, A., Caffrey, M., James, D. C. & Sturrock, R. D. (1973) Lancet 1, 904–907. [DOI] [PubMed] [Google Scholar]
- 22.Geer, L., Terasaki, P. I. & Gjertson, D. W. (1998) in HLA 1998, eds. Gjertson, D.W. & Terasaki, P.I. (Am. Soc. Histocomp. Immunogenet., Mt. Laurel, NJ), pp. 352–353.
- 23.Hande, K. R., Noone, R. M. & Stone, W. J. (1984) Am. J. Med. 76, 47–56. [DOI] [PubMed] [Google Scholar]
- 24.Vazquez-Mellado, J., Morales, E. M., Pacheco-Tena, C. & Burgos-Vargas, R. (2001) Ann. Rheum. Dis. 60, 981–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emmerson, B. T. (1996) N. Engl. J. Med. 334, 445–451. [DOI] [PubMed] [Google Scholar]
- 26.Roujeau, J. C., Bracq, C., Huyn, N. T., Chaussalet, E., Raffin, C. & Duedari, N. (1986) Tissue Antigens 28, 251–254. [DOI] [PubMed] [Google Scholar]
- 27.Roujeau, J. C., Huynh, T. N., Bracq, C., Guillaume, J. C., Revuz, J. & Touraine, R. (1987) Arch. Dermatol. 123, 1171–1173. [PubMed] [Google Scholar]
- 28.Shirato, S., Kagaya, F., Suzuki, Y. & Joukou, S. (1997) Arch. Ophthalmol. 115, 550–553. [DOI] [PubMed] [Google Scholar]
- 29.Chan, S. H. & Tan, T. (1989) Dermatologica 179, 32–33. [DOI] [PubMed] [Google Scholar]
- 30.Chung, W. H., Hung, S. I., Hong, H. S., Hsih, M. S., Yang, L. C., Ho, H. C., Wu, J. Y. & Chen, Y. T. (2004) Nature 428, 486. [DOI] [PubMed] [Google Scholar]
- 31.Mallal, S., Nolan, D., Witt, C., Masel, G., Martin, A. M., Moore, C., Sayer, D., Castley, A., Mamotte, C., Maxwell, D., et al. (2002) Lancet 359, 727–732. [DOI] [PubMed] [Google Scholar]
- 32.Martin, A. M., Nolan, D., Gaudieri, S., Almeida, C. A., Nolan, R., James, I., Carvalho, F., Phillips, E., Christiansen, F. T., Purcell, A. W., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki, Y., Inagi, R., Aono, T., Yamanishi, K. & Shiohara, T. (1998) Arch. Dermatol. 134, 1108–1112. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasula, S. M., Ahmad, M., Ottilie, S., Bullrich, F., Banks, S., Wang, Y., Fernandes-Alnemri, T., Croce, C. M., Litwack, G., Tomaselli, K. J., et al. (1997) J. Biol. Chem. 272, 18542–18545. [DOI] [PubMed] [Google Scholar]
- 35.Abe, R., Shimizu, T., Shibaki, A., Nakamura, H., Watanabe, H. & Shimizu, H. (2003) Am. J. Pathol. 162, 1515–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gati, A., Guerra, N., Gaudin, C., Da Rocha, S., Escudier, B., Lecluse, Y., Bettaieb, A., Chouaib, S. & Caignard, A. (2003) Cancer Res. 63, 7475–7582. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.