SUMMARY
Transient receptor potential (TRP) proteins are a large family of polymodal nonselective cation channels. The TRP vanilloid (TRPV) subfamily consists of six homologous members with diverse functions. TRPV1–TRPV4 are nonselective cation channels proposed to play a role in nociception, while TRPV5 and TRPV6 are involved in epithelial Ca2+ homeostasis. Here we present the cryo-electron microscopy (cryo-EM) structure of functional, full-length TRPV2 at 13.6 Å resolution. The map reveals that the TRPV2 cytoplasmic domain displays a 4-fold petal-like shape in which high-resolution N-terminal ankyrin repeat domain (ARD) structures can be unambiguously fitted. Fitting of the available ARD structures for other TRPV subfamily members into the TRPV2 EM map suggests that TRPV subfamily members have highly homologous structural topologies. These results allowed us to postulate a structural explanation for the functional diversity among TRPV channels and their differential regulation by proteins and ligands.
INTRODUCTION
Transient receptor potential (TRP) proteins are a distinct family of Ca2+-permeable nonselective cation channels originally identified to function in sensory transduction pathways (Venkatachalam and Montell, 2007). Recently, TRP channels also have been implicated in a diverse range of cellular processes, including neuronal development, angiogenesis, cardiac and pulmonary function, and cancer (Preti et al., 2012; Santoni and Farfariello, 2011; Shibasaki et al., 2010; Vennekens, 2011). The mammalian TRP family is subcategorized into six major branches based on amino acid sequence homology: vanilloid (TRPV), canonical, melastatin, ankyrin, mucolipin, and polycystin. They are predicted to function as tetramers, with each monomer consisting of six transmembrane helices (TM1–TM6) and varying sizes of cytoplasmic amino- and carboxy termini (Li et al., 2011; Venkatachalam and Montell, 2007). Most TRP channels are polymodal receptors that are activated and modulated by a variety of chemical and physical stimuli (Baez-Nieto et al., 2011).
The mammalian TRPV subfamily, consisting of six members, is the most extensively characterized subfamily of TRP channels (Vennekens et al., 2008). TRPV channels are predicted to contain six homologous N-terminal ankryin repeats per monomer and a C-terminal TRP box domain (Gaudet, 2008; Ramsey et al., 2006). TRPV1–TRPV4 are nonselective Ca2+-permeable cation channels known as thermoTRPVs due to their putative role in temperature and pain sensation (Venkatachalam and Montell, 2007). TRPV5 and TRPV6 are highly Ca2+-selective channels restricted to epithelial cells (Venkatachalam and Montell, 2007).
Knockout of TRPV5 and TRPV6 validated their essential role in epithelial Ca2+ handling, while genetic ablation of TRPV1 confirmed its role in pain and temperature detection in rodents (Caterina et al., 2000). Genetic deletion of TRPV2–TRPV4 had very little effect on sensory transduction (Huang et al., 2011; Park et al., 2011). Nevertheless, TRPV2 and TRPV4 have been shown to play an important role in skeletal, cardiac, and neuronal physiology, whereas TRPV3 functions in keratinocytes. Despite nearly 50% sequence homology among the TRPV channels, the origin of their functional diversity remains an intriguing question.
One of the first clues toward understanding the functional diversity of TRPV channels came from the structural studies of the N-terminal ankryin repeat domains (ARDs; Phelps et al., 2008). Ankyrin repeats are the most common protein domain structures, consisting of 33 amino acids that form an antiparallel helix-turn-helix motif followed by a β-hairpin loop (Li et al., 2006). ARDs interact with proteins and ligands via a concave ligand binding surface, which consists of the β turn and the first α helix from a varying number of repeats (Binz et al., 2004; Nakamura et al., 2007). Atomic resolution structures of TRPV1, TRPV2, TRPV4, and TRPV6 ARDs have given insight into diverse regulation of TRPV channels by endogenous ligands such as ATP (Lishko et al., 2007; Phelps et al., 2008, 2010). The overall structures of TRPV ARDs are similar; however, TRPV1, TRPV3, and TRPV4 bind ATP at the ARD concave ligand-binding surface, whereas TRPV2, TRPV5, and TRPV6 do not (Phelps et al., 2010). Functional data have shown that ATP sensitizes TRPV1 and TRPV4, prevents sensitization of TRPV3, and has no direct effect on TRPV2 and TRPV6 activity (Al-Ansary et al., 2010; Lishko et al., 2007; Phelps et al., 2010).
In addition, Ca2+ differentially modulates the activity of TRPV channels. TRPV1 and TRPV3 display Ca2+-dependent desensitization, which depends on Ca2+-calmodulin binding to the ARDs and a C-terminal binding site, while TRPV4 activity is potentiated by Ca2+-calmodulin binding at homologous sites (Gaudet, 2009; Lau et al., 2012; Lishko et al., 2007; Phelps et al., 2010; Xiao et al., 2008). Calmodulin is also involved in Ca2+-dependent inactivation of TRPV5 and TRPV6 through direct interaction with the C terminus of the channels (Cao et al., 2013; Derler et al., 2006; Holakovska et al., 2011). Intracellular Ca2+ may also directly and rapidly inhibit TRPV6 through an as yet unknown mechanism (Niemeyer et al., 2001). Interestingly, TRPV2 desensitizes in the presence of Ca2+, but this does not depend on Ca2+-calmodulin. Rather, Ca2+-dependent depletion of phosphoinositides (PIs) contributes to TRPV2 desensitization (Mercado et al., 2010). Overall, these previous studies indicate that, despite sequence and structural homology within the TRPV subfamily, these channels are regulated by common ligands in a diverse manner.
Further structural studies of TRPV channels are needed for additional insight into the functional divergence of the TRPV subfamily. Although moderate-resolution cryo-electron microscopy (cryo-EM) structures of full-length TRPV1 and TRPV4 have been determined (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010), they did not provide an in-depth understanding of the functional diversity between TRPVs. To this end, we determined a structure of full-length TRPV2 at 13.6 Å resolution by cryo-EM through optimization of protein purification, specimen preparation, and employment of several EM image processing packages. In addition, a liposome-reconstituted system was used to determine the single-channel properties of TRPV2, confirming its functionality. Overall, the EM map of functional TRPV2 displayed a “hanging gondola” architecture similar to other TRPV channels (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010). At this resolution, the TRPV2 map also revealed details of the channel domain architecture, most importantly the position and orientation of the N-terminal ARDs in the full-length structure. Fitting of the ARDs into the EM map provided an explanation for the unique regulatory mechanisms of each TRPV family member. The placement of MlotiK1 TM domain crystal structure, a prokaryotic nucleotide-regulated K+ channel, into the TRPV2 EM map confirmed the predicted structural homology between TRP channels and K+ channels (Kalia and Swartz, 2013). Based on this study, we propose that our TRPV2 EM structure provides a model for the general architecture of the TRPV subfamily.
RESULTS
Functional Analysis of Purified TRPV2 Channel
We tested several TRPV channel homologs for structural analysis and found that rat TRPV2 gave the best expression in S. cerevisiae. Through extensive screening of buffers and detergents, we determined that the maltose-neopentyl glycol (MNG) class of detergents stabilizes rat TRPV2 as a functional tetramer (Figure S1A available online). Recently it has been found that these newly synthesized low-critical micelle concentration detergents are optimal for extraction, long-term stabilization, and structure determination of several membrane proteins (Chae et al., 2010). These detergents form a large micelle belt around the TM domain of pure membrane proteins visible in EM micrographs (Jastrzebska et al., 2013; Westfield et al., 2011). In addition, membrane proteins purified in MNG detergents display a higher molecular weight, adding approximately 60 kDa per protein monomer (Jastrzebska et al., 2013). After immunoaffinity purification of TRPV2, size-exclusion chromatography (SEC) and native gel electrophoresis revealed that a homogenous sample was obtained with an approximate molecular weight of 600 kDa (Figure S1A). This mass accounts for the 360 kDa TRPV2 tetramer and 240 kDa of detergent mass. The cryo-EM structures of TRPV1 and TRPV4 were determined using common maltoside detergents, such as decyl-β-D-maltoside (DM) and dodecyl-β-D-maltoside (DDM) (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010). During detergent screens, we observed that TRPV2 purified in DM or DDM exists in multiple aggregating states confirmed by native gel electrophoresis after SEC (Figure S1B), suggesting that TRPV2 purified in these detergents is not suitable for structural analysis.
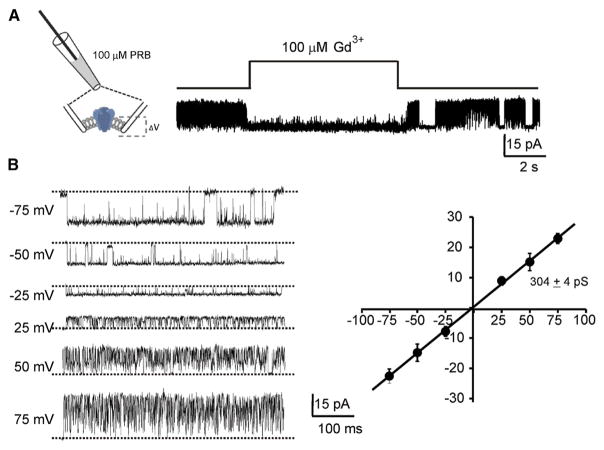
Among the thermoTRPV channels, probenecid, a uricosuric agent, has been shown to specifically activate TRPV2 (Bang et al., 2007), and trivalent cations such as Gd3+ and La3+ are potent blockers of TRPV2-mediated currents (Leffler et al., 2007). To determine the functionality of purified TRPV2, the channel was reconstituted into preformed asolectin liposomes and electrophysiological measurements were made from excised patches in the inside-out configuration. In the presence of 100 μM probenecid in the pipette, robust single-channel currents were observed, displaying very little desensitization. These currents showed a rapid block upon application of 100 μM GdCl3 that was fully reversible during wash-off (Figure 1A). Currents recorded at different holding potentials showed widely differing kinetics for inward and outward currents (Figure 1B). While inward currents were long bursts of high-open probability and infrequent visits to the nonconducting state, the outward currents were characterized by very fast flickers (Figure 1B). A plot of single-channel currents as a function of membrane potential yields a conductance of 304 ± 4 pS. These results demonstrate that purified TRPV2 retains functional properties.
Figure 1. Single-Channel Properties of Liposome-Reconstituted TRPV2.
(A) Excised inside-out patches of proteoliposomes containing TRPV2 channels. The channels were activated by 100 μM probenecid (PRB) in the pipette. Single-channel currents were recorded under symmetrical KCl concentrations (150 mM) and the membrane potential was held at +50 mV. Channel activity was blocked by 100 μM GdCl3 applied using a rapid solution exchanger.
(B) Representative single-channel traces measured at indicated membrane potentials. Current-voltage relationship showed a single-channel conductance of 304 ± 4 pS (n = 4).
Imaging and Map Building
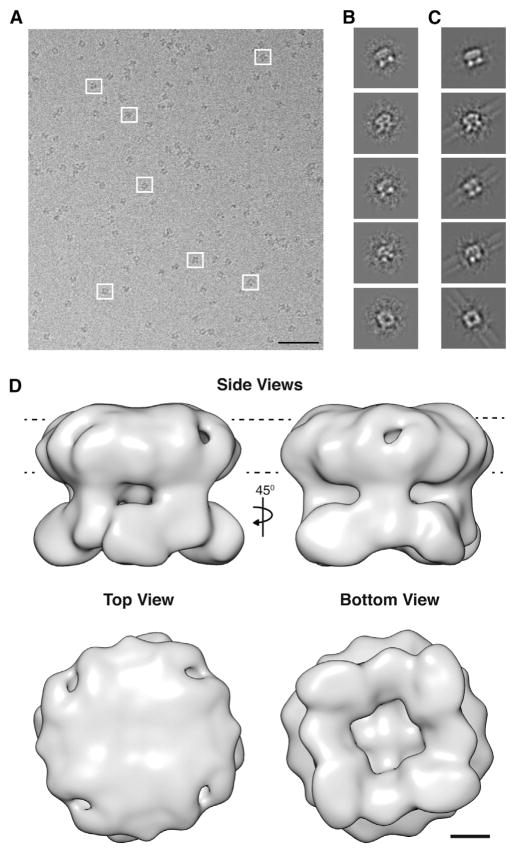
To determine the structure of functional, purified TRPV2, concentrated protein (2.5 mg/ml) was cryogenically frozen on grids and imaged on a Tecnai F20 (200 keV) cryo-electron microscope. Micrographs displayed a monodisperse distribution of TRPV2 (Figure 2A). From a set of 650 micrographs, 23,051 particles were manually picked using the boxer subroutine of EMAN2 (Tang et al., 2007). Reference-free class averages generated with IMAGIC (van Heel et al., 1996) revealed representative top, bottom, and side views of the channel (Figure 2B). Four-fold symmetry was observed in raw images and in reference-free class averages, consistent with the predicted overall channel topology (Figures 2A and 2B). Initial 3D models were generated both with and without imposed symmetry using IMAGIC (Figures S2A and S2B, related to Figure 2). The initial model without imposed symmetry displayed approximate 4-fold symmetry (Figure S2A, related to Figure 2), and reprojections resembled the matching class averages (Figure 2C). Further refinement was performed with FREALIGN (Grigorieff, 2007) and the 3D Fourier Space programs (http://www.sickkids.ca/research/rubinstein; Benlekbir et al., 2012). Topologically similar refined models were obtained using these programs (Figures S2C–S2E, related to Figure 2). The final map has a resolution of 13.6 Å at the 0.143 threshold from the gold-standard Fourier Shell correlation (FSC) curve (Figure 2D; Figure S3, related to Figure 2).
Figure 2. Cryo-EM Imaging and Reconstruction of TRPV2.
(A) Cryo-electron micrograph of monodisperse TRPV2 particles. White boxes represent typical views of TRPV2. Scale bar, 100 nm.
(B) Representative reference-free class averages of TRPV2 generated in IMAGIC (van Heel et al., 1996).
(C) Reprojections of the initial 3D model without imposed symmetry corresponding to the reference-free class averages.
(D) Side views, top view, and bottom view of the cryo-EM reconstruction of TRPV2 at 13.6 Å resolution. The structure shows a well-defined cytoplasmic region that displays a petal-like arrangement. The dashed lines represent the plasma membrane. Scale bar, 25 Å.
Overall Map Features
TRPV2 functions as a tetramer (Li et al., 2011). Each monomer contains 761 residues, with a large N-terminal region of 389 amino acids, a smaller 250-residue TM domain, and a 122-residue C-terminal region. The 3D structure of TRPV2 reveals a “hanging gondola” architecture consistent with the cryo-EM structures of full-length TRPV1 and TRPV4 (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010) (Figure 2D). The TRPV2 structure displays an overall height of 115 Å, which is more compact than the previously determined structures of other thermoTRPV channels with larger molecular masses. The predicted TM domain of TRPV2 is 55 Å in height and 135 Å in width and the cytosolic region is 60 Å in height and 125 Å in width. At this resolution, the overall architecture of the cytoplasmic region of a TRPV channel is clearly resolved, displaying a petal-like shape (Figure 2D). The TRPV2 3D map exhibits a subtle handedness (Figure 2D). A data set of tilt-pair projections, together with the Tiltdiffmulti program (Henderson et al., 2011), validated that the TRPV2 cryo-EM map is displayed with the correct hand (Figure S4, related to Figure 2).
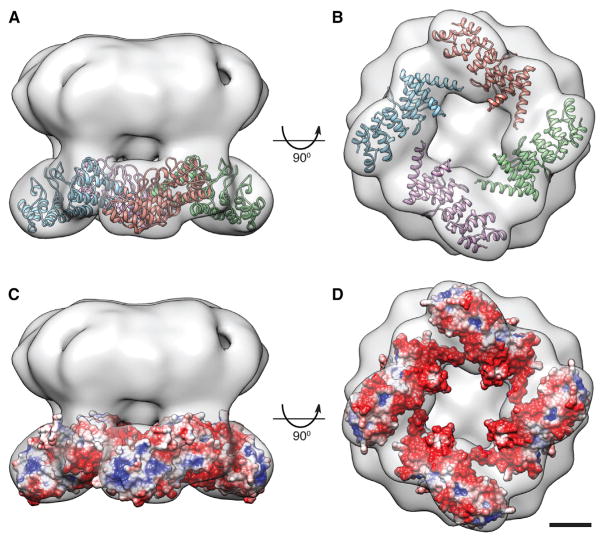
Currently, the only high-resolution structural information available for TRPV2 comes from a crystal structure of the N-terminal ARD (Jin et al., 2006). The rat TRPV2 ARD consists of six ankyrin repeats (residues 75–321), accounting for 63% of the N-terminal region (Jin et al., 2006). To analyze the cytoplasmic region of the TRPV2 3D map, the TRPV2 ARD structure (Protein Data Bank [PDB] ID code: 2ETB) was fitted into the EM map using Colores in the Situs package (Chacón and Wriggers, 2002). The four ARDs unambiguously occupy most of the cytoplasmic density in the TRPV2 EM map (Figure 3A). The best fit for the ARDs as determined by Colores has the β-hairpin loops, which compose the concave ligand-binding surface, facing outward (Figures 3A and 3B). This fit would allow for ligands to interact with the ARDs with limited steric interference. The unoccupied densities on the tips of the petals likely correspond to the first 70 amino acids at the TRPV2 N terminus (Figure 3B). The last 68 residues of the TRPV2 N terminus (linker region) likely occupy a portion of the space between the ARD and the TM domain in the map (Figures 4C and 4D; Figure S5, related to Figure 4).
Figure 3. N-Terminal ARD Arrangement in the TRPV2 Map.
(A and B) Side view (A) and bottom view (B) of the TRPV2 EM map with ARD crystal structures (Jin et al., 2006; PDB ID code: 2ETB) docked by Colores into the cytoplasmic densities. Each color represents an individual subunit.
(C and D) Side view (C) and bottom view (D) of the calculated ARD electrostatic potential (Baker et al., 2001) docked into the TRPV2 map. Positive potentials are shown in blue, negative potentials in red. Scale bar, 25 Å.
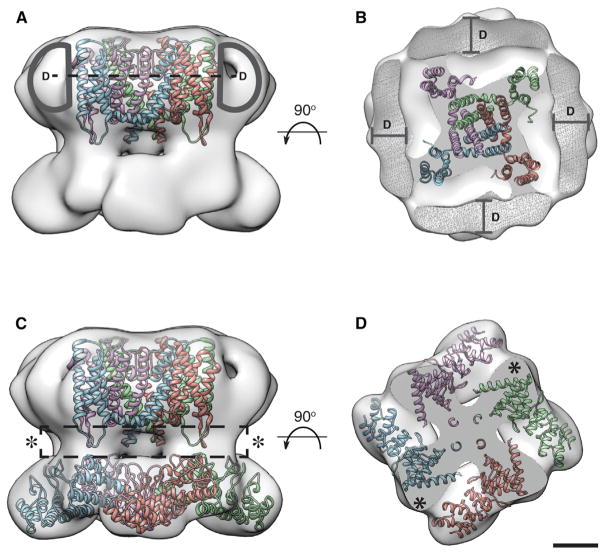
Figure 4. Fitting of Homologous TM Domain Structure and Prediction of the Location of the MNG Detergent Belt, N-Terminal Linker Region, and C Terminus in the TRPV2 Map.
(A) Crystal structure of the MlotiK1 TM domain (Clayton et al., 2008; PDB ID code: 3BEH) docked into the central TM density of the TRPV2 EM map. The extra density around the central TM region outlined in dark gray (labeled as D) represents the uneven 15–20 Å MNG detergent belt. Dashed line represents the cross-section depicted in (B).
(B) A cross-section through the TM domain clearly shows the detergent belt (in mesh labeled as D) relative to the central density where the MlotiK1 TM domain was docked. More cross-sections through the TM region of the density are shown in Figure S5.
(C) Side view of the TRPV2 EM map fitted with the MlotiK1 TM domain and TRPV2 ARD crystal structures. Dashed box represents the possible location of the TRPV2 N terminus linker region and C terminus. A cross-section of this segment is depicted in (D) and marked with asterisks.
(D) A cross-section through the TRPV2 structure showing the possible location of the TRPV2 N terminus linker region and C terminus. More cross-sections through this portion of the EM density are shown in Figure S5. Scale bar, 25 Å.
It has been shown previously that surface charge is important for complex formation between ARDs and other proteins and ligands (Nakamura et al., 2007). Biochemical studies have revealed that the TRPV subfamily members interact with different soluble ligands and proteins (Phelps et al., 2010). The electrostatic surface potential (Baker et al., 2001) of the four TRPV2 ARDs as they were fitted in the EM map shows a cluster of negatively charged residues facing the cytoplasm (Figures 3C and 3D). We hypothesized that the surface charge of the ARDs may be essential for differential ligand and protein binding among TRPVs. When the available structures of the ARDs of TRPV1 (PDB code: 2PNN), TRPV4 in the presence of ATP (PDB code: 4DX2), and TRPV4 in the absence of ATP (Inada et al., 2012; PDB code: 4DX1) were fitted into the TRPV2 EM map using Colores (Figures S6A–S6C, related to Figure 3), all structures displayed reduced overall negative charge compared to the TRPV2 ARDs (Figures 3C and 3D). The TRPV4 ARDs in particular displayed a large positively charged surface facing the cytoplasm (Figures S6B and S6C, related to Figure 3). TRPV6, like TRPV2, displays constitutive activity (Vennekens et al., 2000). Therefore, regulated plasma membrane insertion of these channels is thought to be important for their function (Kanzaki et al., 1999; van de Graaf et al., 2006). Intriguingly, despite only 21% homology between TRPV2 and TRPV6, the TRPV6 ARDs (Phelps et al., 2008; PDB code: 2RFA) fitted into the TRPV2 map displayed a negatively charged surface facing the cytoplasm similar to the TRPV2 ARDs (Figure S6D, related to Figure 3). Previous reports indicate that ATP binds to the ARDs of TRPV1 and TRPV4 and sensitizes the channels, while ATP has no direct effect on TRPV2 and TRPV6 activity (Lishko et al., 2007; Phelps et al., 2010). The negatively charged cytosolic cluster on TRPV2 and TRPV6 likely prevents ATP binding and sensitization of the channel. This analysis suggests that variations in the surface charge of individual TRPV ARDs may contribute to their differences in ligand binding and channel trafficking and regulation.
To analyze the TM domain of the TRPV2 map, we fitted the X-ray structure of the nucleotide-regulated potassium channel MlotiK1 (Clayton et al., 2008; PDB code: 3BEH) into the proposed TM region using Chimera (Figure 4A). It has recently been suggested that the TM domains of TRPV channels more closely resemble the TM structure of MlotiK1 than other K+ channels for which high-resolution structural information is available (Kalia and Swartz, 2013; Shigematsu et al., 2010). The MlotiK1 structure fits well into the observed central densities of the TM region of the EM map (Figures 4A and 4B; see also Figure S5). Moreover, we resolved extra density around the central TM region, which likely corresponds to an MNG detergent micelle belt (Figure 4B; see also Figure S5). The belt forms an uneven 15–20 Å ring around the central TM density in the EM map (Figures 4A and 4B; see also Figure S5). This feature of an MNG detergent belt has been observed in other membrane protein structures determined by cryo-EM (Jastrzebska et al., 2013; Westfield et al., 2011). In addition, this result is consistent with our biochemical data indicating a significant added mass attributed to MNG (see also Figure S1A).
The C-terminal region of TRPV channels is an important modular domain that contains the conserved TRP box sequence (IWKLQK) and interacts with Ca2+-calmodulin and PI(4,5)P2 (Ufret-Vincenty et al., 2011). The structure of a 12-amino-acid C-terminal peptide derived from TRPV1 interacting with Ca2+-calmodulin is the only C-terminal structural information from TRPV channels available to date (Lau et al., 2012). Due to the lack of the structural information on the TRPV2 C terminus, we predicted that the 122 residues of the TRPV2 C terminus would fit into the 3D EM density between the TM region fitted with MlotiK1 structure and the N-terminal ARDs fitted into the cytoplasmic region of the channel, along with the linker region of the N terminus (Figures 4C and 4D; see also Figure S5).
DISCUSSION
Here, we present a 13.6 Å structure of a full-length TRPV2 channel solved by cryo-EM. To produce a suitable cryo-EM specimen, the MNG class of detergents was used during extraction and purification of TRPV2. To confirm that purified TRPV2 was functional, we reconstituted TRPV2 into proteoliposomes and performed patch clamp electrophysiology measurements. This system allowed us to directly measure the single-channel conductance of TRPV2 and show that, in the presence of pro-benecid, TRPV2 opens with high open probability and is blocked by bath application of Gd3+. This system could potentially be used for identification of other pharmacological agents that modulate TRPV2 channel properties.
The cryo-EM structure of TRPV2 allowed us to model the cytoplasmic domain arrangement and fit the TRPV2 ARD crystal structures into the 3D map of the full-length protein. In our model, the TRPV2 ARDs are arranged such that the β-hairpins, which compose the concave ligand-binding surface (Binz et al., 2004; Nakamura et al., 2007), are facing outward and exposed to the cytoplasm (Figure 5A). This arrangement would provide access for cytoplasmic proteins and ligands to interact with the channel without steric hindrance from the other domains of TRPV2 (Perálvarez-Marín et al., 2013). This model indicates that the TRPV2 ARDs do not interact with one another, consistent with previously published reports indicating that TRPV ARDs are not involved in channel oligomerization (Gaudet, 2008; Jin et al., 2006).
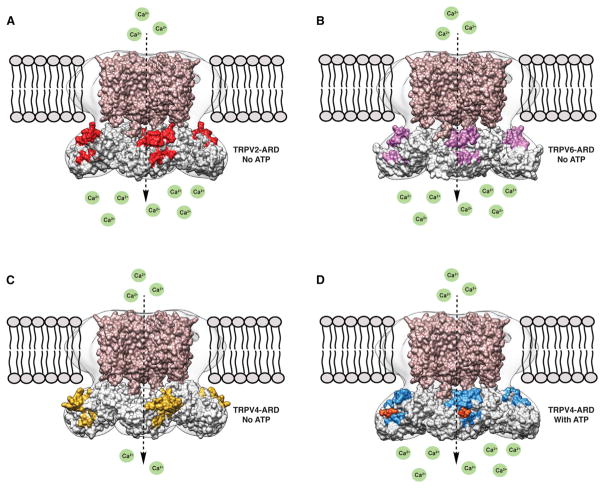
Figure 5. Comparison of TRPV ARD Structures Fitted into the TRPV2 EM Map.
Surface representation of the MlotiK TM domain (brown; PDB ID code: 3BEH) and (A) TRPV2 ARDs (PDB ID code: 2ETB), (B) TRPV6 ARDs (PDB ID code: 2RFA), (C) TRPV4 ARDs in the absence of ATP (PDB ID code: 4DX1), and (D) TRPV4 ARDs in the presence of ATP (PDB ID code: 4DX2), fitted into the EM map. The concave ligand-binding surface on the ARDs is highlighted in red (TRPV2), purple (TRPV6), yellow (TRPV4 ATP-free), and blue (TRPV4 ATP-bound). The ATP molecule is represented in orange and Ca2+ ions are represented in green.
The TRPV2 ARDs, as docked within the 3D map, revealed an acidic cluster facing the cytoplasm (Figures 3C and 3D). This cluster appears to be specific for TRPV2 and TRPV6 ARDs, as the ARDs for TRPV1 and TRPV4 display more positively charged surface potentials facing the cytoplasm (Figure S6). Crystal structures for TRPV1 and TRPV4 ARDs revealed that ATP binds to the ARDs via charged and hydrophobic residues in a unique binding pocket (Lishko et al., 2007). Residues important for ATP binding in TRPV1 are conserved in TRPV2 (Lishko et al., 2007; Phelps et al., 2008). Therefore, some other feature of the TRPV2 ARD likely disrupts ATP binding. We propose that the acidic cluster present in the TRPV2 and TRPV6 ARDs may prevent their interaction with negatively charged ATP. Moreover, TRPV2 and TRPV6 display constitutive activity in the cell and undergo Ca2+-dependent desensitization, which does not depend on Ca2+-calmodulin binding (Cao et al., 2013; Mercado et al., 2010; Niemeyer et al., 2001). The acidic clusters in the TRPV2 and TRPV6 ARDs may facilitate direct interaction with intracellular Ca2+ and regulate the activity of these channels. In addition, different surface charge would allow TRPV2 and TRPV6 to interact with different protein binding partners compared to other TRPV channels, which could affect channel function and trafficking.
ATP binding to the TRPV1 and TRPV4 ARDs has been shown to sensitize the channels and stabilize the ARDs (Inada et al., 2012). Intriguingly, the overall structure of the TRPV4 ARD and its concave ligand-binding surface in the presence of ATP resembles the TRPV2 and TRPV6 ARD structures (Figures 5A, 5B, and 5D). In its ATP-free form, the concave ligand-binding surface of the TRPV4 ARD structure is more extended and resides outside of the TRPV2 EM density compared to the ATP-bound form (Figures 5C and 5D). These results suggest that the conformation adopted by the ARDs of TRPV2, TRPV6, and ATP-bound TRPV4 may confer constitutive channel activity.
Previous moderate-resolution cryo-EM structures of full-length TRPV1 and TRPV4 displayed distinguishable variations in the architecture of their respective cytoplasmic regions (Figure S7; Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010). Here, we have shown that ARDs for the TRPV subfamily fit unambiguously into the cytosolic region of the TRPV2 EM density. Although it is possible that the highly homologous TRPV channels adopt different structural topologies, we propose that the TRPV2 structure presented here represents the general architecture for the TRPV subfamily based on fitting of the ARDs from TRPV family members into the TRPV2 EM density map.
The previously observed structural divergences in the TRPV channel EM maps could be due to structural flexibility of the ARDs, which lacked ATP in sample preparation for TRPV1 and TRPV4 (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010; Figure S7). Computational averaging of the flexible ARDs for TRPV1 and TRPV4 during EM reconstruction may have led to this structural discrepancy. Moreover, based on our experimental observations, MNG detergents preserve structural and functional stability of TRPV channels, whereas commonly used maltoside detergents, DM and DDM, are not as suitable for maintaining long-term TRPV protein integrity (Figure S1B). The presence of possible heterogeneity in TRPV1 and TRPV4 samples after purification in maltoside detergents together with low-contrast imaging potentially affected cryo-EM reconstructions of these channels (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010). The higher contrast and greater overall stability of TRPV2 in MNG detergent likely contributed to the structure determination of full-length TRPV2 at higher resolution.
In summary, our structural and functional analyses of TRPV2 have provided insight into the general architecture for the TRPV subfamily and revealed possible explanations for the functional diversity among subfamily members. Specific higher resolution structural information for each TRPV subfamily member will serve to further delineate the differential modulation by endogenous ligands.
EXPERIMENTAL PROCEDURES
TRPV2 Expression and Purification
Rat TRPV2 with a 1D4-tag was cloned into a YepM plasmid and overexpressed in BJ5457 S. cerevisiae (ATCC). TRPV2 membranes were prepared and solubilized in 0.087% w/v lauryl MNG (Anatrace), 20 mM HEPES (pH 8.0), 150 mM NaCl, 5% glycerol, 1.0 mM dithiothreitol (DTT), and 1 mM phenyl-methanesulfonylfluoride (PMSF) for 1 hr at 4°C. The insoluble fractions were pelleted by ultracentrifugation at 100,000 g, 4°C for 45 min. The soluble fraction containing TRPV2 protein was incubated overnight at 4°C with cyanogen-bromide-activated Sepharose 4B coupled with 1D4 antibody. The column was packed and washed with washing buffer consisting of 0.006% decyl MNG (Anatrace), 20 mM HEPES (pH 8.0), 150 mM NaCl, and 1 mM DTT. The TRPV2 protein was eluted with 3 mg/ml 1D4 peptide (Genescript USA) in washing buffer, concentrated, and subjected to SEC.
Single-Channel Properties of Reconstituted TRPV2
Electrophysiological measurements were made by patch clamp recordings in reconstituted proteoliposomes, as described earlier (Chakrapani et al., 2007; Cortes et al., 2001; Delcour et al., 1989; Velisetty and Chakrapani, 2012). A total of 20 mg soybean polar extract (Avanti polar lipids) in chloroform was dried in nitrogen stream and sonicated in a buffer containing 200 mM KCl and 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) at pH 7.0. The liposomes (at 10 mg/ml concentration) were destabilized by adding 4 mM DDM and were gently agitated for 20 min. Purified TRPV2 was added to the vesicles (in 1:50 [w/w] protein to lipid ratio) and nutated for to 2 hr. Residual detergent was removed by incubation with biobeads (Bio-Rad Laboratories) overnight at 4°C. Channel-incorporated liposome suspension was then centrifuged for 2 hr at 100,000 g and the pellet was resuspended in 60 μl of KCl/MOPS buffer. A drop of the proteoliposome was placed on a glass slide and dried overnight in a desiccator at 4°C. The sample was then rehydrated with 20 μl of buffer, which yielded giant liposomes. This preparation was suitable for patch clamp recordings after ~2 hr. Currents were recorded in 150 mM KCl and 10 mM MOPS at pH 7.0. Recording pipettes were pulled from thin-walled borosilicate glass and heat polished such that they had a bath resistance of 1–2 MΩ. The pipette solution had 100 μM probenecid in a buffer containing 150 mM KCl, 2 mM CaCl2, and 10 mM MOPS, pH 7.0. All measurements in this study were conducted in the inside-out patch configuration with a bath solution containing 150 mM KCl, 10 mM MOPS, and pH 7.0. Experiments were performed at room temperature (20°C–22°C). Pore blockers were applied using RCS-200 fast solution exchanger (Biologic) fed by gravity. Single currents were acquired at 20 kHz and filtered at 5 kHz using Axon 200-B patch-clamp amplifier (Molecular Devices). In the presence of probenecid, most excised patches had multiple overlapping channel activities. Only those recordings with single-open channel were used for further analysis.
Cryo-EM Preparation
Purified TRPV2 was frozen onto Quantifoil R2/1 copper grids (Quantifoil Micro Tools; 400 mesh) using the Vitrobot (FEI). Data were acquired on an FEI Tecnai F20 TWIN microscope operated at 200 keV under low-dose conditions (15 electrons/ Å 2). Images were collected at a nominal magnification of 62,000× at various defocus values between −1.5 and −3 μm and were recorded with a Tietz TemCam-F416 complementary metal oxide semiconductor-based camera (4k × 4k).
Single-Particle Reconstruction and Modeling
The EMAN2 (Tang et al., 2007) boxer routine was used to pick 23,051 particle images from 650 micrographs and to generate a particle stack. The stack was binned by factors of two and four, yielding pixels of 2.7 Å and 5.3 Å, respectively, for image processing. Initial estimates for microscope defocus and astigmatism parameters were calculated with CTFFIND3 (Mindell and Grigorieff, 2003). The bin4 images were filtered, normalized, and band-pass filtered (120–15 Å ) prior to reference-free multivariate statistical analysis classification in IMAGIC (van Heel et al., 1996). The IMAGIC C1-Startup routine was used to generate an initial 3D model without imposed symmetry. As this initial 3D model displayed approximate 4-fold symmetry, a 3D model was also generated with imposed C4 symmetry. The two IMAGIC-generated maps were used as input 3D models for angular search and refinement of the unclassified particle images with FREALIGN (Grigorieff, 2007). The FREALIGN search and refinement procedure was carried out in parallel using either C1 or C4 symmetry and was first performed on the bin4 stack and subsequently on the bin2 stack. The final resolution was 19 Å at the FSC 0.5 threshold for the FREALIGN C1-refined 3D map and 15 Å at the FSC 0.5 threshold for the FREALIGN C4-refined 3D map.
The FREALIGN C4 symmetrized map was used as the input 3D model for angular search and refinement with the 3D Fourier Space programs (http://www.sickkids.ca/research/rubinstein; Benlekbir et al., 2012), which incorporate the gold-standard refinement scheme (Scheres and Chen, 2012). The 3D Fourier Space refinement procedure was carried out in parallel using either C1 or C4 symmetry. The final C1 3D Fourier Space refined map (Figure S2E, related to Figure 2) has a resolution of 20 Å at the 0.143 threshold of the gold-standard FSC curve (25 Å at the 0.5 threshold). For processing with symmetry, C4 symmetric odd and even maps were calculated at the end of each round with FREALIGN in iflag = 0 mode. The 3D Fourier Space cross correlation (CC) values were converted into FREALIGN-style phase residuals with the following equation: phase residual = 100 – CC value. A C4 symmetric final map based on the full data set was calculated at the end of the last refinement round. The final structure is based on all 23,051 particles and has a resolution of 13.6 Å at the 0.143 threshold of the gold-standard FSC curve (19 Å at the 0.5 threshold). The final map is shown sharpened with a B-factor of −1,000 Å 2 and filtered with the Cref filter (Rosenthal and Henderson, 2003). The cryo-EM structure has been deposited in the EM Data Bank (EMD-5688).
The electrostatic potential surfaces for the N-terminal TRPV2 ARDs, TRPV1 ARDS, and TRPV4 ARDs were calculated using Adaptive Poisson-Boltzmann Solver (Baker et al., 2001). Fitting of the TRPV1, TRPV2, TRPV4, and TRPV6 ARD crystal structures was perform using Colores in the Situs package (Chacón and Wriggers, 2002). Visualizations and MlotiK1 crystal structure fitting into the TRPV2 EM map were performed using UCSF Chimera software (Pettersen et al., 2004).
TRPV2 Structure Hand Determination
Tilt-pair cryo-electron micrographs were collected at tilt angles of 0° and 20° using an FEI Titan Krios at 120 keV and magnification of 59,000× with a 10 electron/ Å 2 dose on an FEI 4k ×4k charge-coupled device detector. A total of 186 tilt-pair particle images were selected and binned by a factor of four, yielding a pixel size of 5.4 Å for image processing. The tilt axis, defocus, and astigmatism estimations for the tilted micrographs were calculated using CTFTILT, whereas CTFFIND3 was used for the defocus and astigmatism estimations for the untilted micrographs. The orientational parameters were determined independently for both the 0° and 20° particle images using FREALIGN search and refinement procedures with imposed C4 symmetry and with a resolution range of 150–18 Å, as recommended by Henderson et al. (2011). The parameter files for the 0° and 20° particle images served as input to Tiltdiffmulti, a program designed to determine the best tilt axis and tilt angle relating two views of a particle (Henderson et al., 2011). A tilt-pair parameter plot (TPPP) was generated for TRPV2, and it reveals a cluster of points centered around a tilt axis of 270° and a tilt angle of 20° (Figure S4, related to Figure 2).
A simulated tilt-pair data set was generated using a protein with a known and distinct handedness, Cand1 (PDB ID code: 1U6G; Goldenberg et al., 2004), as described by Williams et al. (2008). The simulated tilt-pair data set was put through the same particle selection, search and refinement, and TPPP analysis scheme as the TRPV2 tilt-pair data set. The simulated data analysis, which was performed with a 3D search model of the correct hand, yielded a TPPP plot with a cluster centered at a tilt axis of 270° and a tilt angle of 20°, as found for TRPV2. The convergence of the clusters for the simulated and experimental data sets suggests that the hand of the TRPV2 map is correct.
Supplementary Material
Acknowledgments
We would like to thank James Love, Brian Kloss, and Teresa Cvetkov for excellent technical suggestions during this project. Additionally, we thank Tamir Gonen for providing access to the electron microscopy facility at Janelia Farm Research Campus (Howard Hughes Medical Institute), Duncan Sousa for collecting a tilt-pair data set at the Biological Science Imaging Resource (Florida State University), and Yufeng Zhou, Irina Serysheva, and Theodore Wensel for support during the early stages of this project. Most of the image processing was performed on the Case Western Reserve University High Performance Computing Cluster. This work was supported by the American Heart Association (NCRP Scientist Development grant 11SDG5280029 to V.Y.M.-B. and M.R.C.).
Footnotes
ACCESSION NUMBERS
The EM Data Bank accession number for the rat TRPV2 structure reported in this paper is EMD-5688.
Supplemental Information includes seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2013.11.008.
References
- Al-Ansary D, Bogeski I, Disteldorf BM, Becherer U, Niemeyer BA. ATP modulates Ca2+ uptake by TRPV6 and is counteracted by isoform-specific phosphorylation. FASEB J. 2010;24:425–435. doi: 10.1096/fj.09-141481. [DOI] [PubMed] [Google Scholar]
- Baez-Nieto D, Castillo JP, Dragicevic C, Alvarez O, Latorre R. Thermo-TRP channels: biophysics of polymodal receptors. Adv Exp Med Biol. 2011;704:469–490. doi: 10.1007/978-94-007-0265-3_26. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang S, Kim KY, Yoo S, Lee SH, Hwang SW. Transient receptor potential V2 expressed in sensory neurons is activated by probenecid. Neurosci Lett. 2007;425:120–125. doi: 10.1016/j.neulet.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Benlekbir S, Bueler SA, Rubinstein JL. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11- Å resolution. Nat Struct Mol Biol. 2012;19:1356–1362. doi: 10.1038/nsmb.2422. [DOI] [PubMed] [Google Scholar]
- Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grütter MG, Plückthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- Cao C, Zakharian E, Borbiro I, Rohacs T. Interplay between calmodulin and phosphatidylinositol 4,5-bisphosphate in Ca2+-induced inactivation of transient receptor potential vanilloid 6 channels. J Biol Chem. 2013;288:5278–5290. doi: 10.1074/jbc.M112.409482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- Chacón P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, et al. Maltoseneopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating II: single-channel currents. J Gen Physiol. 2007;130:479–496. doi: 10.1085/jgp.200709844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc Natl Acad Sci USA. 2008;105:1511–1515. doi: 10.1073/pnas.0711533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour AH, Martinac B, Adler J, Kung C. Voltage-sensitive ion channel of Escherichia coli. J Membr Biol. 1989;112:267–275. doi: 10.1007/BF01870957. [DOI] [PubMed] [Google Scholar]
- Derler I, Hofbauer M, Kahr H, Fritsch R, Muik M, Kepplinger K, Hack ME, Moritz S, Schindl R, Groschner K, Romanin C. Dynamic but not constitutive association of calmodulin with rat TRPV6 channels enables fine tuning of Ca2+-dependent inactivation. J Physiol. 2006;577:31–44. doi: 10.1113/jphysiol.2006.118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R. A primer on ankyrin repeat function in TRP channels and beyond. Mol Biosyst. 2008;4:372–379. doi: 10.1039/b801481g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R. Divide and conquer: high resolution structural information on TRP channel fragments. J Gen Physiol. 2009;133:231–237. doi: 10.1085/jgp.200810137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Henderson R, Chen S, Chen JZ, Grigorieff N, Passmore LA, Ciccarelli L, Rubinstein JL, Crowther RA, Stewart PL, Rosenthal PB. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J Mol Biol. 2011;413:1028–1046. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holakovska B, Grycova L, Bily J, Teisinger J. Characterization of calmodulin binding domains in TRPV2 and TRPV5 C-tails. Amino Acids. 2011;40:741–748. doi: 10.1007/s00726-010-0712-2. [DOI] [PubMed] [Google Scholar]
- Huang SM, Li X, Yu Y, Wang J, Caterina MJ. TRPV3 and TRPV4 ion channels are not major contributors to mouse heat sensation. Mol Pain. 2011;7:37. doi: 10.1186/1744-8069-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada H, Procko E, Sotomayor M, Gaudet R. Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel. Biochemistry. 2012;51:6195–6206. doi: 10.1021/bi300279b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Ringler P, Palczewski K, Engel A. The rhodopsin-transducin complex houses two distinct rhodopsin molecules. J Struct Biol. 2013;182:164–172. doi: 10.1016/j.jsb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Touhey J, Gaudet R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J Biol Chem. 2006;281:25006–25010. doi: 10.1074/jbc.C600153200. [DOI] [PubMed] [Google Scholar]
- Kalia J, Swartz KJ. Exploring structure-function relationships between TRP and Kv channels. Sci Rep. 2013;3:1523. doi: 10.1038/srep01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki M, Zhang YQ, Mashima H, Li L, Shibata H, Kojima I. Translocation of a calcium-permeable cation channel induced by insulin-like growth factor-I. Nat Cell Biol. 1999;1:165–170. doi: 10.1038/11086. [DOI] [PubMed] [Google Scholar]
- Lau SY, Procko E, Gaudet R. Distinct properties of Ca2+-calmodulin binding to N- and C-terminal regulatory regions of the TRPV1 channel. J Gen Physiol. 2012;140:541–555. doi: 10.1085/jgp.201210810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler A, Linte RM, Nau C, Reeh P, Babes A. A high-threshold heat-activated channel in cultured rat dorsal root ganglion neurons resembles TRPV2 and is blocked by gadolinium. Eur J Neurosci. 2007;26:12–22. doi: 10.1111/j.1460-9568.2007.05643.x. [DOI] [PubMed] [Google Scholar]
- Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- Li M, Yu Y, Yang J. Structural biology of TRP channels. Adv Exp Med Biol. 2011;704:1–23. doi: 10.1007/978-94-007-0265-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Mercado J, Gordon-Shaag A, Zagotta WN, Gordon SE. Ca2+-dependent desensitization of TRPV2 channels is mediated by hydrolysis of phosphatidylinositol 4,5-bisphosphate. J Neurosci. 2010;30:13338–13347. doi: 10.1523/JNEUROSCI.2108-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc Natl Acad Sci USA. 2008;105:7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Nakano K, Umehara T, Kimura M, Hayashizaki Y, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure. 2007;15:179–189. doi: 10.1016/j.str.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Niemeyer BA, Bergs C, Wissenbach U, Flockerzi V, Trost C. Competitive regulation of CaT-like-mediated Ca2+ entry by protein kinase C and calmodulin. Proc Natl Acad Sci USA. 2001;98:3600–3605. doi: 10.1073/pnas.051511398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park U, Vastani N, Guan Y, Raja SN, Koltzenburg M, Caterina MJ. TRP vanilloid 2 knock-out mice are susceptible to perinatal lethality but display normal thermal and mechanical nociception. J Neurosci. 2011;31:11425–11436. doi: 10.1523/JNEUROSCI.1384-09.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perálvarez-Marín A, Doñate-Macian P, Gaudet R. What do we know about the transient receptor potential vanilloid 2 (TRPV2) ion channel? FEBS J. 2013;280:5471–5487. doi: 10.1111/febs.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phelps CB, Huang RJ, Lishko PV, Wang RR, Gaudet R. Structural analyses of the ankyrin repeat domain of TRPV6 and related TRPV ion channels. Biochemistry. 2008;47:2476–2484. doi: 10.1021/bi702109w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Wang RR, Choo SS, Gaudet R. Differential regulation of TRPV1, TRPV3, and TRPV4 sensitivity through a conserved binding site on the ankyrin repeat domain. J Biol Chem. 2010;285:731–740. doi: 10.1074/jbc.M109.052548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti D, Szallasi A, Patacchini R. TRP channels as therapeutic targets in airway disorders: a patent review. Expert Opin Ther Pat. 2012;22:663–695. doi: 10.1517/13543776.2012.696099. [DOI] [PubMed] [Google Scholar]
- Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Santoni G, Farfariello V. TRP channels and cancer: new targets for diagnosis and chemotherapy. Endocr Metab Immune Disord Drug Targets. 2011;11:54–67. doi: 10.2174/187153011794982068. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki K, Murayama N, Ono K, Ishizaki Y, Tominaga M. TRPV2 enhances axon outgrowth through its activation by membrane stretch in developing sensory and motor neurons. J Neurosci. 2010;30:4601–4612. doi: 10.1523/JNEUROSCI.5830-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Sokabe T, Danev R, Tominaga M, Nagayama K. A 3.5-nm structure of rat TRPV4 cation channel revealed by Zernike phase-contrast cryoelectron microscopy. J Biol Chem. 2010;285:11210–11218. doi: 10.1074/jbc.M109.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Ufret-Vincenty CA, Klein RM, Hua L, Angueyra J, Gordon SE. Localization of the PIP2 sensor of TRPV1 ion channels. J Biol Chem. 2011;286:9688–9698. doi: 10.1074/jbc.M110.192526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Graaf SF, Chang Q, Mensenkamp AR, Hoenderop JG, Bindels RJ. Direct interaction with Rab11a targets the epithelial Ca2+ channels TRPV5 and TRPV6 to the plasma membrane. Mol Cell Biol. 2006;26:303–312. doi: 10.1128/MCB.26.1.303-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Velisetty P, Chakrapani S. Desensitization mechanism in prokaryotic ligand-gated ion channel. J Biol Chem. 2012;287:18467–18477. doi: 10.1074/jbc.M112.348045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennekens R. Emerging concepts for the role of TRP channels in the cardiovascular system. J Physiol. 2011;589:1527–1534. doi: 10.1113/jphysiol.2010.202077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennekens R, Hoenderop JG, Prenen J, Stuiver M, Willems PH, Droogmans G, Nilius B, Bindels RJ. Permeation and gating properties of the novel epithelial Ca(2+) channel. J Biol Chem. 2000;275:3963–3969. doi: 10.1074/jbc.275.6.3963. [DOI] [PubMed] [Google Scholar]
- Vennekens R, Owsianik G, Nilius B. Vanilloid transient receptor potential cation channels: an overview. Curr Pharm Des. 2008;14:18–31. doi: 10.2174/138161208783330763. [DOI] [PubMed] [Google Scholar]
- Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci USA. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008;16:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Tang J, Wang C, Colton CK, Tian J, Zhu MX. Calcium plays a central role in the sensitization of TRPV3 channel to repetitive stimulations. J Biol Chem. 2008;283:6162–6174. doi: 10.1074/jbc.M706535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.