Abstract
The recent and exciting discovery of germline HOXB13 mutations in familial prostate cancer has brought HOX signaling to the forefront of prostate cancer research. An enhanced understanding of HOX signaling, and the co-factors regulating HOX protein specificity and transcriptional regulation, has the high potential to elucidate novel approaches to prevent, diagnose, stage, and treat prostate cancer. Toward our understanding of HOX biology in prostate development and prostate cancer, basic research in developmental model systems as well as other tumor sites provides a mechanistic framework to inform future studies in prostate biology. Here we describe our current understanding of HOX signaling in genitourinary development and cancer, current clinical data of HOXB13 mutations in multiple cancers including prostate cancer, and the role of HOX protein co-factors in development and cancer. These data highlight numerous gaps in our understanding of HOX function in the prostate, and present numerous potentially impactful mechanistic and clinical opportunities for future investigation.
Keywords: Androgen receptor, HOXB13, HOXB13(G84E), MEIS1, MEIS2, PBX, Prostate, TALE
Abbreviations: ADT, Androgen Deprivation Therapy; AR, Androgen Receptor; HOX, Homeobox; MEIS, Murine Ectopic Integration Site; PIN, Prostatic Intraepithelial Neoplasia; PSA, Prostate-Specific Antigen; TALE, Three Amino Acid Loop Extension
Introduction
Prostate cancer is the most common non-cutaneous cancer and the second leading cause of cancer related mortalities among American men.1 The recent and exciting identification of germline HOXB13 (G84E) mutations within a subset of familial prostate cancers by Isaacs and Cooney in 2012 highlights a novel set of genes and transcriptional signaling pathways to understand prostate tumor etiology and develop new treatment modalities to combat prostate tumor initiation and progression.2 Prior to this discovery, much was already known regarding the expression and function of HOX genes, and their co-factors, in development and cancer. However, there remain significant gaps in our current understanding of HOX biology in prostate development and disease.
The role of HOX genes in organismal development
HOX proteins are highly evolutionarily conserved, homeodomain-containing transcription factors best known for their roles in body axis patterning and tissue differentiation of developing embryos.3, 4 Furthermore, recent studies have shown HOX proteins not only have a role development and organogenesis, but they also contribute to the control of several other processes into adulthood such as cell proliferation, cell cycle, apoptosis, cell differentiation, and cell migration.3, 5, 6 In humans, the 39 HOX proteins are divided into four HOX gene clusters: A, B, C, and D located on chromosomes 7p15, 17q21.2, 12q13, and 2q31 respectively.7 Each cluster is comprised of paralogous genes 1–13 whose 3′ to 5′ organization and expression both follow a pattern of spatial and temporal co-linearity with development, although not every paralog is present in each cluster. The 3′ HOX genes are most highly expressed in the anterior body regions that arise early in development, while the 5′ HOX genes encode more posterior regions that form later in development. The term, “HOX Code,” refers to the phenomenon where tissue specificity is determined by nested and partially overlapping expression of several HOX genes in a given region. The most 5′ HOX gene expressed in a given tissue, however, has dominance in determining a specific tissues' identity compared to the more 3′ HOX gene that may be co-expressed.8 For example, while 36 of the 39 HOX genes are expressed at a detectable level by qRT-PCR in a gross sample of human prostate tissue, it is the 5′ HOX genes like HOXA13 and HOXB13 that are most highly expressed and most significantly confer prostatic identity.9 Several excellent and in-depth reviews have already been published on the general role of HOX genes in development and cancer.3, 6, 10, 11, 12
HOX expression in male reproductive system
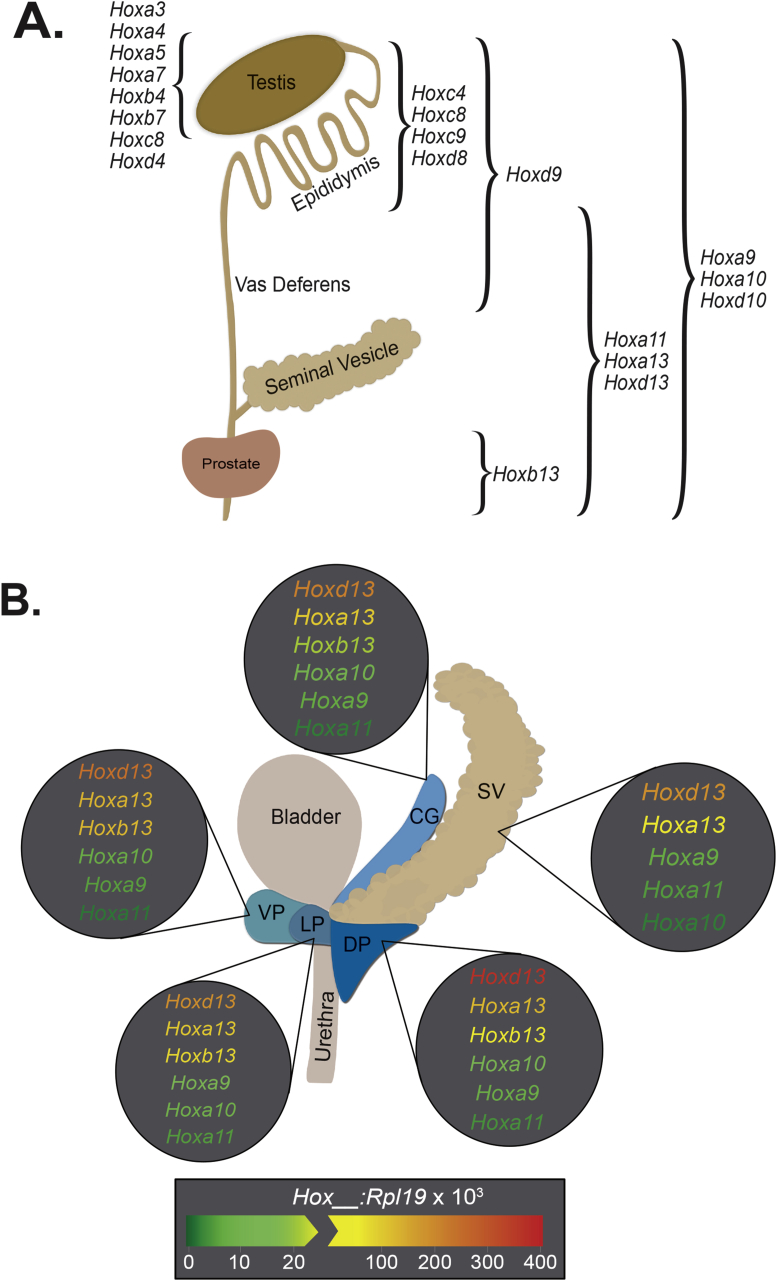
The male reproductive tract is derived from two main developmental structures: the Wolffian (mesonephric) duct, which gives rise to the testis, epididymis, vas deferens, and seminal vesicle; and the urogenital sinus (UGS), which gives rise to the prostate, bulbourethral (Cowper's) glad, bladder, and urethra.13 Given that the reproductive tract is one of the most posterior systems in the body, expression of primarily posterior HOX genes like those in paralog groups 9–13 is most commonly observed (Fig. 1A and B).4, 8, 14 However, several 3′ HOX genes are also expressed in the testis and are thought to have critical roles in spermatogenesis rather than in testis function (Fig. 1A).14
Fig. 1.
Expression patterns of HOX genes in the rodent male reproductive system. A) Depiction of the spatially-restricted pattern of HOX expression in rodent reproductive structures throughout development. Data is compiled from references #4, 8, 16, 18–20, and 73–76. B) Representation of lobe specific, posterior HOX gene expression in the adult rat prostate and seminal vesicle, determined by real time RT-PCR in Huang et al 2007 (reference #4). The seminal vesicle (SV), coagulating gland (CG), ventral prostate (VP), lateral prostate (LP), and dorsal prostate (DP) each have a unique signature of posterior HOX gene expression levels that likely aids in conferring identity. Notably, HOXB13 shows the largest variation in expression between lobes of the prostate and is restricted to urogenital sinus (UGS) derived structures; thus it is absent in the SV. It should also be noted that for studies done in mice, Podlasek et al. demonstrated a different relative expression pattern of HOXA10, HOXA13, and HOXD13 between lobes of the prostate compared to the rat. In their studies, they found that the lowest prostatic expression of HOXA10 was in the CG, rather than VP (reference #19). Additionally, highest expression of HOXD13 was in the SV rather than DP, and followed in order of decreasing expression by the VP, CG, and DP (reference #16). HOXA13 followed a similar pattern as HOXD13, although the CG does not seem to have been analyzed for HOXA13 expression (reference #20). The drawing of the rodent prostate is adapted from reference #77.
Many of the Hox paralogs have redundant and overlapping functions rendering the identification of specific roles for each gene complicated; however, some insight has been gained by observing phenotypes of various Hox gene knockout rodents. For example, while homozygous loss of Hoxa13 (Hoxa13−/−) is considered embryonic lethal due to the perceived role of Hoxa13 in umbilical artery maintenance, examination of Hoxa13−/− fetuses shows severe hypoplasia of the urogenital sinus and arrested or delayed rostral-to-caudal progression of Müllerian ducts.15 Additionally, Hoxd13 deficient mice (Hoxd13−/−) reveal diminished folding in the seminal vesicle stromal sheath, reduced ductal branching and size of the dorsal and ventral prostate lobes, and agenesis of the bulbourethral gland.16 Furthermore, compound homozygous mutants (double Hoxa13−/− and Hoxd13−/−) fetuses have undetectable development of the genital tubercle, nor any distinct hindgut and urogenital sinus, among other deformities.15 In contrast, mice expressing Hoxb13 with a loss-of-function mutation in the homeodomain show no gross morphological defects, but rather have prostate ventral lobe-specific defects in histology and secretory function.17 Histologically, ventral lobe epithelium from Hoxb13 mutant mice are composed of simple cuboidal rather than the tall columnar luminal cells that make up healthy prostate epithelium, and are also devoid of the ventral-specific secretory proteins p12 and p25.17 For a thorough review of reproductive system phenotypes observed with various 5′ Hox gene knockouts, please refer to “Homeobox genes and the male reproductive system” by Rao and Wilkinson.18
In addition to the spatial and temporal patterns of Hox gene expression there is also clear species specificity to the pattern. This is especially well demonstrated when noting the Hox patterns of the prostate in developing mice, rats, and adult humans; however, it should be noted that there is very little data regarding HOX expression in the developing embryonic human prostate. While at a glance, many of the same HOX genes are expressed in all three of these species, the timing, location, and amount of expression can all vary. In murine prostates, Bushman et al found that Hoxa10 expression peaked at embryonic day 19 (E19) and decreased rapidly after birth to near undetectable levels by post-natal day 5 (P5).19 They also showed that Hoxa13 and Hoxd13 expression both peaked around E15 and steadily diminished from there into adulthood; spatially, both Hoxa13 and Hoxd13 had epididymal expression which peaked in the seminal vesicle.20 This observation of Hoxa13 and Hoxd13 expression appears to contrast to the work of Prins et al within the rat prostate demonstrating a postnatal increase in expression that is maintained into adulthood for all three of the previously mentioned genes.4 They also demonstrated that Hoxa13 and Hoxd13 peaked in expression within the dorsal prostate rather than seminal vesicle, and also had a clear anterior boundary at the epididymis.4 Furthermore, in the rat prostate, Prins et al demonstrated Hoxd13 to be the highest expressing Hox gene in each lobe, followed closely by Hoxa13 and Hoxb13, and lastly Hoxa9, Hoxa10, and Hoxa11 each with approximately 10-fold less RNA expression compared to the Hox13 levels.4 In a study evaluating HOX gene expression in a variety of normal adult human organs including prostate, HOXA9, HOXA11, HOXA13, HOXB13, and HOXD9 were all identified as the highest expressing HOX genes with HOXA10 and notably HOXD13 each at a 10-fold lower expression level in the prostate compared to HOXA13 and HOXB13.9 In summary, as expected the 5′ HOX genes (Hoxa-d13) clearly appear to be critical for prostate and GU development, but the timing and location across species is distinct and should be taken into consideration when using animal model systems for HOX biology.
The germline HOXB13-G84E mutation and prostate cancer
The identification of the germline HOXB13(G84E) mutation by Ewing et al within a subset of familial prostate cancers in 2012 brought HOXB13, the genes regulated by HOXB13, and HOX-protein co-factors, into the spotlight of prostate cancer research.2 This discovery highlighted a novel transcriptional regulation pathway that has a key role in prostate development and tumor etiology.2 Patients with the mutation, which substitutes a glutamic acid for glycine at the second position of codon 84, have significantly higher odds for developing prostate cancer than men without the mutation.2 The G84E mutation occurs within the MEIS interaction domain of HOXB13, emphasizing the importance of MEIS-HOX protein interactions in prostate cancer (Fig. 2). Since the initial study, several additional studies have validated the G84E mutation as associated with increased prostate cancer risk (Table 1). It is important to note that the majority of these studies were conducted on Caucasian men of European ancestry, with only 5 of these 22 studies included multiple ethnicities in the study group. In a study conducted by the International Consortium for Prostate Cancer Genetics (ICPCG), they observed a geographical frequency gradient of the G84E mutation across the European continent, with a higher mutation frequency in Nordic countries.21 While multiple studies have corroborated that the G84E mutation is associated with increased prostate cancer risk, the data on the association of G84E with other clinically relevant variables has been mixed. Regarding age of diagnosis, the G84E mutation has been shown to be significantly associated with younger age of diagnosis in the majority of studies,2, 22, 23, 24, 25, 26, 27 with other studies reporting no difference in age of diagnosis.28 A similar pattern has emerged regarding a positive family history of prostate cancer, with all studies reporting a significantly higher odds of the G84E mutation being present in patients with a positive family history or hereditary prostate cancer. In the context of G84E and a potential role in the initiation of more aggressive prostate tumors, Storjberg et al determined that patients carrying the G84E mutation had a significantly higher PSA at diagnosis, higher Gleason score, and a higher likelihood of positive surgical margins at time of radical prostatectomy than non-carriers, implying that the G84E mutation maybe associated with more aggressive prostate cancers.29 However, further analyses are necessary to determine whether mutation of HOXB13 is associated with poor-prognosis prostate tumors. Genetic studies of prostate tumors, however, have documented that in sporadic prostate cancer, HOXB13 is more likely to be amplified but not mutated.30, 31, 32 In summary, the presence of G84E mutation clearly impacts prostate cancer initiation, but data thus far has not strongly implicated the presence of the mutation in contributing to cancer progression and metastasis.
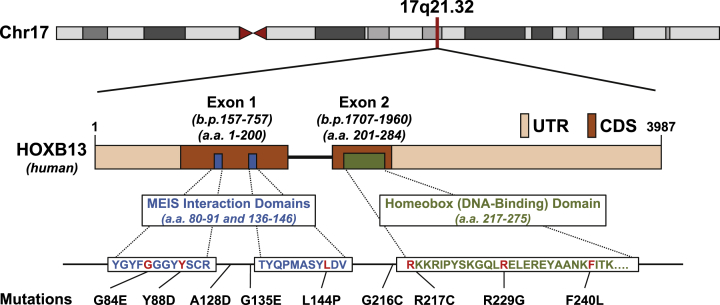
Fig. 2.
Genomic location, domains, and known mutations of human HOXB13. Since the original report of somatic HOXB13(G84E) mutations in a subset of familial cancer, more hereditary mutations conferring increased risk of prostate cancer have been identified (reference #2). The HOXB13 gene is located on human Chromosome 17q21.32 at the 5′ end of the 17q21-22 HOXB cluster, and consists of two exons and three known functional domains (accession number NC_000017/11 and ProtID Q92826). The HOXB13 transcript is 3987 base-pairs (b.p.) long, and Exons 1 and 2 are positioned at 157–757b.p. and 1707–1960b.p, respectively. The regions in beige indicates the untranslated regions (UTR), while the regions in brown indicate coding regions (CDR). The HOXB13 protein is 284 amino acids in length and contains two MEIS-interacting domains (amino acids 80–91 and 136–146) and a single DNA-binding homeobox domain (amino acids 217–275). The two Meis-interaction domains were functionally defined by Williams et al (reference #78 and 79), and the homeodomain was functionally defined in Zeltzer et al (reference #80). Clusters of mutations can be seen within or nearby the two MEIS-interacting domains and the homeodomain.
Table 1.
HoxB13(G84E) mutations in prostate cancer.
| Author | PMID | Study year | Patient population | Age of PrCa Onset G84E Carrierh | Study typei | Genotyping assay | Sample # |
Cancer cases |
Non-cancer controls |
OR (95% CI) | P-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer | Non-cancer | Mutation | Non-mutation | Mutation | Non-mutation | |||||||||
| Akbari | 22781434 | 2012 | Multiple ethnicities, multiple countries | 59.4 | HB | Sanger sequencing | 1853 | 2225 | 10 | 1843 | 2 | 2223 | 5.8 (1.3–26.5) | 0.01 |
| Albitar F | 25874003 | 2015 | USA, Caucasian | NR | HB | Sanger sequencing | 232 | 110 | 2 | 230 | 1 | 109 | 0.95 (0.09–10.6) | 0.97 |
| Beebe-Dimmera | 26108461 | 2015 | Mayo Clinic Biobank, Primarily Caucasian | NR | HB | Taq-Man | 42 | 7218 | 19 | 23 | 1343 | 5875 | 1.99 (1.37–2.90) | <0.0001 |
| Breyer | 22714738 | 2012 | Multiple countries, multiple ethnicities | 53.4 | HB | Taq-Man | 928 | 930 | 20 | 908 | 2 | 928 | 7.9 (1.8–34.5) | 0.0062 |
| Chen | 23393222 | 2013 | Multiple countries, multiple ethnicities | NR | HB | iPLEX MassARRAY | 20 | 3887 | 7 | 13 | 701 | 3186 | RR = 2.45 (1.48–4.07) | 0.01 |
| Ewing* | 22236224 | 2012 | USA, Caucasian | 52.6 | HB | Taq-Man | 5083 | 2662 | 72 | 5011 | 4 | 2658 | 20.1 (3.5–803.3) | 8.50E-07 |
| Gudmundssona | 23104005 | 2012 | Chicago-SPORE, Caucasian | 58.3 | HB | Illumina SNP Chips | 1988 | 1260 | 11 | 1971 | 5 | 1255 | 1.40 (0.49–4.04) | 5.30E-01 |
| Gudmundssonb | 23104005 | 2012 | Iceland, Caucasian | 66.2 | HB | Illumina SNP Chips | 4537 | 54444 | 13 | 4524 | 44 | 54400 | 3.55 (1.91–6.60) | 1.00E-04 |
| Gudmundssonc | 23104005 | 2012 | Netherlands, Caucasian | 63.9 | HB/PB | Illumina SNP Chips | 1520 | 1916 | 23 | 1497 | 4 | 1912 | 7.34 (2.53–21.3) | 3.90E-10 |
| Gudmundssond | 23104005 | 2012 | Spain, Caucasian | NR | HB | Illumina SNP Chips | 717 | 1692 | 1 | 716 | 0 | 1692 | 7.09 (0.29–174.2) | 2.30E-01 |
| Gudmundssone | 23104005 | 2012 | United Kingdom, Caucasian | 61.7 | HB | Illumina SNP Chips | 561 | 1825 | 6 | 505 | 1 | 1824 | 21.67 (2.60–180.4) | 4.40E-03 |
| Gudmundssonf | 23104005 | 2012 | Romanian, Caucasian | 69.4 | HB | Illumina SNP Chips | 722 | 857 | 1 | 721 | 1 | 856 | 1.19 (0.07–19.0) | 9.31E-01 |
| Karlssona | 22841674 | 2014 | Swedish, Caucasian | NR | PB | iPLEX MassARRAY | 2805 | 1709 | 130 | 2675 | 24 | 1685 | 3.4 (2.2–5.4) | 6.40E-10 |
| Karlssonb | 22841674 | 2014 | Swedish, Stockholm-1 group, Caucasian | NR | HB | iPLEX MassARRAY | 2098 | 2880 | 91 | 2007 | 37 | 2843 | 3.5 (2.4–5.2) | 2.00E-11 |
| Kluzniak | 23334858 | 2013 | Polish, caucasian | 67.3 | PB | Taq-Man | 3515 | 2604 | 20 | 3495 | 3 | 2601 | 4.96 (1.47–16.7) | 0.0097 |
| Kote-Jarai | 25595936 | 2015 | United Kingdom, Caucasian | NR | HB | Taq-Man | 8652 | 5252 | 134 | 8518 | 28 | 5224 | 2.94 (1.95–4.42) | <0.0001 |
| Laitinen | 23292082 | 2013 | Finnish, Caucasian | <=55 | HB/PB | Multiple methods | 4571 | 923 | 160 | 4411 | 28 | 895 | 1.15 (0.77–1.74) | 0.47 |
| MacInnis | 23457453 | 2013 | Australian, caucasian | 52.7 | PB | Taq-Man | 1384 | N/A | 19 | 1365 | N/A | N/A | Incidence: 16.4 (2.5–107.2) | N/A |
| Storebjerg | 26779768 | 2016 | Danish | 61.7 | HB | Sanger sequencing | 995 | 1622 | 25 | 970 | 8 | 1614 | 5.12 (0.26–13.38) | 1.30E-05 |
| Stott-Miller | 23129385 | 2013 | USA, Caucasian | NR | PB | Taq-Man | 1457 | 1442 | 18 | 1439 | 5 | 1437 | 3.6 (1.3–9.7) | 0.01 |
| Witte | 23396964 | 2013 | Multiple countries, multiple ethnicities | NR | FB/HB | Taq-Man | 1645 | 1019 | 20 | 1625 | 3 | 1016 | 4.17 (1.24–14.1) | 0.02 |
| Xu | 23064873 | 2013 | Multiple countries, caucasian | 62.8 | FB | iPLEX MassARRAY | 326 | 117 | 154 | 172 | 36 | 81 | 2.01 (1.29–3.16) | 0.002 |
a, b, c, d, e, f: Data from multiple populations present within a single study.
h: Not reported.
i: FB = Family Based; HB = Hospital Based; PB = Population Based.
Other germline HOXB13 mutations associated with prostate cancer risk
Since the discovery of the G84E mutation, there has been greater focus on identifying other novel germline mutations of HOXB13 associated with increased prostate cancer risk. This is of particular importance for non-Caucasian populations, as the risk of prostate cancer associated with the G84E mutation has the highest frequency in European/Caucasian populations. Indeed, new mutations of HOXB13 conferring increased prostate cancer risk have begun to be identified in non-Northern European ancestry. Notably, Lin et al identified the novel G135E mutation to be associated with increased prostate cancer risk in a population of Chinese men, and did not identify the presence of the G84E mutation.33 Similarly, Maia et al identified the A128D and F240L mutations in a population of Portuguese men to associated with prostate cancer risk.34 Ewing et al reported the identification of several rare missense variants of HOXB13 (Y88D, L144P, G216C, R217C, and R229G, Fig. 2 and Table 2) during their initial study of G84E. Of these rare mutations, the R229G and G216C were identified in men with some African ancestry.2 Given the paucity of data, however, on non-G84E mutations of HOXB13, and the lack of study of prostate cancer risk mutations in non-Caucasian populations, continued efforts to identify novel risk mutations of HOXB13 are necessary.
Table 2.
Germline and somatic HoxB13 mutations in cancer.
| Author | PMID | Study year | Cancer primary | Patient population | Study typef | Primary mutation | Germline or somatic | Cancer cases |
Non-cancer controls |
Genotyping assay | OR (95% CI) | P value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutations | Non-mutations | Mutations | Non-mutations | |||||||||||
| Akbari MRa | 23099437 | 2012 | Breast | Canadian Caucasian | HB | G84E | Germline | 2 | 1802 | 1 | 924 | Taq-Man | 1.0 (0.09–11.3) | 0.98 |
| Akbari MRb | 23099437 | 2012 | Breast | Polish Caucasian | HB | G84E | Germline | 5 | 2228 | 3 | 1834 | Taq-Man | 1.37 (0.33–5.75) | 0.67 |
| Akbari MR | 23541221 | 2013 | Colorectal | Canadian, Australian | PB | G84E | Germline | 13 | 2682 | 8 | 4585 | Taq-Man | 2.8 (1.2–6.7) | 0.02 |
| Beebe-Dimmerb | 26108461 | 2015 | Bladder | Primary Caucasian | HB | G84E | Germline | 3 | 23 | 205 | 5875 | Taq-Man | 1.99 (0.84–3.86) | 0.06 |
| Beebe-Dimmerc | 26108461 | 2015 | Leukemia | Primary Caucasian | HB | G84E | Germline | 3 | 23 | 86 | 5875 | Taq-Man | 3.17 (1.35–6.03) | 0.01 |
| Beebe-Dimmerd | 26108461 | 2015 | Sarcoma | Primary Caucasian | HB | G84E | Germline | 1 | 23 | 123 | 5875 | Taq-Man | 1.48 (0.23–3.80) | 0.4 |
| Beebe-Dimmere | 26108461 | 2015 | Testis | Primary Caucasian | HB | G84E | Germline | 1 | 24 | 49 | 5888 | Taq-Man | 2.31 (0.36–5.86) | 0.18 |
| Laitinena | 23292082 | 2013 | Breast | Finnish, Caucasian | HB/PB | G84E | Germline | 16 | 970 | 16 | 1433 | Multiple methods | 1.48 (0.74–2.97) | 0.27 |
| Laitinenb | 23292082 | 2013 | Colorectal | Finnish, Caucasian | HB/PB | G84E | Germline | 7 | 435 | 0 | 459 | Multiple methods | 15.83 (0.90–277.95) | 0.06 |
| Lin | 22718278 | 2013 | Prostate | Chinese | PB | G135E | Germline | 3 | 639 | 0 | 1491 | iPLEX MassARRAY | 16.33 (0.84–316.54) | 0.065 |
| Maia | 26176944 | 2015 | Prostate | Portuguese | FB | A128D/F248L | Germline | 3 | 459 | 0 | 132 | AB 3500 Genetic Analyzer | 2.02 (0.10–39.3) | 0.64 |
| Ewing | 22236224 | 2012 | Prostate | USA, Caucasian | HB | Y88D | Unknowng | N/A | N/A | N/A | N/A | Taq-Man | N/A | N/A |
| Ewing | 22236224 | 2012 | Prostate | USA, Caucasian | HB | L144P | Unknownh | N/A | N/A | N/A | N/A | Taq-Man | N/A | N/A |
| Ewing | 22236224 | 2012 | Prostate | USA, Caucasian | HB | G216C | Germline | 1 | 90 | N/A | N/A | Taq-Man | N/A | N/A |
| Ewing | 22236224 | 2012 | Prostate | USA, Caucasian | HB | R229G | Germline | 1 | 90 | N/A | N/A | Taq-Man | N/A | N/A |
| Xu | 23064873 | 2013 | Prostate | Multiple countries, caucasian | FB | R217C | Germline | 2 | 6420 | 0 | 1902 | iPLEX MassARRAY | 1.48 (0.07–30.9) | 0.8 |
a, b, c, d, e: Data from multiple populations present within a single study.
f: FB = Family Based; HB = Hospital Based; PB = Population Based.
g: Unknown, mutation found in LAPC4 Cell Line.
h: Unknown, mutation found in LNCaP Cell Line.
The function of HOXB13 in the developing and adult prostate
HOXB13 is unique in the prostate because it is highly expressed into adulthood in multiple species, and yet it is the most differentially-expressed HOX protein when comparing between lobes of the rodent prostate, suggesting that it may have more important functions in determining prostatic identity and maintaining organ homeostasis in an adult.4, 35 Within the normal adult human prostate, HOXB13 is localized exclusively in prostate luminal epithelial cells.17, 36 In rodent models, Hoxb13 is most highly expressed in the ventral prostate lobe, has been shown to drive differentiation of prostate luminal epithelial cells, and is also required for the normal secretory function of the ventral prostate.4, 17, 35
An important and somewhat controversial body of data pertains to the relationship between HOXB13 and the Androgen Receptor (AR). This pertains to both the regulation of HOXB13 by AR and cooperation with AR signaling. HOXB13 expression in the prostate is thought to be androgen-independent, as demonstrated by Bieberich et al whereby the steady state mRNA level of HOXB13 in the murine prostate was undiminished 8 days after host castration.36 However, Prins et al observed increased Hoxb13 expression in the rat prostatic ventral lobe upon administration of testosterone, and expression was decreased in the dorsal and lateral lobes upon castration.4 This apparent discrepancy could be accounted for by changes in prostatic cellularity in the context of hormone administration or depletion, since castration results in a significant reduction of HOXB13-positive luminal epithelial cells. In addition to regulation of HOXB13 by androgen signaling, it has been shown that HOXB13 can act as a bivalent regulator of AR chromatin binding and function as either a growth-promoter or growth-suppressor in prostate cancer cells depending on the cellular context.37 For example, in androgen-sensitive prostate cancer cell lines such as LNCaP, increased HOXB13 activity can decrease levels of Cyclin D1 and lead to growth inhibition through reduction of pRb phosphorylation and stabilization of the pRB-E2F complex.5, 6 Conversely, in castration-resistant prostate tumors, HOXB13 overexpression can inhibit p21 and thus act as an oncogene through subsequently promoting E2F activation and cell cycle progression.6 A final noteworthy observation of HOXB13 localization is that, in human radical prostatectomy samples, the nuclear/cytoplasmic ratios of HOXB13 are drastically reduced in prostatic intraepithelial neoplasia (PIN) and prostate cancer when compared to normal glands, indicating much higher cytoplasmic retention and thus lower amounts of functional HOXB13 in the nucleus of tumor cells.5 This suggests a potential mechanism of abrogating the growth-suppressive function of HOXB13 by cytosolic retention. Collectively, these observations highlight numerous important and interesting roles of HOXB13 in the prostate, but also underscore the need for additional mechanistic and functional studies to elucidate the molecular function of HOXB13 within the normal prostate and during prostate tumor initiation.
Germline HOXB13(G84E) in non-prostate tumors
Given the strong relationship between the HOXB13(G84E) mutation and prostate cancer risk, as well as the importance of HOXB13 in development and cancer, several studies have examined the role of HOXB13 mutations in increasing the risk of other tumor types (Table 2). Results between the association of G84E and non-prostate cancer risk have been mixed. Notably, Akbari et al and Beebe-Dimmer et al showed that the G84E mutation was associated with a significantly increased risk of colorectal carcinoma and leukemia, respectively.38, 39 However, Latinen et al showed no significant association between the G84E mutation and colorectal cancer risk, although their results did approach significance.27 The G84E mutation has also been investigated in breast, bladder, testis, and sarcoma, but results have not shown a significant association between the mutation and increased cancer risk among those cancers studied.38 However, it should be noted that a few of these studies approached near significance, and additional studies containing a larger sample size has the potential, in some instances, to establish a significant correlation between the G84E mutation and non-prostate cancer risk.
Deregulation of HOXB13 in non-prostate tumors
Despite its emerging role in prostate cancer, deregulation of HOXB13 expression has been implicated in a variety of human cancers, functioning either as a tumor-promoting factor in some tumor types, or a tumor-repressing factor in others (Table 2). Surprisingly, aberrant expression of HOXB13 has been documented in a variety of non-posterior axis cancers, including thyroid, breast, metastatic melanoma, and oral squamous cell (Table 2). In many instances, however, the functional significance of such expression has yet to be determined. In endometrial, ovarian, melanoma, and breast tumors, increased HOXB13 expression appears to promote tumor progression.40, 41, 42 In endometrial tumors, Yamashita et al demonstrated HOXB13 expression in tumor tissues and demonstrated that HOXB13 over-expression led to increased cellular invasion in vitro.40 In ovarian cancer, Miao et al demonstrated that over-expression of HOXB13 in ovarian cancer cells resulted in increased cell proliferation and survival.41 In melanoma, Maeda et al showed that the expression levels of HOXB13 were significantly higher in patients with metastatic melanoma compared to patients with a non-metastatic primary melanoma.43 In breast cancer, HOXB13 expression is predictive of a poor clinical outcome in tamoxifen-treated breast cancers, indicating that increased HOXB13 could have a prognostic role in breast cancer.44 Furthermore, ectopic expression of HOXB13 in MCF10A breast epithelial cells enhances motility and invasion in vitro, and HOXB13 expression is increased in both pre-invasive and invasive primary breast cancer.44
While the majority of the current literature demonstrates that HOXB13 is generally over-expressed and tumor-promoting in most cancers, several studies support a role for HOXB13 as a tumor-suppressor within other cancer contexts. Jung et al and Kanai et al showed that HOXB13 expression is decreased in primary colorectal adenocarcinoma, and that over-expression of HOXB13 inhibits cell proliferation in colorectal cancer cell lines.44, 45 Furthermore, Cantile et al showed a progressive decrease in HOXB13 nuclear expression in the transition from non-neoplastic thyroid to adenoma to different histologic types of thyroid cancer.46 In bladder cancer, Marra et al found that the loss of nuclear HOXB13 is implicated in shorter disease free survival in non-muscle invasive bladder cancer and decreased nuclear HOXB13 correlates with muscle invasion.47 Thus, it is clear that aberrant expression of HOXB13 plays a key role in the progression of many different cancer types, including both non-genitourinary and genitourinary cancers. Moreover, the context-dependent tumor promoting or repressing functions of HOXB13 further underscore key organ-specific roles of HOXB13 in cancer. Hence, it is the HOXB13-associated binding partners that provide specificity to DNA binding and subsequent gene targets who are the key mediators of HOX-associated tumor initiation and progression. Additional investigation into the function of HOXB13 and its binding partners across various tumors types is thus warranted.
HOX protein binding partners
It has been a long-established paradox that HOX proteins achieve exquisite in vivo gene specificity to program development using simple “AT-rich” gene recognition motifs; such motifs are much too common across the genome to allow HOX proteins working alone to attain such gene specificity (expertly reviewed in Mann et al).48 To accomplish such specificity, HOX proteins rely on multiple co-factors to bind and specify transcriptional activity. The TALE (three amino acid loop extension) proteins are the predominant subtype of homeobox proteins that partner with HOX proteins and specify gene targeting and activity. This family of proteins includes the MEIS, PBX, PKNOX and TGIF homeobox proteins. While they contain the homologous DNA binding domain canonically found in homeobox genes, there are three unique characteristics of the TALE family. First, a three amino acid insertion in their homeodomains allows for cooperative binding to other transcription cofactors.49 It is this ability to create complexes that provide increased binding affinity of homeobox complexes to the DNA. Importantly, not every TALE protein group can bind to every other homeobox gene, increasing specificity of DNA binding depending on the combination of factors present in a complex.49 Second, the regions flanking the homeodomains of TALE proteins are highly conserved across species.50 Third and finally, unlike their spatiotemporally-restricted HOX relatives, TALE factors are more widely expressed across an organism.
While many TALE factors have been implicated in cancers, the recent discovery of the HOXB13 mutations in hereditary prostate cancer to confer a risk for prostate cancer discussed above has sparked an interest in the MEIS proteins in particular.2 Many of the mutations within the HOXB13 gene fall within the MEIS binding domain (Fig. 2). While it is clear, as discussed above, that HOXB13 mutations are strongly associated with increased prostate cancer risk, there are significant gaps of knowledge regarding the mechanism of action of HOXB13 mutations, and how co-factor modulations impact prostate cancer initiation.
The MEIS (murine ecotropic integration site) gene was implicated in cancer based upon the discovery that the MEIS1 gene was the most common location for an ecotropic murine leukemia virus to integrate.51 When the virus integrated, higher expression was noted as the mice developed leukemia, and this was the first indication of MEIS as oncogenes in liquid tumors.51 MEIS proteins function as DNA-binding cofactors with the HOX and PBX families such that the cooperative binding increases DNA binding specificity.48, 52, 53, 54 Our current understanding of MEIS-HOX interactions is that, upon DNA binding of the two, DNA-bound MEIS/HOX complexes recruit collaborator proteins to compile a multimeric protein complex at specific gene promoters.48 It should be noted, however, that TALE proteins have both HOX-dependent and HOX-independent functions and their role in development and disease likely extends beyond regulation of HOX protein DNA specificity.48, 54
Deregulation of MEIS proteins in cancer
While little is known about the MEIS and PBX proteins in the context of prostate cancer, current understanding of functions in other cancer types may provide directions for future work. MEIS proteins have complicated and context-dependent roles in cancer initiation and progression. They are down-regulated in some cancer types, but overexpressed in others, making it unclear if MEIS genes are bona fide oncogenes or tumor suppressors genes. This phenomenon of fluidity between tumor suppression and oncogenesis is not unheard of; in fact, HOX genes display a very similar pattern, as discussed above.12
The most well studied context for the role of MEIS, PBX and HOX proteins in cancer is leukemia, and specifically AML (Acute Myeloid Leukemia) and MLL (Mixed Lineage Leukemia). MEIS1 is required for normal adult bone marrow hematopoiesis, with deletion of MEIS1 leading to stem cell exhaustion and an inability to self-renew.55 MEIS1 alone is not sufficient to transform hematopoietic cells however, as MEIS1 requires the cooperation of HOXA9 to accelerate HOX-induced leukemia.56 There is a common theme across many publications investigating MEIS in leukemia; MEIS proteins can mitigate differentiation while also increasing proliferation, a deadly and oncogenic combination. MEIS1 and HOXA9 are direct targets of the MLL fusion gene57 and MEIS1, in addition to the redundant contributions of PBX2 and 3, appears to be the rate-limiting step in the cell cycle progression of MLL leukemia stem cells.58 In fact, it was shown recently that PBX3 is crucial to help stabilize MEIS1 proteins, and that the dimerization of PBX3 and MEIS1 is required for HOX-induced leukemia.59 In myeloid leukemias, the full length MEIS1-A is able to stop differentiation through G-SCF and promotes proliferation.60
The connection between MEIS1 and the cell cycle, as well as maintenance of a more primitive stem cell state across multiple cell types are likely mechanisms of action that lead to its deregulation in a range of pathological contexts. For example, MEIS1 slows adult and neonatal proliferation in cardiomyocytes by modulating the progression of the cell cycle.61 There are also multiple papers indicating a role for MEIS in Restless Leg Syndrome, and more information on MEIS′ role in this disease can be found in a 2014 review by Garcia-Borreguero et al62 Neuroblastoma displays MEIS1 up-regulation in many cell lines and tumors.63 Neuroblastoma is also the context where many novel, and potentially functionally distinct, MEIS isoforms have been investigated.64 In neuroblastoma SJNB-8 cells, the exogenous expression of MEIS1-E, an isoform lacking a DNA binding domain, induces changes in cell growth proliferation apoptosis, cytoskeleton, long-distance gene regulation, morphogenesis, protein transport, and differentiation markers.64, 65 This analysis, however, did not indicate the direction of change for many of these processes.65 MEIS2 is critical for neuroblastoma cell survival and proliferation by asserting control over M-phase of the cell cycle, again illustrating a cell cycle control function for MEIS in cancer cells.66 Lung adenocarcinomas, in particular those with LKB1 mutations, also show up-regulation of MEIS2, though investigation of the mechanism of action has not been elucidated.67 Thus, in numerous tumor sites, MEIS1 and MEIS2, and potential splice-variants of each, appear to function as promoters of cell cycle progression, and in some instances to maintain cancer cells in a less-differentiated state.
While the majority of cancer-related research into the MEIS and PBX transcription factors has been focused on their overexpression in leukemia, there are many pathological contexts where MEIS proteins appear to function as tumor suppressors. In fact, MEIS proteins can act as a tumor suppressor or oncogene even within a specific organ site of carcinogenesis; however, their roles are restricted to specific molecular subtypes. For example, in the majority of AML cases MEIS1 and HOXA9 act as oncogenes, while within a particular subtype of patients MEIS1 and HOXA9 expression are significantly decreased compared to other AML subtypes where such transcripts are over-represented.68 Patients with the AML-ETO fusion protein show low MEIS1 and HOXA9 mRNA as compared to other AML patients where high MEIS1 expression are typical. MEIS1 down regulation in the AML-ETO patient population is due to methylation at its promoter.68 Additionally, as described above, lung adenocarcinomas with LKB1 mutations display over-expression of MEIS whereas in NSCLC (non-small cell lung cancer) patients, MEIS1 over-expression inhibits cell growth and MEIS1 knock down using siRNA-targeting increases proliferation.69 In this NSCLC context, DNA synthesis is increased when MEIS1 decreased.69 Colorectal adenomas displayed a seven-fold decrease in MEIS transcripts, in particular a homeodomain-truncated splice-variant MEIS1-D.70, 71 Thus, in numerous tumor types, MEIS expression appears to be actively inhibited, either via down-regulation or expression of dominant-negative splice variants.
MEIS and PBX proteins in prostate cancer
MEIS and PBX proteins have been vastly understudied in the context of prostate cancer, and there are numerous avenues of future investigation with clear clinical impact. MEIS1 has been shown to act as an androgen receptor suppressor, where ectopic expression slows LNCaP prostate cancer cell growth.72 MEIS1 can physically interact with the androgen receptor, the most critical driver of prostate cancer and the main target of clinical intervention.72, 73, 74 Moreover, work published by our group demonstrates that higher expression of MEIS1 and MEIS2 provide a survival benefit to men with intermediate and low-grade prostate cancer.73 In the normal prostate, MEIS expression is highest in basal epithelial cells and stromal cells, with a detectable but significantly lower expression in luminal epithelial cells. Tumors with below average MEIS1 and MEIS2 expression convey a significant decrease in patient survival, suggesting a functional role for decreased MEIS expression and the initiation and progression of poor-prognosis prostate tumors.73 Similarly, tumor expression of PBX3 was associated with improved prostate cancer specific survival compared to patients expressing low levels; this study statistically demonstrated that PBX3 expression could be used to potentially predict outcome and enhance tumor staging.75 However, significant additional work is required to more comprehensively understand the function of MEIS and other TALE proteins in prostate cancer.
Conclusions and future directions
Genetic and informatics studies in prostate cancer have clearly implicated a key role for MEIS/HOX signaling in prostate cancer initiation, and have created multiple avenues of potentially fruitful and impactful investigation. Based upon our understanding of MEIS/HOX function in other tumor types and our limited understanding in prostate cancer, several future research questions can be postulated. First, how does the HOXB13(G84E) mutation functionally lead to increased cancer initiation? It is important to elucidate when, during the development and maintenance of the prostate, the G84E mutation manifests itself; that is, to determine whether the prostate of a G84E carrier develops differently or does the G84E mutation impact prostate homeostasis and turnover after puberty and sexual maturity. Second, do other HOXB13 mutations beyond G84E impact prostate function similarly or do they have unique etiologies and function? Third, how does the G84E mutation, and other HOXB13 mutations, functionally modulate MEIS function and MEIS/HOX interactions? Mechanistic studies investigating whether the G84E mutation abrogates or modulates MEIS interaction, and the transcriptional impact of HOXB13 mutations on HOXB13 target genes, will illuminate how the G84E mutation leads to prostate tumor initiation. Fourth and finally, how can MEIS/HOX expression, and their gene targets, be exploited for patient benefit? Efforts to screen and genetically counsel individuals with HOXB13 mutations are clearly warranted; however, mechanistic studies of MEIS/HOX transcriptional function has the high potential to identify targetable pathways for tumor prevention and staging.
Conflicts of interest
None.
Acknowledgments
DOD PCRP PC130587 (Vander Griend); NWU/UC/NSUHS Prostate SPORE (P50 CA180995) the University of Chicago Comprehensive Cancer Center (UCCCC), especially the Cancer Center Support Grant (P30CA014599); H. Brechka and C. Van Opstall were supported by the Cancer Biology Training Grant (T32 CA009594); R. Bhanvadia is supported by a University of Chicago Pritzker School of Medicine Fellowship.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ewing C.M., Ray A.M., Lange E.M. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerda-Esteban N., Spagnoli F.M. Glimpse into Hox and tale regulation of cell differentiation and reprogramming. Dev Dyn. 2014;243:76–87. doi: 10.1002/dvdy.24075. [DOI] [PubMed] [Google Scholar]
- 4.Huang L., Pu Y., Hepps D., Danielpour D., Prins G.S. Posterior Hox gene expression and differential androgen regulation in the developing and adult rat prostate lobes. Endocrinology. 2007;148:1235–1245. doi: 10.1210/en.2006-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamid S.M., Cicek S., Karamil S. HOXB13 contributes to G1/S and G2/M checkpoint controls in prostate. Mol Cell Endocrinol. 2014;383:38–47. doi: 10.1016/j.mce.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Rezsohazy R., Saurin A.J., Maurel-Zaffran C., Graba Y. Cellular and molecular insights into Hox protein action. Development. 2015;142:1212–1227. doi: 10.1242/dev.109785. [DOI] [PubMed] [Google Scholar]
- 7.Lappin T.R., Grier D.G., Thompson A., Halliday H.L. HOX genes: seductive science, mysterious mechanisms. Ulst Med J. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- 8.Prins G.S., Putz O. Molecular signaling pathways that regulate prostate gland development. Differentiation. 2008;76:641–659. doi: 10.1111/j.1432-0436.2008.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi Y., Hamada J., Murakawa K. Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Exp Cell Res. 2004;293:144–153. doi: 10.1016/j.yexcr.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Favier B., Dolle P. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. doi: 10.1093/molehr/3.2.115. [DOI] [PubMed] [Google Scholar]
- 11.Mallo M., Wellik D.M., Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 13.Sadler T.W. Urogenital system. In: Sadler T.W., editor. Langman's Medical Embryology. 12th ed. Lippincott Williams & Wilkins; Baltimore, MD: 2012. pp. 232–259. [Google Scholar]
- 14.Rao M., Wilkinson M.F. Homeobox genes and the male reproductive system. In: Robaire BaH B.T., editor. The Epididymis: From Molecules to Clinical Practice. Springer; United States: 2002. pp. 269–283. [Google Scholar]
- 15.Warot X., Fromental-Ramain C., Fraulob V., Chambon P., Dolle P. Gene dosage-dependent effects of the Hoxa-13 and Hoxd-13 mutations on morphogenesis of the terminal parts of the digestive and urogenital tracts. Development. 1997;124:4781–4791. doi: 10.1242/dev.124.23.4781. [DOI] [PubMed] [Google Scholar]
- 16.Podlasek C.A., Duboule D., Bushman W. Male accessory sex organ morphogenesis is altered by loss of function of Hoxd-13. Dev Dyn. 1997;208:454–465. doi: 10.1002/(SICI)1097-0177(199704)208:4<454::AID-AJA2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 17.Economides K.D., Capecchi M.R. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development. 2003;130:2061–2069. doi: 10.1242/dev.00432. [DOI] [PubMed] [Google Scholar]
- 18.Rao M., Wilkinson M.F. Homeobox genes and the male reproductive system. In: Robaire B., Hinton B.T., editors. The Epididymis: From Molecules to Clinical Practice: A Comprehensive Survey of the Efferent Ducts, the Epididymis and the Vas Deferens. Springer US; Boston, MA: 2002. pp. 269–283. [Google Scholar]
- 19.Podlasek C.A., Seo R.M., Clemens J.Q., Ma L., Maas R.L., Bushman W. Hoxa-10 deficient male mice exhibit abnormal development of the accessory sex organs. Dev Dyn. 1999;214:1–12. doi: 10.1002/(SICI)1097-0177(199901)214:1<1::AID-DVDY1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Podlasek C.A., Clemens J.Q., Bushman W. Hoxa-13 gene mutation results in abnormal seminal vesicle and prostate development. J Urol. 1999;161:1655–1661. [PubMed] [Google Scholar]
- 21.Xu J., Lange E.M., Lu L. HOXB13 is a susceptibility gene for prostate cancer: results from the International Consortium for Prostate Cancer Genetics (ICPCG) Hum Genet. 2013;132:5–14. doi: 10.1007/s00439-012-1229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbari M.R., Trachtenberg J., Lee J. Association between germline HOXB13 G84E mutation and risk of prostate cancer. J Natl Cancer Inst. 2012;104:1260–1262. doi: 10.1093/jnci/djs288. [DOI] [PubMed] [Google Scholar]
- 23.Gudmundsson J., Sulem P., Gudbjartsson D.F. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson R., Aly M., Clements M. A population-based assessment of germline HOXB13 G84E mutation and prostate cancer risk. Eur Urol. 2014;65:169–176. doi: 10.1016/j.eururo.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Kluzniak W., Wokolorczyk D., Kashyap A. The G84E mutation in the HOXB13 gene is associated with an increased risk of prostate cancer in Poland. Prostate. 2013;73:542–548. doi: 10.1002/pros.22594. [DOI] [PubMed] [Google Scholar]
- 26.Kote-Jarai Z., Mikropoulos C., Leongamornlert D.A. Prevalence of the HOXB13 G84E germline mutation in British men and correlation with prostate cancer risk, tumour characteristics and clinical outcomes. Ann Oncol. 2015;26:756–761. doi: 10.1093/annonc/mdv004. [DOI] [PubMed] [Google Scholar]
- 27.Laitinen V.H., Wahlfors T., Saaristo L. HOXB13 G84E mutation in Finland: population-based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22:452–460. doi: 10.1158/1055-9965.EPI-12-1000-T. [DOI] [PubMed] [Google Scholar]
- 28.Stott-Miller M., Karyadi D.M., Smith T. HOXB13 mutations in a population-based, case-control study of prostate cancer. Prostate. 2013;73:634–641. doi: 10.1002/pros.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storebjerg T.M., Hoyer S., Kirkegaard P. Prevalence of the HOXB13 G84E mutation in Danish men undergoing radical prostatectomy and its correlations with prostate cancer risk and aggressiveness. BJU Int. 2016;118:646–653. doi: 10.1111/bju.13416. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A., Coleman I., Morrissey C. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasso C.S., Wu Y.M., Robinson D.R. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research N The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X., Qu L., Chen Z. A novel germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate. 2013;73:169–175. doi: 10.1002/pros.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maia S., Cardoso M., Pinto P. Identification of two novel HOXB13 germline mutations in Portuguese prostate cancer patients. PLoS One. 2015;10:e0132728. doi: 10.1371/journal.pone.0132728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMullin R.P., Mutton L.N., Bieberich C.J. Hoxb13 regulatory elements mediate transgene expression during prostate organogenesis and carcinogenesis. Dev Dyn. 2009;238:664–672. doi: 10.1002/dvdy.21870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sreenath T., Orosz A., Fujita K., Bieberich C.J. Androgen-independent expression of hoxb-13 in the mouse prostate. Prostate. 1999;41:203–207. doi: 10.1002/(sici)1097-0045(19991101)41:3<203::aid-pros8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 37.Norris J.D., Chang C.Y., Wittmann B.M. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405–416. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akbari M.R., Anderson L.N., Buchanan D.D. Germline HOXB13 p.Gly84Glu mutation and risk of colorectal cancer. Cancer Epidemiol. 2013;37:424–427. doi: 10.1016/j.canep.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beebe-Dimmer J.L., Hathcock M., Yee C. The HOXB13 G84E mutation is associated with an increased risk for prostate cancer and other malignancies. Cancer Epidemiol Biomarkers Prev. 2015;24:1366–1372. doi: 10.1158/1055-9965.EPI-15-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Yamashita T., Ishikawa M. Regulation of tumor invasion by HOXB13 gene overexpressed in human endometrial cancer. Oncol Rep. 2005;13:721–726. [PubMed] [Google Scholar]
- 41.Miao J., Wang Z., Provencher H. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci USA. 2007;104:17093–17098. doi: 10.1073/pnas.0707938104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez R., Garrido E., Pina P. HOXB homeobox gene expression in cervical carcinoma. Int J Gynecol Cancer. 2006;16:329–335. doi: 10.1111/j.1525-1438.2006.00350.x. [DOI] [PubMed] [Google Scholar]
- 43.Maeda K., Hamada J., Takahashi Y. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer. 2005;114:436–441. doi: 10.1002/ijc.20706. [DOI] [PubMed] [Google Scholar]
- 44.Ma X.J., Wang Z., Ryan P.D. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Kanai M., Hamada J., Takada M. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep. 2010;23:843–851. [PubMed] [Google Scholar]
- 46.Cantile M., Scognamiglio G., La Sala L. Aberrant expression of posterior HOX genes in well differentiated histotypes of thyroid cancers. Int J Mol Sci. 2013;14:21727–21740. doi: 10.3390/ijms141121727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marra L., Cantile M., Scognamiglio G. Deregulation of HOX B13 expression in urinary bladder cancer progression. Curr Med Chem. 2013;20:833–839. [PubMed] [Google Scholar]
- 48.Mann R.S., Lelli K.M., Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang C.P., Shen W.F., Rozenfeld S., Lawrence H.J., Largman C., Cleary M.L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 50.Burglin T.R. The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes? Dev genes Evol. 1998;208:113–116. doi: 10.1007/s004270050161. [DOI] [PubMed] [Google Scholar]
- 51.Moskow J.J., Bullrich F., Huebner K., Daar I.O., Buchberg A.M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moens C.B., Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 53.Mann R.S., Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 54.Mukherjee K., Burglin T.R. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- 55.Ariki R., Morikawa S., Mabuchi Y. Homeodomain transcription factor Meis1 is a critical regulator of adult bone marrow hematopoiesis. PLoS One. 2014;9:e87646. doi: 10.1371/journal.pone.0087646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kroon E., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar A.R., Li Q., Hudson W.A. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong P., Iwasaki M., Somervaille T.C., So C.W., Cleary M.L. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia-Cuellar M.P., Steger J., Fuller E., Hetzner K., Slany R.K. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox-induced murine leukemia. Haematologica. 2015;100:905–913. doi: 10.3324/haematol.2015.124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calvo K.R., Knoepfler P.S., Sykes D.B., Pasillas M.P., Kamps M.P. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci USA. 2001;98:13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahmoud A.I., Kocabas F., Muralidhar S.A. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia-Borreguero D., Williams A.M. An update on restless legs syndrome (Willis-Ekbom disease): clinical features, pathogenesis and treatment. Curr Opin Neurol. 2014;27:493–501. doi: 10.1097/WCO.0000000000000117. [DOI] [PubMed] [Google Scholar]
- 63.Jones T.A., Flomen R.H., Senger G., Nizetic D., Sheer D. The homeobox gene MEIS1 is amplified in IMR-32 and highly expressed in other neuroblastoma cell lines. Eur J Cancer. 2000;36:2368–2374. doi: 10.1016/s0959-8049(00)00332-4. [DOI] [PubMed] [Google Scholar]
- 64.Geerts D., Revet I., Jorritsma G., Schilderink N., Versteeg R. MEIS homeobox genes in neuroblastoma. Cancer Lett. 2005;228:43–50. doi: 10.1016/j.canlet.2005.01.047. [DOI] [PubMed] [Google Scholar]
- 65.Geerts D., Schilderink N., Jorritsma G., Versteeg R. The role of the MEIS homeobox genes in neuroblastoma. Cancer Lett. 2003;197:87–92. doi: 10.1016/s0304-3835(03)00087-9. [DOI] [PubMed] [Google Scholar]
- 66.Zha Y., Xia Y., Ding J. MEIS2 is essential for neuroblastoma cell survival and proliferation by transcriptional control of M-phase progression. Cell Death Dis. 2014;5:e1417. doi: 10.1038/cddis.2014.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fernandez P., Carretero J., Medina P.P. Distinctive gene expression of human lung adenocarcinomas carrying LKB1 mutations. Oncogene. 2004;23:5084–5091. doi: 10.1038/sj.onc.1207665. [DOI] [PubMed] [Google Scholar]
- 68.Lasa A., Carnicer M.J., Aventin A. MEIS 1 expression is downregulated through promoter hypermethylation in AML1-ETO acute myeloid leukemias. Leukemia. 2004;18:1231–1237. doi: 10.1038/sj.leu.2403377. [DOI] [PubMed] [Google Scholar]
- 69.Li W., Huang K., Guo H., Cui G. Meis1 regulates proliferation of non-small-cell lung cancer cells. J Thorac Dis. 2014;6:850–855. doi: 10.3978/j.issn.2072-1439.2014.06.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sabates-Bellver J., Van der Flier L.G., de Palo M. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 71.Crist R.C., Roth J.J., Waldman S.A., Buchberg A.M. A conserved tissue-specific homeodomain-less isoform of MEIS1 is downregulated in colorectal cancer. PLoS One. 2011;6:e23665. doi: 10.1371/journal.pone.0023665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cui L., Li M., Feng F. MEIS1 functions as a potential AR negative regulator. Exp Cell Res. 2014;328:58–68. doi: 10.1016/j.yexcr.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 73.Chen J.L., Li J., Kiriluk K.J. Deregulation of a Hox protein regulatory network spanning prostate cancer initiation and progression. Clin Cancer Res. 2012;18:4291–4302. doi: 10.1158/1078-0432.CCR-12-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jung C., Kim R.S., Zhang H.J., Lee S.J., Jeng M.H. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004;64:9185–9192. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 75.Ramberg H., Grytli H.H., Nygard S. PBX3 is a putative biomarker of aggressive prostate cancer. Int J Cancer. 2016;139:1810–1820. doi: 10.1002/ijc.30220. [DOI] [PubMed] [Google Scholar]