Abstract
Biomass deconstruction to small simple sugars is a potential approach to biofuels production, however the highly recalcitrant nature of biomass limits the economic viability of this approach. Thus, research on efficient biomass degradation is necessary to achieve large-scale production of biofuels. Enhancement of cellulolytic activity by increasing synergism between cellulase enzymes holds promise in achieving high-yield biofuels production. Here we have inserted cellulase pairs from extremophiles into hyper-stable α-helical consensus ankyrin repeat domain scaffolds. Such chimeric constructs allowed us to optimize arrays of enzyme pairs against a variety of cellulolytic substrates. We found that endocellulolytic domains CelA (CA) and Cel12A (C12A) act synergistically in the context of ankyrin repeats, with both three and four repeat spacing. The extent of synergy differs for different substrates. Also, having C12A N-terminal to CA provides greater synergy than the reverse construct, especially against filter paper. In contrast, we do not see synergy for these enzymes in tandem with CelK (CK) catalytic domain, a larger exocellulase, demonstrating the importance of enzyme identity in synergistic enhancement. Furthermore, we found endocellulases CelD and CA with three repeat spacing to act synergistically against filter paper. Importantly, connecting CA and C12A with a disordered linker of similar contour length, shows no synergistic enhancement, indicating that synergism results from connecting these domains with folded ankyrin repeats. These results show that ankyrin arrays can be used to vary spacing and orientation between enzymes, helping to design and optimize artificial cellulosomes, providing a novel architecture for synergistic enhancement of enzymatic cellulose degradation.
Keywords: Cellulase, ankyrin, scaffold, DNS, artificial cellulosome, biofuels
Introduction
Large-scale dependence on fossil fuels for liquid fuel production is a major concern. Though modest progress is being made in some sectors, the global dependence on crude oil for fuel for transportation remains high. Moreover there is a major environmental concern associated with the fossil fuel dependence related to the increase in atmospheric CO2 concentration, and climate change. Biomass is a promising alternative for biofuels and chemical production, since its seasonal generation removes CO2 from the atmosphere making it a carbon neutral energy source (1, 2).
Production of fuels from biomass, in particular cellulose, (a polymer composed of β-D-glucose -- the main structural component of plants) helps in solving these problems, since biomass constitutes one of the most available and renewable energy sources, and since the CO2 produced by biomass fuel combustion is captured from atmospheric CO2 by photosynthesis. However, there are major barriers to the cost-effective production of biofuel that must be overcome. Chief among these problems is the low activity of the cellulase enzymes used in this process, and high production costs of the enzymes.
Cellulose breakdown to glucose requires different types of enzyme activities to achieve high synergism. There are three different types of cellulases: endocellulases, exocellulases and β-glucosidases. Endocellulases bind and hydrolyze the cellulose chain internally, exocellulases bind and hydrolyze the cellulose chain from the ends, and β-glucosidases bind and hydrolyze cellobiose to produce glucose (3, 4). Current cellulolytic methods involve the use of cellulase cocktails with different types of activities (endocellulases, exocellulases and β-glucosidases) (5–7).
In anaerobic microorganisms, cellulose is degraded efficiently by tethering cellulases together in a large (estimated >2MDa) extracellular complex (8–10). A scaffolding protein assembles various cellulases with complementary activities, promoting a synergistic enhancement of cellulose degradation (11–13). Cellulose catalytic domains are attached via dockerin domains to the cohesin domain within the scaffolding protein. However, within a species, the cohesin-dockerin interaction is not specific, resulting in heterogenous cellulosomes, where identities and spatial distances of the various catalytic domains differ (14, 15). Furthermore, there is variation within the scaffoldins in the number of residues between cohesin domains and these linker regions are not structured, allowing for variation in cellulosome structure from an expanded to a collapsed state (16–18).
Purified cellulosome complexes have low specific activities, which seems to be related to the exact composition and mode of action of the enzymes present and to the large size of these complexes. Therefore, several attempts to engineer cellulosomes of reduced complexity (and greater homogeneity) have been made using mini-scaffoldins (19–24). In addition, different, more structurally rigid scaffolds can be used to build cellulase arrays. For example, Mitsuzawa et al. fused an eighteen subunit circular complex to cohesins, in order to bind cellulase dockerin units. The assembly increased the cellulolytic activity relative to the free enzymes in solution. However, like the cellulosome, this engineered cohesion-dockerin approach lacks specificity in cellulase identity and placement (25).
A specific order, spacing, and orientation between enzymes can be achieved by expressing several cellulase catalytic domains in a single polypeptide chain with a structured linker in between. Here we use ankyrin repeats as a structured scaffold to embed cellulase catalytic domains. Ankyrin repeats are 33 residues and form a helix-loop-helix, with each repeat connecting to the next with an extended beta-hairpin loop (26–28). Ankyrin repeats have approximately 11 Å linear spacing, which is about the size of a cellobiose molecule (2 glucose units). Each repeat is highly stabilized by the interface formed with adjacent repeats (29, 30). This is particularly true for consensus ankyrin repeats used here, which have very high thermostabilities. We have found previously that we can embed cellulase catalytic domains with a range of N to C termini distances within consensus ankyrin scaffolds, and that these ankyrin-embedded cellulases maintain and (in some cases exceed) the activities of the free enzymes (31).
Here, we have cloned, expressed and purified the catalytic domains of four different cellulases, and have embedded pairs of these catalytic domains into consensus ankyrin repeat scaffolds at different repeat spacings (and N to C-terminal register). As in our previous study, we have also embedded single catalytic domains into consensus ankyrin domains, to quantify the degree to which synergy is provided when catalytic domains are in cis. Our goal is to identify constructs for which the product of one enzyme can readily be used as the substrate of the next enzyme, promoting a greater efficiency in cellulose breakdown. We find that some cellulase pairs have enhanced activity compared to the corresponding trans mixture, demonstrating the ankyrin fusion approach to be a viable approach to increasing cellulose activity. We find that the spacing and the orientation of the inserted enzymes influence the cellulolytic activity of the designed constructs, and that the observed differences are substrate dependent. Spacing and orientation also influences the thermal unfolding of these constructs.
Materials and Methods
Cloning
Cellulase catalytic domains were cloned by PCR from genomic DNA from Clostidium thermocellum (ATCC 27405) and Thermotoga maritima (ATCC 43589). To clone isolated cellulase catalytic domains sequences without flanking ankyrin repeats, we used PCR standard protocol. CA and C12A catalytic domains were cloned as before (31). CelD catalytic domain was defined from Y139 to A569 and CK catalytic domain was defined from D213 to A809. Resulting PCR products were inserted into the expression vector pET-24a at BamHI and XhoI sites, using an In-Fusion kit (Clontech). The disordered RAM polypeptide was cloned from human Notch 1, using Gibson assembly (NEB E2611S), and includes residues 1758 to 1888.
To create an expression construct that inserts cellulose coding sequences catalytic domains into consensus ankyrin domains, we used a single consensus repeat (R) cloned into the expression vector pET-15b+ described by Aksel et al. (29), and modified the repeat sequence by QuikChange (Agilent Technologies) to insert AgeI and KpnI restriction sites and form R13 (Figure 1). Cellulase catalytic domain sequences were amplified using PCR (primers designed according to In-Fusion kit instructions). Resulting PCR products were cloned into the R13 consensus ankyrin repeat using an In-Fusion kit. This insertion splits R13 into two flanking half-repeats, which we term R0.5-Cel-R0.5. For catalytic domains that had restriction sites that would be used downstream in the cloning protocol, silent mutations were inserted to remove those sites through QuikChange. RAM was amplified for Gibson assembly into pet-15b+ using primers that introduce flanking BamHI and BglII cloning sites. Additional ankyrin repeats were added on both the 5′ and 3′ sides of the catalytic domain genes using BamHI and BglII sites present on each side of the repeats, along with a distal HindIII site (29, 32). For the double constructs each R0.5-Cel- R0.5 was treated as a repeat unit. All DNA sequences were confirmed by DNA gel electrophoresis and DNA sequencing.
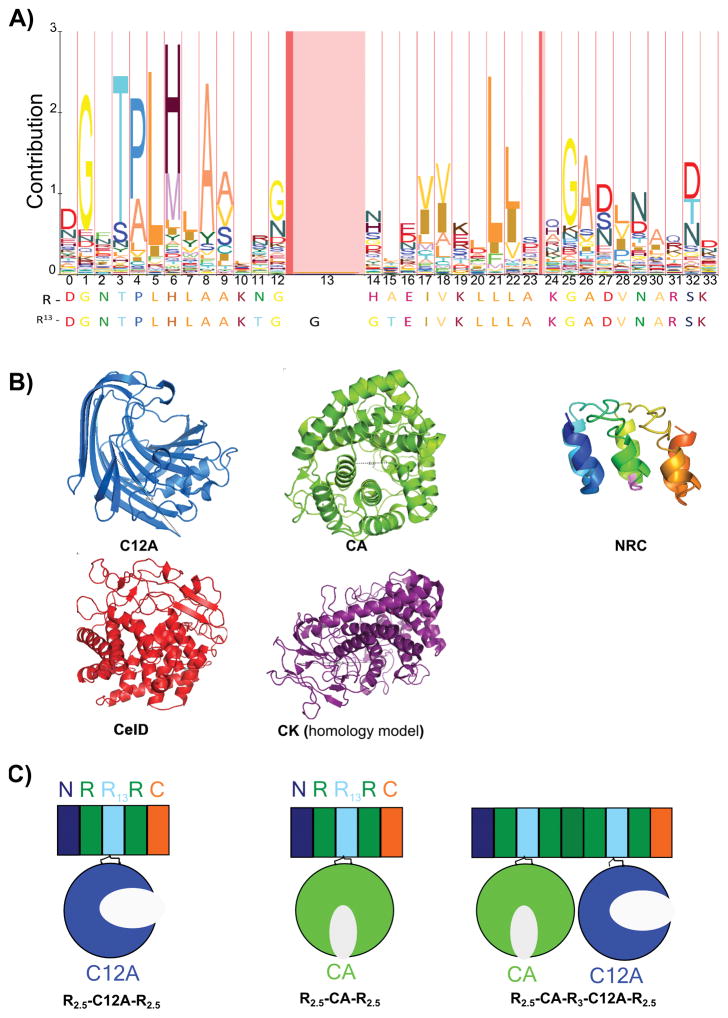
Figure 1. Sequence and structural features of the ankyrin-cellulase scaffolding strategy.
A) HMM logo representing the ankyrin repeat family. B) The structures of the CA catalytic domain from Clostridium thermocellum (green, PDB id 1IS9), the C12A catalytic domain of Thermotoga maritima (blue, PDB id 3AMH), the three consensus ankyrin repeat construct NRC (rainbow, PDB id 2L6B), CelD catalytic domain from Clostridium thermocellum (red, PDB id 1CLC), and homology model of CK catalytic domain from Clostridium thermocellum (purple, from PDB id 1RQ5). CA and CelD are (α/α̇6 barrels, with distances between the N and C termini of about 8 Å. The active sites are opposite to the termini. C12A is a β-sandwich, with a distance between the N and C termini of 30.8 Å. The active site of C12A catalytic domain lies in the concave cleft of the β-sandwich. NRC shows the typical ankyrin fold, with a helix-turn-helix within each repeat, and an extended loop connecting each repeat. The insertion site is in the turn between the two α-helices of each repeat, depicted in pink (position 13, panel A). CK is an (α/α̇6 barrel, with a distance between the N and C termini of about 30 Å. The active site is on the side of the termini. C) Cartoon representation of a five repeat consensus ankyrin (rectangles) fused with CA (R2.5-CA-R2.5), C12A (R2.5-C12A-R2.5) and both CA and C12A in the double construct R2.5-CA-R3-C12A-R2.5. The linker region is composed of five residues on each side is black.
Protein expression and purification
Expression vectors were transformed into E. coli BL21 (DE 3). Cells were grown at 37°C in autoinduction media for 24h (33). Cells were collected by centrifugation and stored at -80°C. Cells were resuspended in 300mM NaCl and 50mM NaH2PO4 pH 6.5 (CA constructs), or 300mM NaCl 50mM Tris-HCl pH 8.0 (all other constructs). Cells were lysed using an Avestin EmulsiFlex C3 and were then treated with DNAse I at 4°C for 1h with stirring. The lysed cells were pelleted, and cell pellets were resuspended in the same buffers with 8M Urea. Resuspended pellets were loaded onto a Ni column (QIAGEN; Valencia, CA). The column was washed with the same buffered urea solution, and then with 100mM NaCl and 50mM NaH2PO4 pH 6.5 (CA constructs) or 100 mM NaCl 50mM HCl-Tris pH 8 (all other constructs) to refold on the column.
Proteins were eluted with 250mM imidazole in the same salt and buffer conditions, in the smallest volume possible. Proteins eluted from nickel columns were diluted two- to three-fold with 50mM of the corresponding buffer to decrease salt concentration. Diluted proteins were then loaded on ion exchange columns (Q-sepharose for CA constructs and SP-sepharose for C12A constructs; QIAGEN; Valencia, CA). After washing the bound protein with 100mM NaCl and 50mM of the corresponding buffer, the proteins were eluted with a sharp buffered 1M NaCl step.
Circular dichroism spectroscopy
Circular dichroism measurements were made with an AVIV Model 400 Circular Dichroism spectropolarimeter (Aviv Associates, Lakewood, NJ). Far-UV circular dichroism spectra were collected with a 1nm step size, with 30 seconds averaging time in a 1 mm quartz cuvette. Protein concentration ranged from 5 μM to 20 μM. Thermal unfolding transitions were monitored at 222 nm, with 2°C step size within the range of 20 to 98°C, in a thermal resistant 10 mm quartz cuvette. Each temperature step had a temperature equilibration time of 3 minutes, and a 30 second signal averaging time. Protein concentration ranged from 1 to 2 μM. All thermal denaturations were at pH 8, as thermal denaturations at pH 4.75 did not show a sigmoidal transition, which complicates the interpretation of the results (data not shown). Despite extensive efforts to obtain reversibility, all thermal denaturations shown here are irreversible; thus thermal unfolding midpoints (Tm) reflect both protein stability and thermal inactivation kinetics (34).
Cellulase assays
Cellulase activity was measured on three substrates: CMC (soluble chemically modified cellulose), RAC (prepared by acid-base treatment of insoluble microcrystalline cellulose to form an emulsion) and FP (5 mm hole punch of Whatman No. 1 filter paper) (35) . For preparation of low viscosity CMC solution, powdered CMC (DP=400; DS=0.65–0.9; Sigma-Aldrich) was dissolved to 2 percent w/v in 1M NaCl 50mM NaAcetate, pH 4.7, the buffer conditions of the cellulose degradation reactions.
Cellulase activities were measured through a previously described modified version of the 3,5-dinitrosalycilic acid (DNS) method described by Ghose (31, 36). The degradation step was carried out for 30 minutes (CMC and RAC) or for 16 hours (FP) in a thermally resistant plate sealed with sticky aluminum foil sheets (Corning, NY). Plates were incubated in a Veriti thermocycler (Applied Biosystems, CA) at temperatures ranging from 20°C to 95°C (results presented here were carried out at 75°C). The lid was kept 2°C above the well temperature to avoid condensation. The final concentration of CMC used was 1%. All assays were carried out with 4 pmol of each catalytic domain, whether in cis or in trans. After the degradation step, 140 μL of DNS solution (36) was added and the samples were subsequently incubated at 100°C for 10 minutes in the thermocycler, with a lid temperature of 105°C. After cooling to room temperature, 100 uL each sample was transferred to an optically clear plate and the absorbance was read in GloMax Modulus Microplate Reader (Promega, WI) at 510 nm. Each measurement was made in triplicate in the same plate, and repeated in a minimum of two separate plates. Absorbances were corrected using values from undigested, identically processed substrate (i.e., substrates without enzyme). We included a set of glucose standards on each plate. The scaling factor between absorbance at 510 nm and μmol of glucose hydrolyzed in our assay volume is 5.23 AU/μmol (31).
Results
Selection of ankyrin insertion sites and cellulase domains
To identify viable insertion sites for cellulase catalytic domains within the ankyrin consensus sequence, we used the ankyrin HMM sequence profile from Pfam (37). Position 13 (boxed in pink in the sequence logo, Figure 1A, representing the most probable sites for insertion) was chosen to insert cellulase catalytic domains. Each catalytic domain was first inserted into R13, giving R0.5-Cel-R0.5. The R13 embedded catalytic domain was then fused to consensus ankyrin repeats on both sides, and was capped by N- and C-terminal capping motifs (30). Since the driving force for ankyrin repeat folding is interface formation, two repeats are needed to adopt a stable fold (29). To maintain solubility and create stable interfaces between repeats, all constructs include N- and C- terminal caps, and have a minimum of two full ankyrin repeats, plus one (terminal) or two (internal) half repeats (Figure 1C). These half repeats result from catalytic domain insertion into R13. We use the nomenclature Rx-Cel-Rx, where x is the number of repeats (x= 2.5, 3, 4 or 5 repeats). For example, in R2.5-CA-R2.5, the CA catalytic domain is flanked on both termini by 2.5 repeats. In R2.5-CA-R3-C12A-R2.5, an N terminal N-cap is followed by 1.5 R repeats, the CA catalytic domain, three R repeats (beginning at helix 2 and ending at helix 1), the C12A catalytic domain, 1.5 R repeats and a C-terminal C-capping repeat (Figure 1C). We also used the human Notch 1 RAM linker to test the effect of having an unstructured polypeptide connecting two cellulase catalytic domains. This 131 residues linker has been shown to be disordered and expanded in solution (38–40).
We built ankyrin fusion constructs using three endocellulases: CelA (CA, α/α6) and CelD (α/α6) from Clostridium thermocellum, and Cel12A (C12A, β-jelly) from Thermotoga maritima (Figure 1B). We also used CelK (CK, α/α6), which is classified as an exocellulase from Clostridium thermocellum (3, 41–45). Although CK has not been crystallized it has high similarity to CbhA from Clostridium thermocellum, suggesting that it may have arisen from a gene duplication event (46). CbhA has been reported to be an exocellulase (3, 45, 47), although Wilson and colleagues have also detected endocellulase activity (48).
All Clostridium thermocellum cellulases are part of the cellulosome complex and tend to have other domains attached such as the dockerin and/or the cellulose binding modules (CBMs). CA and C12A have both been shown to be well behaved when singly inserted into ankyrin repeats and remain active against both CMC and RAC (31). We also cloned and expressed the catalytic domains of three exocellulases from Clostridium thermocellum: CbhA (D213 to A809), CK (D213 to A809) and CelS (CS, G37 to F661). Of these we chose exocellulase CK to embed in tandem with endocellulase ankyrin arrays, because CK expressed well, could be purified, and had the highest activity compared to the domains of CbhA and CS. Moreover, CK showed the greatest of synergistic enhancement in trans to the endocellulase catalytic domains (data not shown).
Effects on cellulolytic activity of embedding multiple cellulase catalytic domains into consensus ankyrin repeats
To measure cellulolytic activity we used a high-throughput DNS plate assay as described (31). We tested the cellulolytic activity of each ankyrin-inserted catalytic domain, both as single and double constructs, against CMC, RAC and filter paper (FP) substrates. Though DNS assays are typically performed at 50°C, we found these enzymes to have higher activities at 75°C, increasing the precision of activity measurements. Thus, we report activities and synergistic enhancements at 75°C, though the general trends and synergistic enhancement of activities are maintained at 50°C.
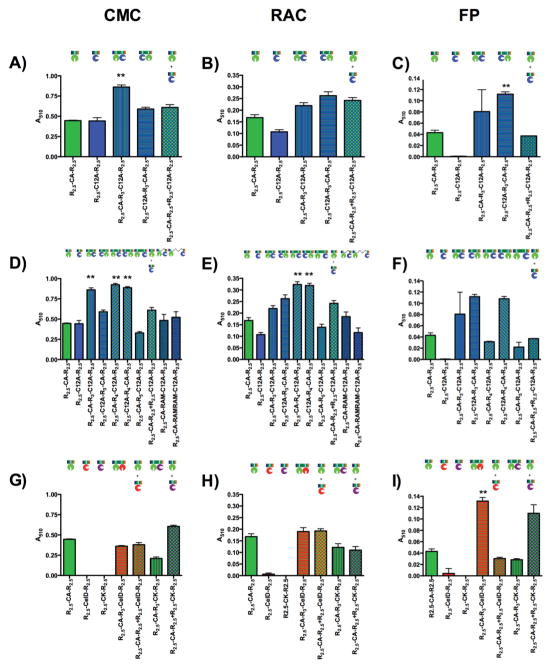
We first tested for a synergistic enhancement in trans by adding two R2.5-Cel-R2.5 constructs in a single reaction, relative to the sum of the activities in separate reactions. We saw a synergistic enhancement in trans for R2.5-CA-R2.5 and R2.5-CK-R2.5 against FP and CMC, but not RAC (Figure 2). To determine the contribution of tethering multiple cellulase domains in cis in consensus ankyrin arrays, we compare the activities of double constructs with the activities of 1:1 mixtures of the same two cellulase domains in trans (Figure 2; Table 1). The cis construct R2.5-CA-R3-C12A-R2.5 shows a statistically significant activity increase relative to the trans mixture of R2.5-CA-R2.5 and R2.5-C12A-R2.5, against the substrate CMC (Figure 2A). Although the swapped 1 double construct R2.5-C12A-R3-CA-R2.5 does not show statistically significant activity increase relative to the trans mixture against CMC, there is a statistically significant enhancement in activity against FP (Figure 2C).
Figure 2. Cellulolytic activities of the ankyrin-cellulase constructs.
The double asterisk (**) indicates a calculated p-value < 0.05 between the activity of the double construct and the corresponding trans mixture, determined by an unpaired t-test with Welch’s correction (62), corresponding to statistically significant synergy in cis. (A-C) Cellulolytic activity of CA and C12A catalytic domains scaffolded with three repeats spacing. Statistically significant synergy is seen for R2.5-CA-R3-C12A-R2.5, against CMC (A) and FP (C), but not RAC (B). (D-F) Cellulolytic activity of CA and C12A catalytic domains scaffolded with different number of spacing repeats and with one and two disordered RAM sequences. Statistically significant synergy is seen for R2.5-CA-R4-C12A-R2.5 and R2.5-C12A-R4-CA-R2.5, against CMC (D), but not RAC (E) Statistically significant synergy is seen for R2.5-C12AR4-CA-R2.5, against FP (F). (G–I) Cellulolytic activity of CelD and CK catalytic domains scaffolded with CA, with three repeats spacing. No synergy is observed in cis for either CMC (G), or RAC (H). Statistically significant synergy is seen for R2.5-CA-R4-CelD-R2.5 against FP (I). In contrast, the trans mixture of R2.5-CA-R2.5 + R2.5-CK-R2.5 shows an enhancement in cellulolytic activity against CMC (G) and FP (I), but not RAC (H). The scaling factor between absorbance at 510 nm and μmol of glucose hydrolyzed is 5.23±0.05 Au/μmol.
Table 1.
Synergism of R2.5-Cel1-Rx-Cel2-R2.5
| Enzyme | CMC | RAC | FP |
|---|---|---|---|
| R2.5-CA-R3-C12A-R2.5 | 1.41** | 0.91 | 2.17 |
| R2.5-C12A-R3-CA-R2.5 | 0.97 | 1.08 | 3.01** |
| R2.5-CA-R4-C12A-R2.5 | 1.51** | 1.33** | 0.85 |
| R2.5-C12A-R4-CA-R2.5 | 1.45** | 1.32** | 2.91** |
| R2.5-CA-R5-C12A-R2.5 | 0.54 | 0.57 | 0.59 |
| R2.5-CA-R3-CelD-R2.5 | 0.95 | 0.99 | 4.33** |
| R2.5-CA-R3-CK-R2.5 | 0.35 | 1.11 | 0.26 |
| R2.5-CA-RAM-C12A-R2.5 | 0.79 | 0.76 | --- |
| R2.5-CA-RAMRAM-C12A-R2.5 | 0.85 | 0.48 | --- |
Synergism values were calculated as the ratio of the activity of the double construct R2.5-Cel1-Rx-Cel2-R2.5 relative to the activity of the corresponding R2.5-Cel1-R2.5 and R2.5-Cel2-R2.5 trans mixtures. The double asterisk (**) represents the statistically significant enhancements (p<0.05) determined by a t-test determined by an unpaired t-test with Welch’s correction.
To investigate whether spacing influences synergy in cis, we inserted an additional (fourth) ankyrin repeat between cellulase catalytic domains. The addition of a fourth repeat between the two catalytic domains of CA and C12A augments cellulolytic activity in the double construct R2.5-CA-R4-C12A-R2.5, both against CMC and RAC, but not against FP (Figure 2D, E and F respectively). Moreover, the swapped double construct R2.5-C12A-R4-CA-R2.5 shows statistically significant activity increase, relative to the trans mixture of R2.5-CA-R2.5 and R2.5-C12A-R2.5, against all substrates tested (Figure 2D, E and F). In contrast, the addition of a fifth repeat between CA and C12A catalytic domains in R2.5-CA-R5-C12A-R2.5 decreases cellulolytic activity to levels comparable to that of CA catalytic domain alone in all substrates tested (Figure 2D, E and F)2.
In contrast, constructs in which CA and C12A are connected with either one or two copies of the unstructured RAM polypeptide show no synergy, relative to in trans mixtures for all substrates tested. For CMC, activities of the two RAM-linked constructs are indistinguishable from the trans mixture (Figure 2D). For RAC, activities are somewhat lower (Figure 2E), whereas for FP, the RAM linked cis constructs show no detectable activity (data not shown). Taken together, these data demonstrate that both spacing and orientation play a role in the synergistic enhancement of cellulolytic activity with CA and C12A catalytic domains and that synergistic enhancement is dependent also on the substrate used.
To further test the role of enzyme identity in synergistic activity enhancement of the double constructs, we tethered CA with the catalytic domains of CelD with three repeats spacing. The double construct shows statistically significant activity increase against FP, relative to the trans mixture of R2.5-CA-R2.5 and R2.5-CelD-R2.5 (Figure 2I, G and H). In contrast, cellulolytic activity of R2.5-CA-R3-CelD-R2.5 against RAC and CMC is equivalent to the activity of having the two endocellulases in trans (and also to the activity for R2.5-CA-R2.5, since R2.5-CelD-R2.5 has no measurable activity on its own against these substrates in the conditions tested). Thus, the synergistic enhancement observed for CA depends on the identity of the other endocellulases embedded with it in the consensus ankyrin scaffold.
Synergism is traditionally seen when mixing enzymes with different activities, such as endocellulases and exocellulases. To test the role of tethering one endocellulase and one exocellulase with the consensus ankyrin array, we used both CA and CK catalytic domains. The trans mixture of R2.5-CA-R2.5 with the putative exocellulase construct R2.5-CK-R2.5 shows enhanced activity relative to the sum of the activities of R2.5-CA-R2.5 and R2.5-CK-R2.5, against both CMC and FP (Figure 2G and I). However, the double construct containing exocellulase CK, R2.5-CA-R3-CK-R2.5, shows no statistically significant activity increase relative to the trans mixture of R2.5-CA-R2.5 and R2.5-CK-R2.5, against either substrate (Figure 2G, H and I), indicating either steric hindrance between the cellulases or substrate occlusion by the geometry of the construct. It will be important to extend this finding to other confirmed endocellulase/exocellulase pairs.
Effects of embedding multiple cellulase catalytic domains into consensus ankyrin repeats on thermal denaturation
As a test for potential hindrance between catalytic domains, and to test whether activity changes may be related to changes in the extent of folding and thermostability, we measured the denaturation of cellulase-ankyrin fusions by circular dichroism spectroscopy. Far-UV circular dichroism spectra of all constructs at pH 8 have shapes expected for α-helical structured proteins, with double minima at 208 and 222 nm (data not shown).
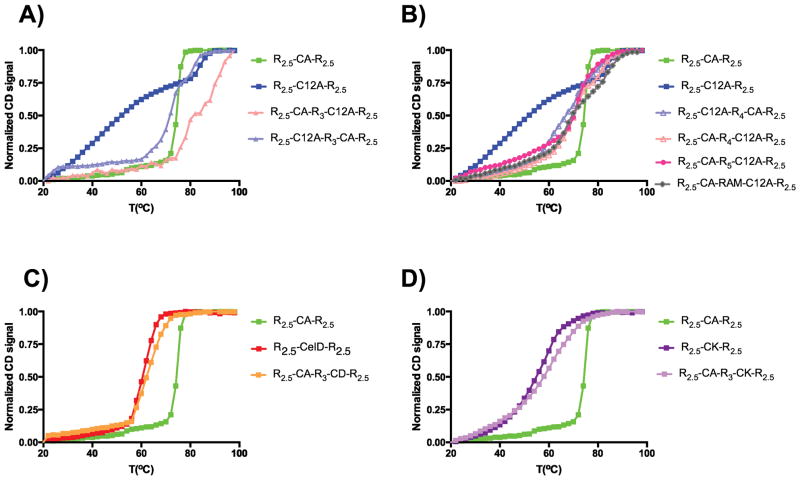
Thermal denaturations by circular dichroism of the constructs containing two endocellulases were measured, and were compared with their single counterparts inserted into a five consensus repeat scaffold. We have previously found that the thermal stabilities of CA and C12A catalytic domains are not significantly perturbed when singly inserted into consensus ankyrin arrays (31). To test whether the two cellulases remain stably folded when in tandem, we compared the thermal denaturations of R2.5-CA-R2.5 and R2.5-C12A-R2.5 with the thermal denaturations of double constructs R2.5-CA-R3-C12A-R2.5 and R2.5-C12A-R3-CA-R2.5 (Figure 3A). We find the CA catalytic domain to be stabilized in both double constructs. The significantly broadened transition seen for R2.5-C12A-R2.5 is not seen in any of the other cellulase-consensus fused constructs. R2.5-CA-R3-C12A-R2.5 shows the highest thermal denaturation melting point of all double constructs studied. A two stage thermal unfolding is clearly seen for the swapped R2.5-C12A-R3-CA-R2.5, and the lower temperature transition seems to coincide with the R2.5-CA-R2.5 transition. The observation that the two double constructs show different thermal denaturation profiles highlights the importance of the order of the catalytic domains in these tandem constructs.
Figure 3. Thermal denaturations of ankyrin-cellulase catalytic domain fusions.
Thermal denaturations were followed by circular dichroism at 222 nm at pH 8 for single and double constructs. R2.5-CA-R2.5 thermal denaturation is plotted in all panels (green squares). R2.5-C12A-R2.5 thermal denaturation is plotted in panels A–B (blue squares). A) R2.5-CA- R3-C12A-R2.5 (pink triangles), R2.5-C12A- R3-CA-R2.5 (light blue triangles) B) R2.5-CA-R4-C12A-R2.5 (pink open triangles) and R2.5-C12A-R4-CA-R2.5 (light blue open triangles), R2.5-CA-R5-C12A-R2.5 (dark pink circles) and R2.5-CA-RAM-C12A-R2.5 (grey squares) C) R2.5-CelD-R2.5 (red squares) and R2.5-CA-R3-CelD-R2.5 (orange squares) D) R2.5-CK-R2.5 (dark purple squares) and R2.5-CA-R3-CK-R2.5 (light purple squares).
Unlike the constructs with catalytic domains separated by three repeats, the thermal denaturations of R2.5-CA-R4-C12A-R2.5 and of R2.5-C12A-R4-CA-R2.5 are very similar (Figure 3B). Both thermal denaturations show two distinct transitions; though these transitions are not resolved by a flat baseline, they are separated by an inflection point at 76°C. The two transitions have midpoints at 66°C, and 82°C. These results suggest that the addition of a fourth repeat provides enough separation between the CA and C12A catalytic domains that their stabilities are no longer coupled. Consistent with this interpretation, these transitions overlay with that of R2.5-CA-RAM-C12A-R2.5 construct, for which we fully expect uncoupled stabilities between the CA and C12A domains. As expected, addition of a fifth repeat between CA and C12A catalytic domains affords no additional thermal stabilization.
In contrast to the CA-C12A double constructs, the thermal denaturation profiles of R2.5-CA-R3-CelD-R2.5 and R2.5-CA-R3-CK-R2.5, resemble those of the single ankyrin-embedded CelD and CK catalytic domains (Figure 3C and D). Specifically, the tandem constructs are thermally denatured with the same profile as the least stable single catalytic domain (CelD, Figure 3C, and CelK, Figure 3D), indicating that either of these unfolded domains in cis may be sufficient to unfold the CelA catalytic domain and ankyrin host.
Discussion
Enzymatic cellulose degradation for biofuels production is currently cost prohibitive. The high costs of this step are related to enzyme lifetime and stability, as well as low rates of catalysis. We have previously investigated the fundamental parameters for enzyme (cellulases CelA and C12A) stability through reversible unfolding and protein engineering, providing means to understanding the best approaches to stabilize and increase cellulase lifetime (31). To improve the enzymatic degradation of cellulosic biomass here, we have built and characterized constructs in which multiple cellulase catalytic domains are arrayed in a single polypeptide of defined spacing and orientation. Synergistic enhancement can be achieved by having enzymes that catalyze subsequent steps in close proximity (Figure 4). If the product of the first enzyme is the substrate of the second, the probability of capture by the second enzyme increases inversely with distance, being highest when the active sites are in contact (31). Alternatively, synergy may result from pairs of enzymes with complementary catalytic activities (e.g. endo/endo pairs), if substrate has a repetitive polymeric structure. In such cases, synergy may result from co-localization or from effects on accessibility of neighboring sites.
Figure 4. Model for synergistic enhancement of cellulolytic activity.
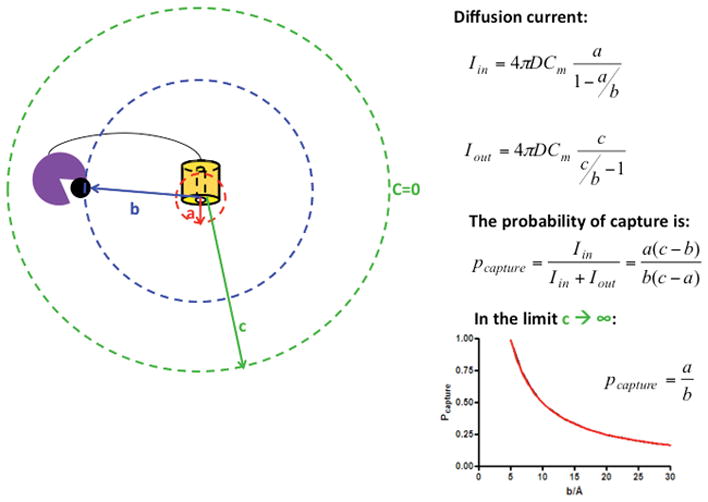
Synergistic enhancement of distinct activities depends on the distance between the two cellulases with distinct activities. Diffusion current inwards (Iin) and outwards (Iout) depends on the diffusion coefficient, the maximum concentration of substrate (Cm) and the distances a, b and c (radius of enzyme a, distance between enzyme a and b and distance at which the substrate diffuses out to the medium respectively). The probability of capture is given by the ratio of the current Iin/(Iin+Iout), and is maximum when b=a (in the Pcapture plot a is taken to be 5Å).
For domains CA and C12A we varied the spacing by increasing the number of repeats between domains, starting from a three-repeat linker. Based on the structure of ankyrin repeat proteins, the spacing between catalytic domains termini is expected to range from 33 to 55 Å. We also swapped the order of the cellulolytic domains to probe the role of the order in cellulolytic activity. In addition, we fused CelD and CK with CA to probe the roles of enzyme identity in cellulolytic activity and test for synergy with an endo/exo pair. This strategy provides a greater degree of control over spacing, orientation and enzyme identity than in the cellulosome and analogous recent efforts to build more precisely defined cellulosomes (18, 49–54).
Cellulolytic activity CA and C12A catalytic domains scaffolded with three repeats spacing
In our previous work, the consensus ankyrin domain was demonstrated to be compatible with cellulolytic activity of CA catalytic domain, and to enhance the cellulolytic activity of C12A catalytic domain (31). Here, we inserted both of these catalytic domains in tandem into the consensus ankyrin domain, in both orientations, and tested the cellulolytic activity of these constructs against a variety of model substrates (CMC, RAC and FP; Figure 2).
When these two ankyrin-embedded enzymes are added in trans or connected by an unstructured polypeptide, the measured activity is no greater than (and is sometimes less than) the sum of the activities of the isolated constructs, regardless of the substrate. In contrast, when CA and C12A catalytic domains are combined in cis within a single consensus ankyrin repeat array, the measured activity of R2.5-CA-R3-C12A-R2.5 exceeds that of the trans mixture against CMC by a factor of 1.4 (Figure 2A). Swapping the two catalytic domains in cis (R2.5-C12A-R3-CA-R2.5) results in an activity against FP that is increased relative to the trans mixture by a factor of 3.0 (Figure 2C). Against RAC, the cellulolytic activities remain about the same with all constructs. The activity enhancements against CMC and FP show that, tethering these enzymes into proximity plays an important role in the synergism between CA and C12A catalytic domains. This is particularly notable given that both CA and C12A are endocellulases.
Cellulolytic activity CA and C12A catalytic domains scaffolded with increased repeat spacing
For CA and C12A, cellulolytic activity against CMC and RAC increases relative to the trans mixture, when an additional repeat is added (Figure 2D and E). Furthermore, R2.5-C12AR4-CA-R2.5 (but not R2.5-CA-R4-C12A-R2.5) shows a statistically significant enhancement in activity relative to the trans mixture, against FP (Figure 2F). This enhancement in activity with increased spacing is especially strong when C12A is on the N terminal side of CA. This spacing enhancement may be due to lower steric hindrance, as the additional repeat should provide more space to accommodate the two catalytic domains. The order between the catalytic domains also clearly plays a role, most notably against FP: the construct with C12A on the N terminal side, shows an enhancement in cellulolytic activity whereas the swapped construct does not. The effect of increasing spacing on activity of tandem catalytic domains can be seen by directly comparing the three- and four- repeats spaced constructs.
In contrast, the addition of a fifth repeat between the CA and C12A catalytic domains, does not show an enhancement in synergy or overall activity (Figure 3D, E and F). Taken together, these results suggest that the optimal spacing between CA and C12A catalytic domains is four repeats (about 44Å).
One possible contribution to the synergy observed between CA and C12A is the difference in substrate specificity. The catalytic domain of CA can accommodate chain length up to 5-glucosyl units and has lower activity against smaller sugars such as cellotetraose or cellotriose. C12A is restricted to a chain length of 4-glucosyl units (cellotetraose), with highest activity against smaller sugars (41, 55–57). Thus the product from CA may serve as substrate for C12A. These differences in substrate specificity may account for the unexpected synergy seen between these two endoglucanases (for three and four repeats spacing), much like the synergy seen with cellulase catalytic domains from different EC classes (such as endocellulases, exocellulases and β-glucosidases). The difference in synergy observed for different catalytic domain order may result from fixed orientations imparted by the ankyrin domain scaffold. Such orientation biases may affect both distance of the active sites (which may influence reactivity as shown in Figure 4), and the relative accessibility to their complex, polymeric substrates.
Cellulolytic activity of scaffolded CA and CelD catalytic domains with three repeats spacing
To probe how enzyme identity affects synergy in the scaffolded constructs, we cloned the endoglucanase domain of CelD in tandem with the CA catalytic domain. Although R2.5-CelD-R2.5 has the lowest activity of all the endoglucanases tested and shows no trans synergy with R2.5-CA-R2.5 against any substrates (Figure 2G, H and I), the cis construct R2.5-CA-R3-CelD-R2.5, shows a sizeable synergistic enhancement against FP, comparable to that of R2.5-C12A-R3-CA-R2.5, and is 4.3 times greater than the trans mixture (Figure 2I).
As with CA and C12A catalytic domains, the synergism observed between CA and CelD may result from the different substrate specificities, with CelD showing highest activity against cellotriose (58). These data show that enzyme identity plays an important role in synergistic enhancement, as the single domains have distinct activities in the different substrates. It is noteworthy that the highest activity enhancements observed were against FP, the most recalcitrant (and most insoluble) material.
Cellulolytic activity of scaffolded CA and CK catalytic domains with three repeats spacing
Since synergism is usually seen with cellulases of different EC classes, we cloned the exoglucanase domain of CK in tandem with the endoglucanase domain of CA (3). Although the single R2.5-CK-R2.5 shows no measurable activity, the trans mixture R2.5-CK-R2.5 and R2.5-CA-R2.5 has greater activity against CMC and FP than the sum of the activities of the isolated constructs (Figure 2G and I). However, the cis construct R2.5-CA-R3-CK-R2.5 has less activity than the trans mixture, showing no more activity than R2.5-CA-R2.5. CK is a large domain (597 residues), therefore, it is possible that with a three repeat spacing there is significant steric hindrance between the CK and CA catalytic domains, inhibiting the activity of both3. Another possibility is that the on-rates of the CA endocellulase domain for sites on FP are significantly faster than that of CK exocellulase domain, sequestering it from the cellulose chain termini.
Thermostability of scaffolded catalytic domains
In a previous study (31), we have obtained, analyzed, and assigned the unfolding transitions of single catalytic domain constructs (Rx-Cel-Rx). By comparing the stabilities of the R2.5-Cel-R2.5 with the stabilities of R2.5-Cel1-R3-Cel2- R2.5, we can resolve the contributions of each domain to thermostability of the double construct. The double constructs with CA and C12A catalytic domains with three ankyrin repeat spacing show different thermal unfolding transitions depending on the order of the two domains. Thus, the order influences both stability (Figure 3A) and activity (Figure 2A, B and C).
Adding repeats between CA and C12A catalytic domains alters the thermal denaturation profile. Both constructs with four repeat spacing have similar unfolding curves, indicating that the stability coupling interaction seen at three repeat spacing is lost with the addition of a fourth repeat (Figure 3B). Furthermore, the construct with RAM connecting CA and C12A shows a similar unfolding curve, consistent with decoupling of unfolding of the two catalytic domains connected by four ankyrin repeats. The five repeat CA C12A tandem is less stable than five repeat consensus ankyrin domain (Tm=84°C, data not shown), suggesting that the unfolding of R2.5-CA-R5-C12A-R2.5 is driven by the unfolding of the CA catalytic domain (Figure 3B).
Thermal denaturation of R2.5-CelD-R2.5 is similar to that of R2.5-CA-R2.5, consistent with the similar N to C termini spacing (8.9 Å) in the crystal structure of these two catalytic domains. In contrast, R2.5-CK-R2.5 shows a broader transition consistent with a larger N to C termini spacing. These results suggest that the ankyrin repeats can interact across an embedded cellulase domain if the distance between the termini is small enough (Figure 3C and 3D). The thermal denaturation of both double constructs with CelD and CK in tandem with CA, are similar to their single counterparts (R2.5-CelD-R2.5 and R2.5-CK-R2.5 respectively), suggesting that CelD and CK destabilize the CA catalytic domain and the consensus ankyrin repeats (Figure 3C and 3D).
It should be noted that in some instances, unfolding midpoints are lower than the temperature at which enzymatic activities are measured. It is possible that cellulase catalytic domains are stabilized by substrate (59, 60). For irreversible thermal unfolding, the ligand can preserve the active site, impacting the conversion to the thermally inactive form (61). Although added cellulose substrates interfere with the detection of thermal unfolding by circular dichroism, it may be possible to test for substrate stabilization by fluorescence or calorimetric methods.
Conclusions
In this study, we found that the consensus ankyrin domain can host multiple cellulase catalytic domains, that the domains remain thermally stable and active, and that the ankyrin domain remains folded. Enzyme pairs arrayed within the consensus ankyrin domain show synergistic enhancement of activity over the corresponding trans mixtures. These synergies are dependent on catalytic domain identity, spacing, orientation, and substrate identity. Four repeat spacing provides greater synergy then three or five repeat spacing when C12A catalytic domain is N terminal to CA, both against CMC and RAC. For FP, spacing plays a smaller role than orientation, with C12A/CA constructs having higher activities than CA/C12A constructs. For the endoglucanase CelD, the highest enhancement is seen against FP, with proximity playing an important role (4x enhancement relative to trans). Surprisingly, CK did not show an enhancement in activity in cis relative to the trans mixture, but shows considerable enhancement in trans relative to isolated constructs. The present system provides an efficient means to test the generality of this observation in this scaffolding format.
Acknowledgments
We thank Prof. Bertrand Garcia-Moreno E., Prof. Jan Hoh, Prof. Brendan Cormack, Prof. Mario Amzel, Prof. Richard Cone, Prof. Juliette Lecomte, and Prof. Brian Barr for prolific discussions during the development of the work. We also thank Dr. Tural Aksel for providing the consensus ankyrin repeat constructs and the modified pET-15b+ vector and Dr. Kate Sherry for the RAM construct. This work was supported from an NIH Grant (1 R01-GM068462) to DB, from the Environment, Energy, Sustainability, and Health Institute at Johns Hopkins, and from generous support from the Office of the Provost at Johns Hopkins University ESC was supported by a Predoctoral Fellowship from the Portuguese FLAD/Fulbright and from the Portuguese Foundation for Science and Technology (SFRH / BD / 36119 / 2007), and from a training grant from the Department of Energy (DE-FG02-04ER25626).
Footnotes
By swapped, we mean that the order of the two catalytic domains from N- to C- terminus is reversed.
The double construct R2.5-C12A-R5-CA-R2.5, did not express well in E. coli; thus we were not able to measure activity
Construct R2.5-CA-R4-CK-R2.5 did not express well in E. coli.
References
- 1.Report (2013) The outlook for energy: a view to 2040. (ExxonMobil).
- 2.Guerriero G, Hausman J-F, Strauss J, Ertan H, Siddiqui KS. Lignocellulosic biomass: Biosynthesis, degradation, and industrial utilization. Engineering in Life Sciences. 2016;16(1):1–16. [Google Scholar]
- 3.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83(1):1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 5.Sukumaran RK, Singhania RR, Mathew GM, Pandey A. Cellulase production using biomass feed stock and its application in lignocellulose saccharification for bio-ethanol production. Renewable Energy. 2009;34(2):421–424. [Google Scholar]
- 6.Beckham GT, et al. Molecular-level origins of biomass recalcitrance: decrystallization free energies for four common cellulose polymorphs. J Phys Chem B. 2011;115(14):4118–4127. doi: 10.1021/jp1106394. [DOI] [PubMed] [Google Scholar]
- 7.Himmel ME, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 8.Beguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31(3):201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 9.Doi RH. Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann N Y Acad Sci. 2008;1125:267–279. doi: 10.1196/annals.1419.002. [DOI] [PubMed] [Google Scholar]
- 10.Bayer EA, Morag E, Lamed R. The cellulosome--a treasure-trove for biotechnology. Trends Biotechnol. 1994;12(9):379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 11.Fujino T, Beguin P, Aubert JP. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J Bacteriol. 1993;175(7):1891–1899. doi: 10.1128/jb.175.7.1891-1899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leibovitz E, Beguin P. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J Bacteriol. 1996;178(11):3077–3084. doi: 10.1128/jb.178.11.3077-3084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunnoo M, et al. Nanoscale Engineering of Designer Cellulosomes. Advanced materials. 2016 doi: 10.1002/adma.201503948. [DOI] [PubMed] [Google Scholar]
- 14.Boisset C, et al. Digestion of crystalline cellulose substrates by the clostridium thermocellum cellulosome: structural and morphological aspects. Biochem J. 1999;340( Pt 3):829–835. [PMC free article] [PubMed] [Google Scholar]
- 15.Lytle B, Myers C, Kruus K, Wu JH. Interactions of the CelS binding ligand with various receptor domains of the Clostridium thermocellum cellulosomal scaffolding protein, CipA. J Bacteriol. 1996;178(4):1200–1203. doi: 10.1128/jb.178.4.1200-1203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Alvarez B, et al. Molecular architecture and structural transitions of a Clostridium thermocellum mini-cellulosome. J Mol Biol. 2011;407(4):571–580. doi: 10.1016/j.jmb.2011.01.060. [DOI] [PubMed] [Google Scholar]
- 17.Fontes CM, Gilbert HJ. Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem. 2010;79:655–681. doi: 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- 18.Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 19.Perret S, Belaich A, Fierobe HP, Belaich JP, Tardif C. Towards designer cellulosomes in Clostridia: mannanase enrichment of the cellulosomes produced by Clostridium cellulolyticum. J Bacteriol. 2004;186(19):6544–6552. doi: 10.1128/JB.186.19.6544-6552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You C, Zhang X-Z, Zhang Y-HP. Mini-scaffoldin enhanced mini-cellulosome hydrolysis performance on low-accessibility cellulose (Avicel) more than on high-accessibility amorphous cellulose. Biochemical Engineering Journal. 2012;63:57–65. [Google Scholar]
- 21.Ding SY, et al. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science. 2012;338(6110):1055–1060. doi: 10.1126/science.1227491. [DOI] [PubMed] [Google Scholar]
- 22.Fierobe HP, et al. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277(51):49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 23.Cha J, et al. Effect of multiple copies of cohesins on cellulase and hemicellulase activities of Clostridium cellulovorans mini-cellulosomes. J Microbiol Biotechnol. 2007;17(11):1782–1788. [PubMed] [Google Scholar]
- 24.Stern J, et al. Significance of relative position of cellulases in designer cellulosomes for optimized cellulolysis. PLoS One. 2015;10(5):e0127326. doi: 10.1371/journal.pone.0127326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsuzawa S, et al. The rosettazyme: a synthetic cellulosome. J Biotechnol. 2009;143(2):139–144. doi: 10.1016/j.jbiotec.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24(8):311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 27.Zweifel ME, Barrick D. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 2. Solution stability and cooperativity of unfolding. Biochemistry. 2001;40(48):14357–14367. doi: 10.1021/bi011436+. [DOI] [PubMed] [Google Scholar]
- 28.Tripp KW, Barrick D. Enhancing the stability and folding rate of a repeat protein through the addition of consensus repeats. J Mol Biol. 2007;365(4):1187–1200. doi: 10.1016/j.jmb.2006.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aksel T, Majumdar A, Barrick D. The contribution of entropy, enthalpy, and hydrophobic desolvation to cooperativity in repeat-protein folding. Structure. 2011;19(3):349–360. doi: 10.1016/j.str.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aksel T, Barrick D. Analysis of repeat-protein folding using nearest-neighbor statistical mechanical models. Methods Enzymol. 2009;455:95–125. doi: 10.1016/S0076-6879(08)04204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunha ESHCL, Barrick D. Insertion of endocellulase catalytic domains into thermostable consensus ankyrin scaffolds: effects on stability and cellulolytic activity. Appl Environ Microbiol. 2013;79(21):6684–6696. doi: 10.1128/AEM.02121-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci U S A. 1999;96(7):3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Jackson WM, Brandts JF. Thermodynamics of protein denaturation. A calorimetric study of the reversible denaturation of chymotrypsinogen and conclusions regarding the accuracy of the two-state approximation. Biochemistry. 1970;9(11):2294–2301. doi: 10.1021/bi00813a011. [DOI] [PubMed] [Google Scholar]
- 35.Zhang YH, Cui J, Lynd LR, Kuang LR. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules. 2006;7(2):644–648. doi: 10.1021/bm050799c. [DOI] [PubMed] [Google Scholar]
- 36.Ghose T. Measurement of cellulase activities. Pure Appl Chem. 1987;59(2):257–268. [Google Scholar]
- 37.Schuster-Bockler B, Schultz J, Rahmann S. HMM Logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherry KP, Johnson SE, Hatem CL, Majumdar A, Barrick D. Effects of Linker Length and Transient Secondary Structure Elements in the Intrinsically Disordered Notch RAM Region on Notch Signaling. J Mol Biol. 2015;427(22):3587–3597. doi: 10.1016/j.jmb.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertagna A, Toptygin D, Brand L, Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochemical Society transactions. 2008;36(Pt 2):157–166. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- 40.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch 1. Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278(23):21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 41.Alzari PM, Souchon H, Dominguez R. The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure. 1996;4(3):265–275. doi: 10.1016/s0969-2126(96)00031-7. [DOI] [PubMed] [Google Scholar]
- 42.Cheng YS, et al. Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima. Proteins. 2011;79(4):1193–1204. doi: 10.1002/prot.22953. [DOI] [PubMed] [Google Scholar]
- 43.Kataeva IA, Seidel RD, 3rd, Li XL, Ljungdahl LG. Properties and mutation analysis of the CelK cellulose-binding domain from the Clostridium thermocellum cellulosome. J Bacteriol. 2001;183(5):1552–1559. doi: 10.1128/JB.183.5.1552-1559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kataeva I, Li XL, Chen H, Choi SK, Ljungdahl LG. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J Bacteriol. 1999;181(17):5288–5295. doi: 10.1128/jb.181.17.5288-5295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubot FD, et al. Structural basis for the exocellulase activity of the cellobiohydrolase CbhA from Clostridium thermocellum. Biochemistry. 2004;43(5):1163–1170. doi: 10.1021/bi030202i. [DOI] [PubMed] [Google Scholar]
- 46.Zverlov VV, Velikodvorskaya GA, Schwarz WH, Kellermann J, Staudenbauer WL. Duplicated Clostridium thermocellum cellobiohydrolase gene encoding cellulosomal subunits S3 and S5. Appl Microbiol Biotechnol. 1999;51(6):852–859. doi: 10.1007/s002530051473. [DOI] [PubMed] [Google Scholar]
- 47.Yaniv O, et al. A single mutation reforms the binding activity of an adhesion-deficient family 3 carbohydrate-binding module. Acta crystallographica. Section D, Biological crystallography. 2012;68(Pt 7):819–828. doi: 10.1107/S0907444912013133. [DOI] [PubMed] [Google Scholar]
- 48.McGrath CE, Wilson DE. Endocellulolytic activity of the Clostridium thermocellum Cel9C (formerly CbhA) catalytic domain. Industrial Biotechnology. 2008;4(1):99–104. [Google Scholar]
- 49.Bayer EA, Lamed R, White BA, Flint HJ. From cellulosomes to cellulosomics. Chem Rec. 2008;8(6):364–377. doi: 10.1002/tcr.20160. [DOI] [PubMed] [Google Scholar]
- 50.Bayer EA, Shimon LJ, Shoham Y, Lamed R. Cellulosomes-structure and ultrastructure. J Struct Biol. 1998;124(2–3):221–234. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 51.Bayer EA, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8(5):548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 52.Vazana Y, Morais S, Barak Y, Lamed R, Bayer EA. Designer cellulosomes for enhanced hydrolysis of cellulosic substrates. Methods Enzymol. 2012;510:429–452. doi: 10.1016/B978-0-12-415931-0.00023-9. [DOI] [PubMed] [Google Scholar]
- 53.Morais S, et al. Cellulase-xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. MBio. 1(5) doi: 10.1128/mBio.00285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fierobe HP, et al. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J Biol Chem. 2005;280(16):16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- 55.Cheng YS, et al. Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop. Appl Microbiol Biotechnol. 95(3):661–669. doi: 10.1007/s00253-011-3791-4. [DOI] [PubMed] [Google Scholar]
- 56.Cheng YS, et al. Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima. Proteins. 79(4):1193–1204. doi: 10.1002/prot.22953. [DOI] [PubMed] [Google Scholar]
- 57.Petre J, Longin R, Millet J. Purification and properties of an endo-beta-1,4-glucanase from Clostridium thermocellum. Biochimie. 1981;63(7):629–639. doi: 10.1016/s0300-9084(81)80061-2. [DOI] [PubMed] [Google Scholar]
- 58.Juy M, et al. Three-dimensional structure of a thermostable bacterial cellulase. Nature. 1992;357(6373):89–91. [Google Scholar]
- 59.Schmid RD. Stabilized soluble enzymes. Springer; 1979. [Google Scholar]
- 60.Iyer PV, Ananthanarayan L. Enzyme stability and stabilization—aqueous and non-aqueous environment. Process Biochemistry. 2008;43(10):1019–1032. [Google Scholar]
- 61.Ladero M, et al. Thermal and pH inactivation of an immobilized thermostable Î2-galactosidase from Thermus sp. strain T2: Comparison to the free enzyme. Biochemical Engineering Journal. 2006;31(1):14–24. [Google Scholar]
- 62.Welch BL. The generalization of “Student’s” problem when several different population variances are involved. Biometrika. 1947;34(1–2):28–25. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]