Abstract
Hydrogel microcapsules provide miniaturized and biocompatible niches for three-dimensional (3D) in vitro cell culture. They can be easily generated by droplet-based microfluidics with tunable size, morphology, and biochemical properties. Therefore, microfluidic generation and manipulation of cell-laden microcapsules can be used for 3D cell culture to mimic the in vivo environment towards applications in tissue engineering and high throughput drug screening. In this review of recent advances mainly since 2010, we will first introduce general characteristics of droplet-based microfluidic devices for cell encapsulation with an emphasis on the fluid dynamics of droplet breakup and internal mixing as they directly influence microcapsule’s size and structure. We will then discuss two on-chip manipulation strategies: sorting and extraction from oil into aqueous phase, which can be integrated into droplet-based microfluidics and significantly improve the qualities of cell-laden hydrogel microcapsules. Finally, we will review various applications of hydrogel microencapsulation for 3D in vitro culture on cell growth and proliferation, stem cell differentiation, tissue development, and co-culture of different types of cells.
Graphical Abstract
This critical review discusses microfluidic generation and manipulation of cell-laden hydrogel microcapsules, and their applications for in vitro cell culture.
1. Introduction
In native tissues and organs, cells exist in 3D microenvironments with intricate cell-cell and cell-extracellular matrix (ECM) interactions, and complex support and regulatory systems for metabolism 1–3. The standard 2D monolayer in vitro culture approaches, commonly employed, does not adequately represent or replicate the characteristics of cell’s in vivo condition. This renders 2D monolayer approaches unreliable and mostly inaccurate for drug screening and other tissue engineering applications 4–6. To this end, 3D in vitro culture, which allow cells to grow, develop, and communicate in all three spatial dimensions within artificial or synthetic ECMs, was proposed to mimic cells in vivo 7–10. Compared to 2D culture, it not only facilitates cell aggregation and tissue formation via long term cell culture 11, 12, but also modulates cell morphology, behavior, and functionality via regulating gene and protein expressions, proliferation, differentiation, and migration in 3D physiologically relevant milieus 13–15. Therefore, 3D cell culture approaches are expected to be more accurate than 2D monolayer approaches, particularly for drug toxicology and pharmacokinetics to reduce or even eliminate animal test subjects during preclinical trials 16–19.
Various approaches have been developed for 3D cell culture, such as ultra-low attachment plates 6, rotating bioreactors 20, hanging drops 21, micropatterned surfaces 22, magnetic levitation 23, porous scaffolds 24, and 3D bioprinting 25. But these culture methods either cannot construct 3D ECM microenvironment within predefined space (ultra-low attachment plate, bioreactors, hanging drops, and magnetic levitation), or are incapable of controlling and modifying ECM biophysical and biochemical properties (micropatterned surfaces and porous scaffolds). In addition, their macro sizes impose severe challenges for effective transportations of nutrients, oxygen, and wastes, which would induce interior hypoxia and cellular toxicity. Recently, encapsulation of cells within hydrogel microcapsules has been used as a novel platform for 3D in vitro culture 10, 26. It usually first disperses cells into precursor solution, followed by the breakup of cell suspension into discrete droplets and the polymerization of the precursor droplets into hydrogel microcapsules 27, 28. The miniaturized size of microcapsules help avoid problems associated with mass transport due to the enlarged surface-to-volume ratio, allowing for optimum cell metabolism, growth, and functions 29, 30.In addition, the biocompatible nature of hydrogel matrices can simulate natural ECM with tunable structures and properties to achieve biomimetic cell culture and tissue engineering 31–33. Besides individual microcapsules, cells or microtissues can also be conveniently encapsulated in continuous microfibers as long as meters by one-phase microfluidics 34, 35, but their elongated morphology not only imposes severe barriers in cell handling, especially for assembly and injection, but also mostly restricts the cell interaction and tissue formation in one dimension. Therefore, this review would mainly focus on hydrogel microcapsules rather than microfibers for 3D in vitro culture.
Cell-laden hydrogel microcapsules could be fabricated in multiple ways 36. Electrospraying, which takes advantage of electric fields and Rayleigh-Plateau instability, is conventionally used to generate cell-laden microdroplets and hydrogel microcapsules 37, 38. Nevertheless, microcapsules produced by this method have high size polydispersity due to the unstable breakup in jetting mode and irregular morphologies due to the entry impact of microcapsules into the solution of crosslinking agents 39–42. An alternative method to generating hydrogel microcapsules is to incorporate 2D array of wells and/or pneumatically-driven vibrators into microfluidic feeding platforms, but their intermittent working characteristics limit their throughput 43, 44. Recently, microfluidic approaches to generate droplets (droplet-based microfluidics) have attracted more attention due to their potential to continuously produced highly monodisperse hydrogel microcapsules 45–47. Droplet-based microfluidics employs two immiscible fluids, a dispersed aqueous phase (suspended with cells) and a carrier oil phase, to generate microdroplets and hydrogel microcapsules in microchannels. The size, components, structure, and properties of the cell-laden microcapsules can be tuned via multiphase microfluidic dynamics 48–51. In addition, various on-chip manipulation strategies, such as fission, fusion, and separation, could be streamlined together on miniaturized devices to improve the qualities of microcapsules 48, 52.
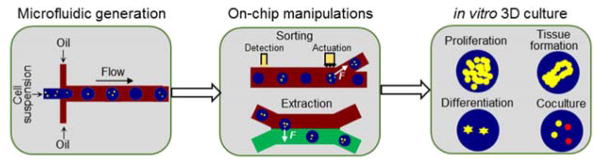
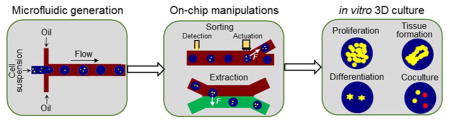
In what follows, we review the recent progresses of cellular hydrogel microencapsulation by droplet-based microfluidics for 3D in vitro culture. We summarize the concepts we discuss in this paper in Fig. 1. We first introduce general characteristics of microfluidic devices (Section 2) for cell encapsulation, and summarize the fluid dynamics of droplet breakup and interior mixing as they directly govern the size and morphology of microcapsules produced by such devices. Then we discuss two on-chip manipulation strategies (Section 3) for cell-laden microcapsules, sorting and extraction from oil into aqueous phase. We have omitted other strategies of droplet-based microfluidics, such as fission 53, fusion 54, disruption 55, trapping 56, and storage 57, 58, as they are still not widely applied in the context of 3D in vitro culture. We then introduce and discuss applications of cell-laden hydrogel microcapsules as 3D culture platforms (Section 4) to study cell growth and proliferation, stem cell differentiation, tissue development, and cell co-culture. While many other biomedical applications of hydrogel microcapsules, such as drug delivery and release 59, 60, cell preservation 39, 61–63, cell therapy 64–67, and tissue regeneration 35, 68, 69 exist, they are not within the scope of this current review. Finally, we give a brief discussion of current challenges and research prospects in this field (Section 5).
Fig. 1.
The organization of the content of this review. It is divided into three major sections, generation of hydrogel microcapsules by droplet-based microfluidics, on-chip manipulation strategies for cell-laden microcapsules, and off-chip long-term 3D cell culture for various applications.
2. Generation of cell-laden hydrogel microcapsules
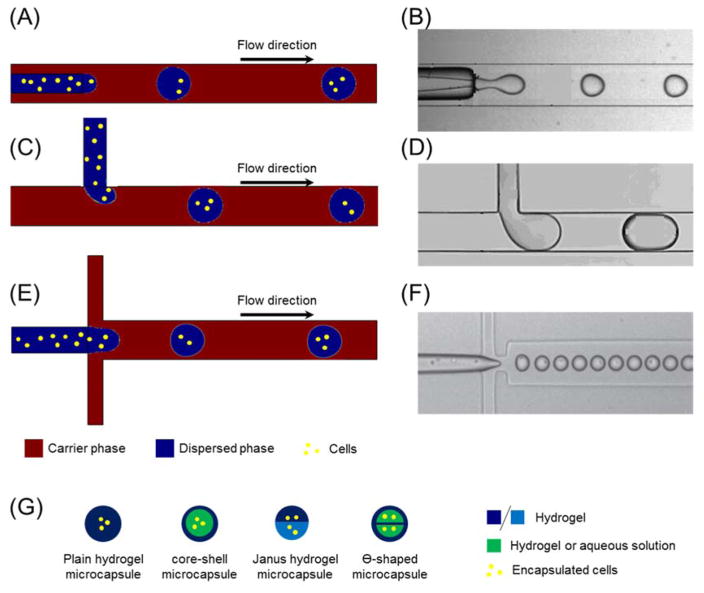
Droplet-based microfluidics for cell encapsulation can be classified into three basic types according to the configuration of the microchannels: the co-axial microchannel, the T-junction, and the flow-focusing junction (FFJ) (Fig. 2). Co-axial microfluidics aligns an inner tube of aqueous solution and suspended cells within an outer channel filled with carrier oil in parallel (Fig. 2(A–B)) 70. The T-junction allows the dispersed phase and carrier phase to merge perpendicularly at the joint point (Fig. 2(C–D)) 71, while the FFJ arranges the dispersed and carrier solution symmetrically at a cross such that the dispersed phase is focused in the center of the microchannel (Fig. 2(E–F)) 72. Despite the differences in architecture, the underlying principal for all these devices is that dispersed aqueous solution including suspended cells would be pinched into microcapsules due to the interfacial tension between the aqueous and the oil phases when these two immiscible fluids encounter at the junctions. The resulting microcapsules tend to be spherical in shape to minimize the free energy, however, other morphologies can also be created by further manipulating device structures and experimental conditions 73–75. Besides these three basic configurations, other modified devices were proposed for specific applications, such as the microcapillary coaxial glass devices to generate multiple emulsions 76, 77 and FFJ with two dispersed aqueous fluids to produce core-shell structured microcapsules 78, 79. Some common types of hydrogel microcapsules with different internal fabrics, such as core-shell structured 80, Ө-shaped 81, and Janus 82 microcapsules, are illustrated in Fig. 2(G).
Fig. 2.
Schematics of the generation of cell-laden hydrogel microcapsules by droplet-based microfluidics. There are three basic types of microfluidic devices for cell encapsulation: (A–B) Coaxial; (C–D) T-junction; (E–F) Flow-focusing junction. (B), (D), and (F) are experimental images, and they are reprinted and recreated with permission from reference 70, 71, and 72, respectively. Cells are suspended in aqueous solution prior droplet breakup at the junctions. Then they are encapsulated in compartmented individual droplets and hydrogel microcapsules post downstream gelation. (G) Basic types of cell-laden hydrogel microcapsules with various internal structures generated by droplet-based microfluidics.
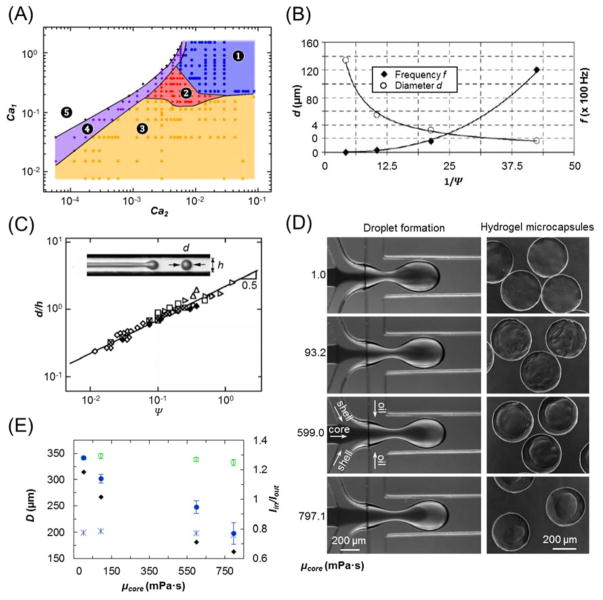
The fluid dynamics of microcapsule breakup and transport in droplet-based microfluidics has been extensively studied experimentally and numerically 70, 76, 83–89. Generally, three major forces, i.e. inertia force, viscous force, and surface or interfacial tension, govern the flow in multiphase droplet-based microfluidics. Their relationships can be effectively represented by various dimensionless parameters (Reynolds number Re, Capillary number Ca, and Weber number We) as shown in Table 1. Many distinctive patterns of microcapsule generation would emerge under various flow conditions (Fig. 3(A)) 83, but only dripping and jetting modes can stably and continuously produce microcapsules. Dripping mode produces droplets by either pressure difference or shear force near the junction where the interfacial tension tries to hold the dispersed phase back to prevent breakup 90, 91. In contrast, the jetting mode pinches off the droplets far downstream the junction by the well-known Rayleigh-Plateau instability driven by the interfacial tension 92, 93. The transition between dripping and jetting modes is usually determined by those dimensionless parameters although differences of criteria exist among various devices 87, 94. Of note, microcapsules produced by the Rayleigh-Plateau instability in jetting mode are usually less homogeneous than those generated in dripping mode 76, which renders dripping preferable to jetting for cell encapsulation. In addition, the size and generation frequency of microcapsules are also controlled by those dimensionless parameters, either in dripping or jetting mode, as exemplified by Fig. 3(B–C) 83, 95. Furthermore, it is usually assumed that the presence of suspended cells in the solution does not affect the fluid flow because the cell movement is almost dictated by the fluid viscous force given the cell Reynolds number (the characteristic length being cell diameter) is much smaller than one 96, 97. Therefore, the number of cells per microcapsules is mainly dictated by the cell density in aqueous suspension and the size of microcapsules, and principally follows the Poisson distribution 72, 98.
Table 1.
Dimensionless parameters of fluid dynamics in droplet-based microfluidics
| Dimensionless parameter | Mathematical definition | Physical implication | |
|---|---|---|---|
| Reynolds number (Re) |
|
The ratio of inertia forces to viscous forces | |
| Capillary number (Ca) |
|
The ratio of viscous force to surface or interfacial tension | |
| Weber number (We) |
|
The ratio of inertia forces to surface or interfacial tension | |
| Flow rate ratio (Ψ ) |
|
The flow rate ratio between dispersed and carrier phases | |
| Viscosity ratio (Ω ) |
|
The viscosity ratio between dispersed and carrier fluids |
Symbol ρ : fluid density; L : characteristic length; V : characteristic velocity; μ : dynamic viscosity of fluids; σ : surface tension or interfacial tension between two fluid phases; Q : flow rate. Subscript 1: dispersed phase; 2: carrier phase.
Fig. 3.
Generation of microcapsules on droplet-based microfluidics. (A) Operating diagrams of a flow-focusing microfluidic devices based on Capillary numbers of dispersed (Ca1) and carrier (Ca2) phases. Five regimes: ➊ threading, ➋ jetting, ➌ dripping, ➍ tubing and ➎ viscous displacement, were identified. But only jetting and dripping can continuously generate microcapsules. Image is reprinted and recreated with permission from reference 83.(B) Diameters and frequencies of microcapsules generated in dripping mode. Ψ is the flow rate ratio between dispersed and carrier phases. Image is reprinted and recreated with permission from reference 95. (C) Diameters of microcapsules generated in jetting mode. Image is reprinted and recreated with permission from reference 83. (D–E) Mixing between core and shell aqueous fluids during droplet formation that would alter the core size and composition. Images are reprinted and recreated with permission from reference 81. (D) Phase contrast images of droplet breakup on a non-planar FFJ and resultant core-shell hydrogel microcapsules with core fluid of different viscosities. (E) Calculated mixing degree between core and shell fluids (Iin/Iout) using a semi-empirical equation from reference 81 and measured microcapsule sizes. Symbols: ◆, Iin/Iout; ○, shell (outer) diameter; ■, core diameter; *, theoretical core diameter assuming no fluid displacement or mixing.
Besides microcapsule size and cell number, the morphology and structure of microcapsules are also critical for optimizing the cell culture microenvironment. These critical factors are largely controlled by the fluid flow both in and around the microcapsules after their generation. Internal fluid circulation within a microcapsule is induced as it travels along straight or serpentine channels due to the non-slip boundaries of the microchannel wall, 99–101. Although this mixing effect is considered beneficial for rapid chemical reaction or particle synthesis applications, it poses a daunting challenge for the fabrication of cell-laden microcapsules with delicate structures due to the chaotic advection within microcapsules. Moreover, fluid displacement within core-shell structured microcapsules was identified during droplet breakup at FFJ microfluidics, which transports a portion of the shell fluid into the central region of microcapsules. As a result, Ө-shaped distribution of shell fluid and resultant hydrogel microcapsules can be formed post gelation (Fig. 2(D)) 81. In addition, fluid displacement and associated mixing can alter the core size and constitution of 3D core-shell structured microcapsules (Fig. 3(D)). The degree of fluid displacement and mixing are dependent on not only Re and Ca, but also the flow rate ratio Ψ and viscosity ratio Ω between dispersed and carrier fluids. For instance, with the increase of viscosity of aqueous core solution, the mixing degree would decrease and the apparent core size can be less inflated from to their theoretical values assuming no mixing (Fig. 3(E)) 81. These phenomena should be borne in mind during the design and operation of droplet-based microfluidics for cell encapsulation as the component and structure of microcapsules directly determine the microenvironment and thus, phenotype of encapsulated cells.
After the generation of microcapsules at microfluidic junctions, they need to be gelled to form hydrogel scaffold for 3D in vitro cell culture. Due to the fragility of most cells, the gelation of cell-laden microcapsules should be mild and gentle 102. Three approaches have been widely used for this purpose: photo crosslinking, chemical agents, and thermal assembly. Many polymers, such as dextran hydroxyethyl methacrylate 103, polyethylene glycol diacrylate 104, collagen-gelatin 105, and ethocylated trimethylolpropane triacrylate 106, can be readily crosslinked into hydrogel matrix by ultra-violet (UV) or blue light, because most current microfluidic devices are made of optically clear polydimethylsiloxane (PDMS) or glass. Particularly, both collagen and gelatin hydrogel particles can be obtained and strengthened by irreversible riboflavin-mediated crosslinking under blue light irradiation 105, 107. A major challenge with this method is the restriction of exposure time and intensity to minimize injury to the encapsulated cells 108, 109. Another approach is to deliver chemical crosslinking agents into aqueous microdroplets of precursor solution to trigger the polymerization and gelation 110–112. The most notable example of this method is the alginate hydrogel microcapsules gelled from sodium alginate droplets via the diffusion of Ca2+ or Ba2+ from carrier phase or in situ generation of those bications from previously infused nano particles 79, 113–115. Many other hydrogel microcapsules, such as collagen, agarose, and poly-N-isopropyl acrylamide, can be obtained from the self-assembly of thermal responsive monomers in aqueous microdroplets by changing temperatures 43, 116–119. The hydrogel materials for cell encapsulation, and their structures and properties have been extensively reviewed in other publications 110, 120, 121.
3. On-chip manipulations for cell-laden hydrogel microcapsules
After the generation and polymerization of aqueous droplets in droplet-based microfluidics, the cell-laden hydrogel microcapsules need further processing to improve their conditions in many scenarios. For instance, an important challenge during microencapsulation of pancreatic islets is to separate islet-laden microcapsules from empty ones before injection into patients to treat type I diabetes 122. A similar problem exists for single-cell encapsulation where empty or multiple-cell microcapsules make up at least 63% of total microcapsules according to the Poisson distribution. These non-single cell microcapsules should be removed before any downstream applications 123. Moreover, because microcapsules are usually produced in cell-incompatible oil phase, they should be transferred timely into aqueous phase to minimize cellular injuries 124, 125. Therefore, it is essential to incorporate on-chip manipulation strategies such as sorting and removal of carrier oil, into droplet-based microfluidics for cell-laden hydrogel microcapsules to be viable and reliable platforms.
3.1. Sorting of cell-laden hydrogel microcapsules
Microfluidic sorting of hydrogel microcapsules enables segregation of subpopulations of microcapsules of particular interest, purification of heterogeneously mixed microcapsules, and manipulation of individual microcapsules 126, 127. Sorting techniques are classified as either “active” or “passive”. Active sorting relies on external power, such as electric fields, magnetic fields, acoustic waves, and optical forces, for the detection and deflection of microcapsules of interests 52. Passive sorting, on the other hand, is performed without any extra power source, usually by exploiting the dynamic interactions between the fluid flow field and suspended microcapsules under specific experimental conditions 128.
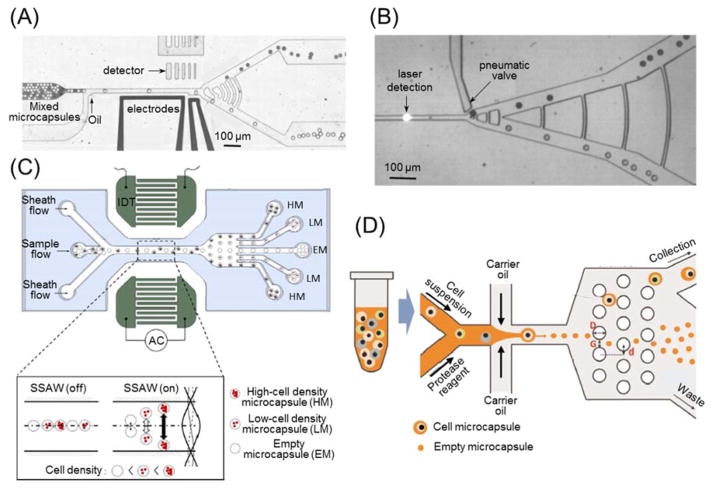
Active sorting for fluorescent microcapsules can be accomplished by combining laser detection and dielectrophoresis (DEP) or pneumatic membrane for deflection 129, 130. As illustrated in Fig. 4(A), when fluorescence-activated microcapsules pass through a detection region, the laser detector senses a dramatic change in signal intensity, which triggers the actuation of the electrodes to generate a non-uniform electric field in the microchannel 131. Therefore, a lateral DEP force is imposed on microcapsules to deflect their trajectories into upper exit of the branched outlet. On the other hand, microcapsules without fluorescence cannot activate the DEP and they would follow the streamlines into the lower exit. A mixture of E. coli cells that express either enzyme β-galactosidase reporter or inactive variants, were encapsulated and sorted at rates of ~ 300 microcapsules per second via this method 130. Furthermore, when two types of cells were stained with different fluorescent dyes, the microcapsules containing two different cells could be sorted when the signals of both fluorescent signals were detected positively 132. The DEP force for deflection can be readily replaced by other forces such as a mechanical push from gas-powered elastic membrane valve (Fig. 4(B)) 129.Principally, this approach can be employed for the aforementioned isolation of pancreatic islet-laden microcapsules from massive numbers of empty ones. Another example of active sorting utilizes acoustic wave to separate microcapsules of various sizes or densities 133, 134. Standing surface acoustic waves (SSAWs) could be readily generated by placing patterned interdigitated transducers (IDTs) along microchannels, and would drive microcapsules of different cell numbers onto different lateral locations in the microchannel according to the difference of their densities (Fig. 4(C)) 135. Thus, microcapsules of different cell densities were sorted and collected at the branched outlets.
Fig. 4.
Strategies of on-chip sorting for hydrogel microcapsules. (A) Dielectrophoresis (DEP)-based sorting for fluorescent microcapsules with laser detectors. Image is reprinted and recreated with permission from reference 131. (B) Pneumatic membrane valve-based sorting for fluorescent microcapsules with laser detector. Image is reprinted and recreated with permission from reference 129. (C) Standing surface acoustic wave (SSAW)-based sorting for microcapsules of various cell densities. Image is reprinted and recreated with permission from reference 135. (D) Size-dependent sorting of single cell microcapsules via deterministic lateral displacement. Image is reprinted and recreated with permission from reference 147.
Passive sorting of microcapsules can also be achieved by multiple approaches. In inertial microfluidics where channel Reynolds number falls in the range between ~100 and ~102, the nonlinear convection term in the Naive-Stokes equations cannot be neglected and the inertial force of suspended microcapsules becomes significant compared to viscous force 136, 137. Therefore, the microcapsules experience an inertial lift force, and migrate towards certain lateral equilibrium positions that are usually proportional to the particle Reynolds number (characteristic length being the microcapsule diameter) 138–140. As a result, microcapsules of different sizes can be sorted by inertial microfluidics 141. In addition, Dean flow caused by spiral microchannel or centrifugal force caused by rotating device can be coupled with inertial microfluidics to separate microcapsules of slightly different sizes or densities with improved accuracy and efficiency 142, 143. Mass or size dependent sorting was also achieved by gravity-driven microfluidics 144. However, to best of our knowledge, these sorting strategies has not been used for cell-laden hydrogel microcapsules, though in principal they could be applied. Moreover, deterministic lateral displacement (DLD) is well established for size-dependent separation through an array of cylindrical posts 145, 146. As shown in Fig. 4(D), cell-laden microcapsules generated in jetting mode were bigger than those empty ones, which could be readily purified and enriched by downstream DLD separation 147. Overall, these active and passive sorting strategies provide a variety of selection options to separate various kinds of microcapsules as summarized in Table 2.
Table 2.
On-chip manipulation strategies for hydrogel microcapsules
| Purpose | Method | Mechanism | Notes | References |
|---|---|---|---|---|
| Sorting | Fluorescence activated microcapsule sorting | Apply dielectrophoresis (DEP) or mechanical force on specific microcapsules based on previous detection of fluorescence intensity | With detection and actuation systems; High accuracy; High throughput | 129–132 |
| Acoustic wave - based sorting | Acoustic waves drive microcapsules of different sizes or densities onto different lateral locations | Active sorting without detection system; High throughput | 133–135 | |
| Inertial microfluidics | Microcapsules of different sizes would migrate to different equilibrium locations by inertial lift force | Laminar flow with Re > 1; Dean flow can be incorporated; High throughput | 136, 137, 139, 142 | |
| Sedimentation-based sorting | Microcapsules of different size or mass would sediment on different locations under gravity or centrifugal force | High throughput; Low accuracy; Has not utilized for cell-laden microcapsules | 143, 144 | |
| Deterministic lateral displacement-based sorting | Microcapsules of different sizes would choose different trajectories as they pass through an array of microposts | High throughput; High accuracy | 145–147 | |
| Extraction from oil into aqueous phase | Spontaneous phase separation | Microcapsules enclosed by a thin layer of oil can transfer into aqueous solution by the spontaneous dewetting of oil phase | The oil shell prevents microcapsule gelation by chemical crosslinking agents | 151 |
| Depletion of carrier oil | The carrier oil of microcapsules is depleted and replaced by aqueous solution via multiple side channels | Complex channel network; Unstable oil-aqueous interface | 125, 149 | |
| Mechanical filter gate | Microcapsules can not pass through mechanical filter gates that are smaller than their sizes, and sink into aqueous chamber. | With extra fabrication of aqueous chamber; Potential blockage issue | 116 | |
| Interfacial tension-based extraction | Microcapsules in carrier oil would migrate into aqueous solution once they contact oil-aqueous interface by interfacial tension | Require stable oil-aqueous interface; Mechanical stiffness and size constraints | 80, 97, 195 | |
| DEP-based extraction | Microcapsules would experience a DEP force towards aqueous solution for extraction under non-uniform electric filed | Combine interfacial tension in extraction; No stiffness or size constraints | 152 |
3.2. Extraction of microcapsules from oil into aqueous phase
As discussed above, cell-laden hydrogel microcapsules are usually generated in nutrient and oxygen-depleted environment and/or crosslinking agent-enriched oil phase. Therefore, the exposure to carrier oil should be minimized to reduce cellular injuries during the generation, operation, and collection of hydrogel microcapsules 148, 149. Conventionally, microcapsules are transferred from carrier oil into aqueous solution by multiple steps of off-chip centrifugation and washing, which however, would result in aggregation of microcapsules, residual oil on microcapsules surface, low retrieval efficiency, and compromised cell viability 116, 125, 150. Therefore, various on-chip approaches have been proposed to timely extract hydrogel microcapsules from carrier oil into aqueous phase. Such on-chip extraction not only effectively prevent aggregate formation as microcapsules would not be crowded in oil phase at the collection, but also enhance cell survival due to the shortened lingering period in carrier oil. Accordingly, the integration of such functional units into microfluidic encapsulation platforms is essential to improve both viability of encapsulated cells and overall quality of encapsulation. Various extraction approaches of hydrogel microcapsules from oil into aqueous phase have been developed as recapitulated in Table 2.
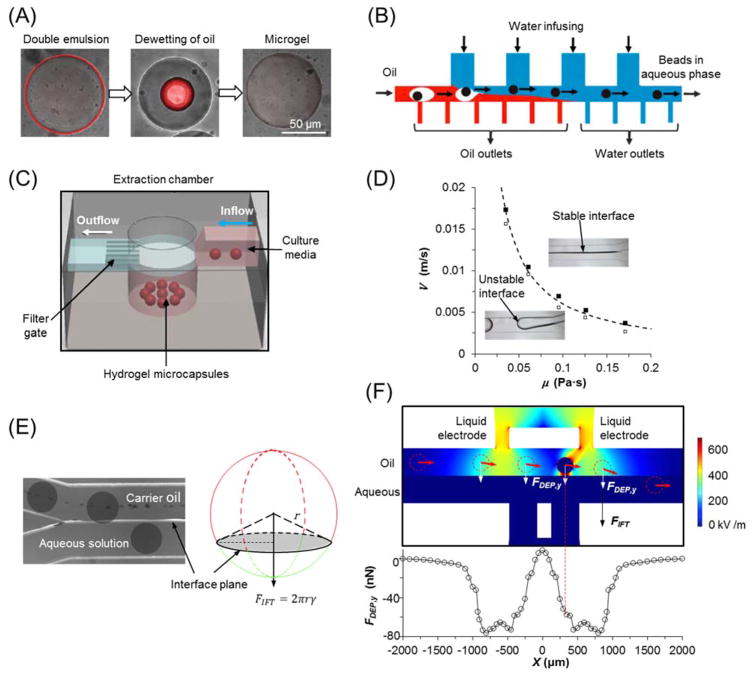
An extraction method for double emulsion microcapsules (water-in-oil-in-water) utilizes a sacrificial thin layer of oil that spontaneously dewets from the hydrogel microcapsules in aqueous solution (Fig. 5(A)) 151. Nevertheless, the presence of oil shell prevents the diffusion of chemical crosslinking agents and limits the gelation methods for aqueous microcapsules. In addition, extraction can be achieved by the fluid exchange of carrier oil phase based on oil depletion and aqueous solution infusion with multiple side channels (Fig. 5(B)) 125. Through the fabrication of filter gates in the microchannel, suspended hydrogel microcapsules can be blocked and transferred into an aqueous chamber for extraction (Fig. 5(C)) 116. However, these approaches require further manufacturing of specific devices or structures, increase the system complexity and operational difficulty and thus, compromise their reliability and reproducibility.
Fig. 5.
Strategies of on-chip extractions for hydrogel microcapsules from oil into aqueous phase. (A) Double emulsion with a sacrificial ultra-thin oil shell that can spontaneously dewet from hydrogel microcapsules. Image is reprinted and recreated with permission from reference 151. (B) Fluid exchange of carrier phase based on oil depletion and water infusion with multiple side branches. Image is reprinted and recreated with permission from reference 125. (C) Extraction chamber with mechanical filter gate to block the pass of microcapsules. Image is reprinted and recreated with permission from reference 116. (D) The criterion of stable oil-aqueous interface in microchannel for continuous extraction. (E) Interfacial tension (FIFT) based extraction of hydrogel microcapsules. Images (D–E) are reprinted and recreated with permission from reference 97. (F) Dielectrophoretic force (FDEP) based extraction for mechanical stiffness-independent microcapsules. Image is reprinted and recreated with permission from reference 152.
Recently, it was found that a stable interface between carrier oil phase and exaction aqueous phase can be established in microchannels if the viscous force overcomes the instability caused by interfacial tension between these two immiscible fluids (Fig. 5(D)) 97. In addition, the suspended microcapsules is brought towards the interface when they enter the extraction channel due to the disappearance of non-slip boundary on the interface side. Upon contacting the stable interface, the hydrogel microcapsules are exposed to an interfacial tension force that immediately drags them into the aqueous extraction solution (Fig. 5(E)) 97. However, this method demands the hydrogel microcapsules to satisfy certain criteria of size and mechanical strength. To this end, DEP force was introduced to achieve size- and stiffness-independent extraction by applying non-uniform electric field in extraction channel (Fig. 5(F)). Furthermore, the electric field used in DEP applications would not harm encapsulated cells due to the Faraday Cage effect of the electrically conductive hydrogel microcapsules 152.
4. Hydrogel microencapsulation for 3D cell culture
As discussed above, cell-laden hydrogel microcapsules can be produced with well-defined structures and properties by droplet-based microfluidics due to its versatile generation and manipulation strategies. Because of the hydrophilic and biocompatible nature of hydrogels, encapsulated cells can maintain high viability and normal functions for extended period of time 28, 153–155. Their homogenous size and controllable morphology also improve in vivo injectability and facilitate 3D imaging 51, 156. Moreover, they provide biomimetic 3D in vitro microniches to investigate cell growth and proliferation, stem cell differentiation, tissue development, and cell co-culture which we will discuss in the following.
4.1. Cell growth and proliferation
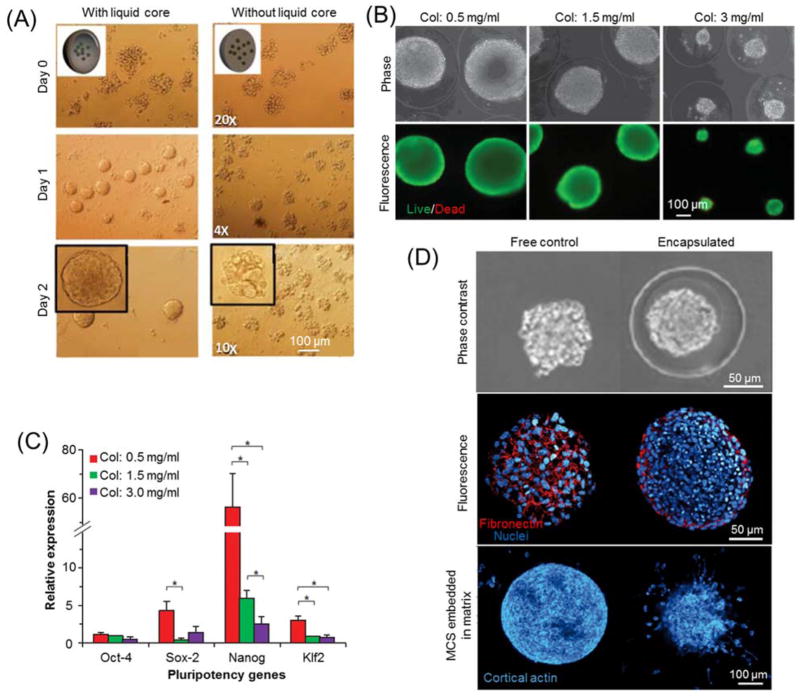
The growth and proliferation of cells in hydrogel microcapsules depend on many factors, such as cell type, culture medium, and biochemical and biomechanical properties of microcapsules. For example, when cells are encapsulated in calcium-alginate hydrogel microbeads, they form small satellite clusters of bumpy shapes as shown in Fig. 6(A), probably due to the transport barrier for cell migration and aggregation in the highly viscoelastic microenvironment 78. On the contrary, they are ready to proliferate and aggregate to form a single spheroid in the liquid core of core-shell structured microcapsules 78, 157, 158. Similarly, embryonic stem cells (ESCs) in the core of collagen core-alginate shell microcapsules expand faster and express more pluripotency genes with lower collagen concentration (Fig. 6(B–C)) 80. However, human mesenchymal stem cells (MSCs) in low (1.7%) or high (2.5%) concentration of alginate and fibroblasts L929 in poly(2-methacryloyloxyethyl phosphorylcholine-co-n-butyl methacrylate-co-p-vinylphenylboronic acid) (PMBV) - poly(vinyl alcohol) (PVA) hydrogel microbeads do not proliferate at all, which could be used as models to control cell number or suppress excessive cell proliferation without losing cell function 159, 160. These studies demonstrated diverse destinies of cell expansions in hydrogel microcapsules, mainly depending on the close interaction between encapsulated cells and their microenvironment.
Fig. 6.
Hydrogel microencapsulation for cell growth and proliferation. (A) Typical bright field images of mouse embryonic carcinoma cells encapsulated in alginate hydrogel microcapsules with or without liquid core. Image is reprinted and recreated with permission from reference 78. (B) Phase contrast and fluorescence images of mouse embryonic stem cells (ESCs) encapsulated in collagen core-alginate shell microcapsules with various collagen concentrations. (C) Quantitative RT-PCR of ESC spheroids formed in various core ECM. Images (B–C) are reprinted and recreated with permission from reference 80. (D) Phase contrast and Immunofluorescence staining micrographs of multicellular spheroids (MCS) in liquid core-alginate shell microcapsules and unencapsulated spheroids as free controls. Image is reprinted and recreated with permission from reference 173.
Although traditional 3D in vitro culture methods, such as bioreactors, hanging drops and bioprinting, can be used for cell expansion and aggregation, they cannot produce cell aggregates or tissues with uniform and controllable size and morphology at high throughput 5, 161. By creating core-shell structured microcapsules with liquid or degradable core and hydrogel shell, homogeneous aggregates can be continuously produced in the core zone 162, 163. Furthermore, the size and morphology of the resultant aggregates can be predefined by the core space 80, 164. In addition, the core-shell structure of microcapsules can not only eliminate cell protrusion in microcapsules and improve surface smoothness 165, 166, but also prevent the cell escape or leakage from microcapsules during cell expansion 167.
Hydrogel microencapsulation has also been utilized to investigate cell viability, phenotype, mobility, and drug response under various artificial or synthetic ECMs. For example, the mechanical stiffness of microcapsules can influence the extent of spreading for encapsulated cells 105. The incorporation of Arg-Gly-Asp (RGD) peptide in alginate or polycarprolactone nanofibers in polyethylene glycol (PEG) hydrogel microcapsules can promote cell attachment and spreading in 3D milieus 168, and the fabrication of microgrooved surfaces can improve cell adhesion and induce cell alignment along the groove direction 169–171. Furthermore, MSCs encapsulated in alginate-matrigel mixed hydrogel microcapsules demonstrate stronger drug resistance to vincristine than conventional monolayer cultured cells 172, and alginate hydrogel microcapsules of MSCs can convert inflammatory macrophages to alternative M2 macrophage phenotype 159. In addition, multicellular spheroids encapsulated in liquid core-alginate shell microcapsules undergo dramatic cellular reorganization when they reach the boundary between core and shell, and the peripheral cells can significantly improve their motility in comparison to unconfined free multicellular spheroids (Fig. 6(D)) 173. This 3D biomimetic culture of tumor spheroids in hydrogel microcapsules with controllable properties provides a convenient platform to investigate the interplay mechanisms between tumor metastasis and surrounding tissues and to screen anticancer drugs 174, 175.
4.2. Stem cell differentiation
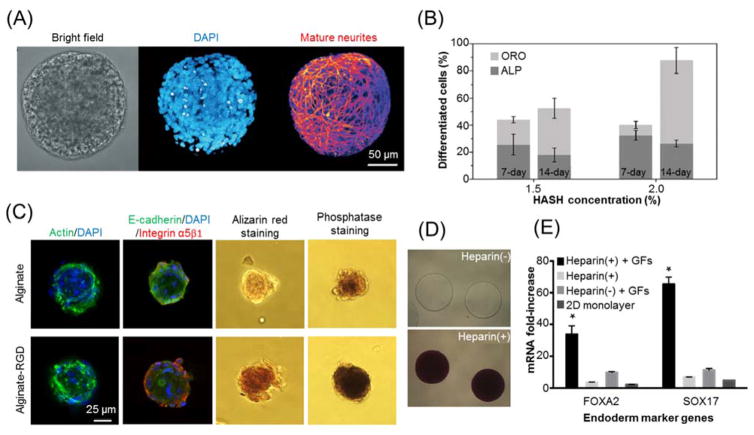
Hydrogel microcapsules can not only maintain and even improve the pluripotency of encapsulated stem cells 79, 115, 160, but also direct stem cell differentiation via their tunable physical and chemical properties, such as stiffness, porosity, affinity, and degradation speed 176. Stem cells or induced pluripotent cells (iPSCs) in microcapsules interact with a 3D ECM which could trigger biochemical and biomechanical cues to promote or inhibit specific differentiation routes 177. For example, human neural stem cells encapsulated in the core of matrigel core-alginate shell microcapsules or mouse embryonic carcinoma cells in alginate hydrogel beads are prone to neuron differentiation (Fig. 7(A)) 56, 178. Human MSCs in thiol-modified hyaluronic acid hydrogel prefer adipogenic to osteogenic differentiation after 14-day culture (Fig. 7(B)) 179. They also can be guided into osteogenic differentiation when encapsulated in fibrous collagen or poly lactic-co-glycolic acid (PLGA) core-alginate shell microcapsules or even alginate hydrogel beads 180–182. When bovine MSCs encapsulated in hyaluronic acid modified methacrylate alginate hydrogel, their chondrogenic gene expression and the superficial zone chondrocyte phenotype are upregulated 183. MSCs can also undergo myogenic differentiation with successful formation of multinucleated myotubes if they are encapsulated in fibrin hydrogel embedded with fast degradable microbeads 184. In addition, encapsulated ESCs can differentiate into not only hepatocytes or pancreatic islet-like cells in alginate microcapsules 185, 186, but also beating cardiomyocytes in liquid core-alginate shell microcapsules 79, 187. These examples demonstrate that hydrogel microcapsules can guide various stem cells into divergent pathways of differentiations in 3D well-defined microniches.
Fig. 7.
Hydrogel microencapsulation for stem cell differentiation. (A) Typical images of neuron differentiation of human neural stem cells in matrigel core-alginate shell hydrogel microcapsules. Image is reprinted and recreated with permission from reference 178. (B) Percentages of osteogenic (ALP staining) and adipogenic (ORO staining) differentiations of human mesenchymal stem cells (MSCs) in thiol-modified hyaluronic acid (HASH) hydrogel of different concentrations. Image is reprinted and recreated with permission from reference 179. (C) Osteogenic differentiation of human MSCs in alginate hydrogel microcapsules with or without RGD modification. Image is reprinted and recreated with permission from reference 157. (D–E) Enhanced endodermic differentiation of PEG-encapsulated ESCs with heparin modification. Images are reprinted and recreated with permission from reference 190. (D) Toluidine blue staining of PEG hydrogel microcapsules with (+) or without (-) heparin. (E) Definitive endoderm marker expression of encapsulated mouse ESCs. The heparin-containing PEG hydrogel microcapsules promote endodermic differentiation with a single dose of growth factors due to the sequestration effect of heparin.
Hydrogel microcapsules can be modified to further improve their modulatory effects on stem cell differentiation. RGD incorporation into alginate hydrogel microcapsules can not only promote cell attachment and spreading as discussed above, but also upregulate osteogenic differentiation for encapsulated human MSCs (Fig. 7(C)) 157, 188. Gelatin modification of alginate microcapsules can enhance adipogenic differentiation for human adipose-derived stem cells (ADSCs) 189. Because heparin can absorb multiple growth factors into hydrogel microcapsules due to charge interactions (Fig. 7(D)), the incorporation of heparin in PEG microcapsules enhances the endodermic differentiation of encapsulated ESCs with reduced concentrations of growth factors (Fig. 7(E)) 190. In addition, by alternately repeating electrospinning and microfluidic process, rat bone marrow MSCs and bone morphogenetic protein-2 could be simultaneously encapsulated in alginate hydrogel microcapsules to form multilayer scaffold, which demonstrated improved osteogenic differentiation and accelerated ectopic bone formation 191. Overall, these engineering strategies of hydrogel microcapsules can extensively enhance their capabilities for guided stem cell differentiation.
4.3. Tissue development
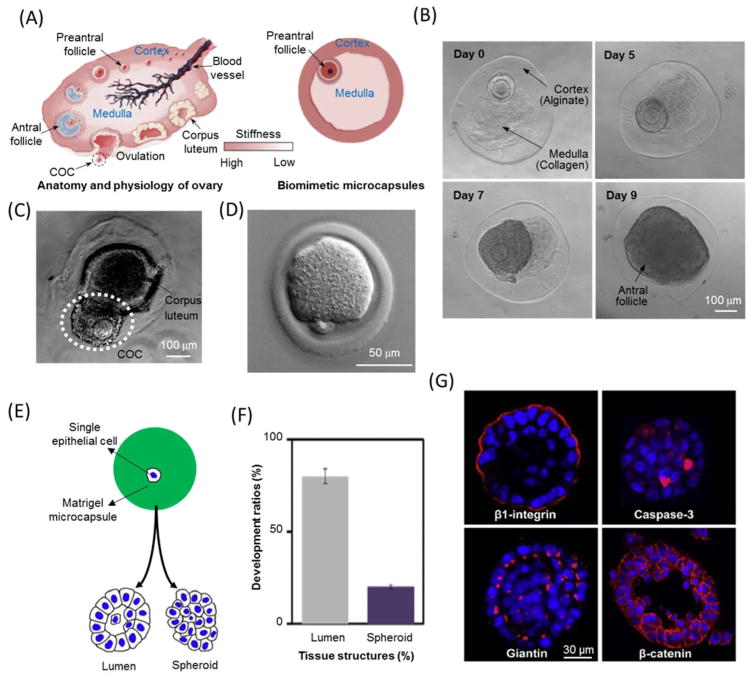
Besides expansion, aggregation, and differentiation, hydrogel microcapsules are also ideal for tissue development. Guided by various biochemical and biomechanical stimuli provided by the hydrogel, encapsulated cells could follow their development roadmaps to produce specific tissues or organs 192, 193. For example, the heterogeneity of mechanical stiffness of the ovarian structure is regarded to be critical for ovary function and evolution 194. Therefore, biomimetic microcapsules with soft core to resemble natural medulla and stiff shell to mimic cortex was fabricated by droplet-based microfluidics (Fig. 8(A)). Preantral follicles were encapsulated at the boundary between soft core (made of collagen) and stiff shell (made of alginate) so that they can experience mechanical heterogeneity as in native microenvironment. Therefore, they could develop into antral follicles with much higher efficiency than those of 2D monolayer culture or homogenous microcapsules (Fig. 8(B)) 195. In addition, the developed antra follicles could perform in vitro ovulation in the biomimetic microcapsules without the supplementation of growth factors as shown in Fig. 8(C) 195, and metaphase II (MII) oocytes could be obtained (Fig. 8(D)) 80. The effects of various core and shell materials on the development and ovulation of preantral follicles were further screened 80. This 3D in vitro culture and development platform for preantral follicles may be an attractive strategy to preserve and restore the fertilities of women (especially who experienced aggressive medical treatment or environmental biohazards) and endangered animals 196, 197.
Fig. 8.
Hydrogel microencapsulation for tissue development. (A–D) Biomimetic encapsulation of mouse preantral follicles for in vitro development and ovulation in soft core-stiff shell hydrogel microcapsules. Images (A–C) and (D) are reprinted and recreated with permission from reference 195 and 80, respectively. (A) A schematic of mouse ovarian structure and engineered biomimetic microcapsules with mechanical heterogeneity. (B) In vitro development of preantral follicle in biomimetic ovarian microcapsules. (C) In vitro ovulation of an antral follicle without using luteinizing hormone and epidermal growth factor. (D) Metaphase II (MII) oocytes obtained from the antral follicles with first polar body and mitotic spindle. (E–G) The development of single epithelial cell toward acinus in matrigel microcapsules. Images are reprinted and recreated with permission from reference 198. (E) A schematic of the development of an encapsulated single epithelial cell into lumen or spheroid. (F) Percentages of encapsulated single epithelial cell developing into lumen and spheroid over 7-day culture. (G) Immunofluorescence stainings of lumen formation in hydrogel microcapsules.
Another example of engineered tissues by hydrogel microencapsulation is the formation of an acinus from a single epithelial cell in matrigel microcapsules. According to Fig. 8(E–F), single prostate epithelial cell in matrigel bead is more likely to form lumen rather than cellular spheroid over 7-day culture 198. The active-caspase-3 immunostaining in Fig. 8(G) reveals that lumen formation is associated with the apoptosis of interior cells of the spheroids. Furthermore, isolated single microcapsule of single cell in a single well without any cell interaction or communication could also develop into acinus 198. Compared to conventional 3D culture of epithelial cells in microwell plates 199, 200, this method significantly improves the purity, homogeneity, and throughput of 3D acini formation. Furthermore, it enables microscopic observations and analyses with high temporal and spatial resolutions to probe the mechanisms of acinar development and carcinogenesis. In addition, this approach was also used to achieve 3D in vitro kidney epithelialization by encapsulating Madin Darby canine kidney cells in geltrex microgel 201.
4.4. Cell co-culture
Cell-cell communication and interactions are crucial to maintain normal structure and functionality of in vivo and in vitro tissues 202–204. Droplet-based microfluidics offers a practical platform to co-encapsulate and co-culture two or more different types of cells with arbitrary ratios of cell numbers within miniaturized microenvironment. For example, metastatic breast cancer cells MDA-MB-231 and non-tumorigenic breast cells MCF-10A were encapsulated with a ratio of 1:1 in the core of matrigel core-alginate shell microcapsules 162. Over 7-day co-culture, the initially random mixture of cells evolved into a core-shell structured organization with MDA-MB-231 cells enclosing MCF-10A cells in the central region (Fig. 9(A)). This cell segregation is probably due to the differential E-cadherin expression of these two types of cells, which would induce spontaneous cell reorganization to minimize overall interfacial free energy according to the differential adhesion hypothesis 205, 206. Likewise, fibroblasts encapsulated in peptide-based self-assembly hydrogel microcapsules could be co-cultured with keratinocytes binding on the surface of hydrogel microcapsules 207, while epithelial and mesenchymal microtissues were co-cultured based on the complementary sequence of templating DNA 208. Janus hydrogel microcapsules (Fig. 2(D)) are also excellent carriers for cell co-culture if different types of cells seeded in different sections of microcapsules 209. Compared to conventional co-culture approaches (e.g. transwell plate) 210–213, hydrogel microcapsules not only offers robust cell microniches with tunable structures and ECM properties, but also significantly reduces the requirement of cell number for co-culture compared to the bulk co-culture approaches due to decreased spatial distance and enhanced cellular interaction in miniaturized environment of microcapsules.
Fig. 9.
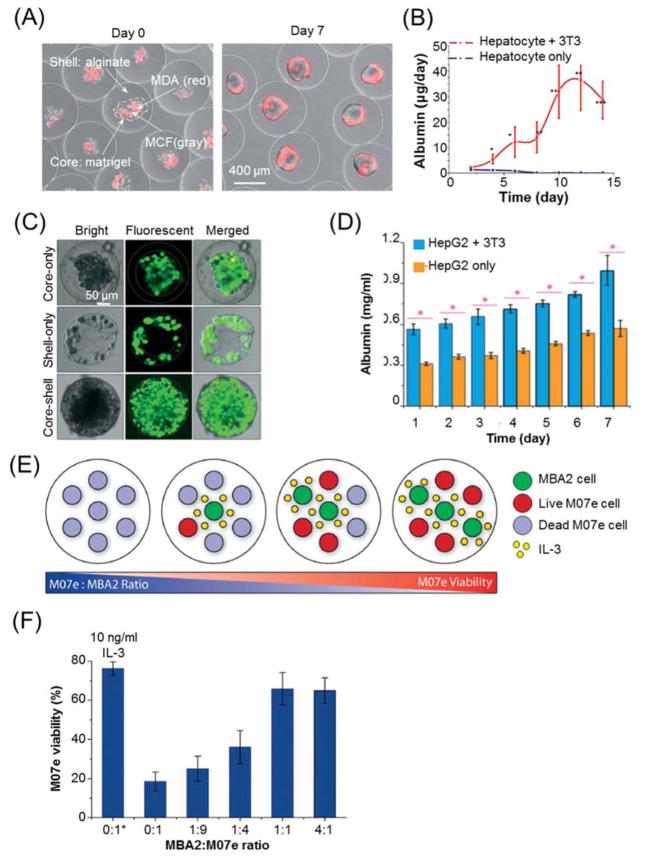
Hydrogel microencapsulation for 3D cell co-culture. (A) Co-culture of breast epithelial cell lines (MDA, stained with red fluorescence) and MCF-10A cells, in the core of matrigel core-alginate shell hydrogel microcapsules. (B) Co-culture of hepatocytes and fibroblasts 3T3-J2 in the core of collagen core-alginate shell microcapsules to enhance the albumin secretion from encapsulated hepatocytes. Images (A–B) are reprinted and recreated with permission from reference 162. (C–D) Co-culture of liver hepatocellular carcinoma (HepG2) and fibroblasts NIH-3T3 in liquid core and alginate hydrogel shell, respectively. Images are reprinted and recreated with permission from reference 215. (C) Distributions of HepG2 and fibroblasts in core-shell structured microcapsules. (D) Albumin secretion of HepG2 over 7-day culture. (E–F) Co-culture of MBA2 supporting cells and M07e megakaryoblastic leukemia cells in agarose hydrogel microcapsules. Images are reprinted and recreated with permission from reference 216. (E) A schematic of MBA and M07e co-culture at various ratios of cell number. (F) The viability of M07e cells under different cell co-culture ratios. * indicates treatment with 10 ng/ml IL-3 as a positive control.
Hydrogel microcapsules also provide miniaturized platforms for 3D in vitro cell co-culture to demonstrate cellular functionalities. One notable example would be the co-culture of hepatocytes and fibroblasts that testifies the beneficial effects of fibroblasts on the viability and functionalities of hepatocytes. When hepatocytes were co-cultured with 3T3 fibroblasts in the ECM core of core-shell structured microcapsules 162, or with endothelial progenitor cells in alginate-collagen mixed hydrogel microcapsules 214, their long-term viability, spheroid formation, and albumin secretion can be significantly enhanced compared to the those without co-culture (Fig. 9(B)). Similarly, if liver hepatocellular carcinoma HepG2 and 3T3 fibroblasts were co-encapsulated respectively in the liquid core and alginate shell of hydrogel microcapsules (Fig. 9(C)) 215, the liver function per microcapsules would be augmented by the co-culture with fibroblasts as indicated by Fig. 9(D). Another example would be the co-encapsulation of MBA2 supporting cells and other functional cells. When M07e leukemia cells and MBA2 cells were encapsulated by agarose microcapsules with various cell ratios (Fig. 9(E)) 216. The paracrine secretion of IL-3 from MBA2 cells can modulate the viability of M07e cells (Fig. 9(F)). In addition, co-encapsulation and co-culture of MBA2 and umbilical cord blood (UCB) cells were also performed by the same system, and differential responsiveness of UCB subpopulations to paracrine signals from MBA2 were revealed 216. Of note, the immunomodulatory effects of MSC on inflammation was also demonstrated by co-culturing hydrogel encapsulated MSC and macrophage or organotypic hippocampal slice cultures 37, 64, 159, though those encapsulations were not performed by droplet-based microfluidics due to its relatively low throughput compared to electrospray.
5. Discussion
During past decades, hydrogel microencapsulation for 3D in vitro cell culture by droplet based-microfluidics has achieved tremendous advances. As summarized before, one of the biggest advantages of hydrogel microencapsulation via droplet-based microfluidics is the tunable geometric, mechanical, electrical, chemical, and biological properties of microcapsules. Highly homogeneous microcapsules of various morphologies and sizes could be readily generated by adjusting the geometry of microchannels, fluid properties, and flow rates. Their biophysical and biochemical properties, such as mechanical stiffness, chemical affinity, and biological compatibility, could also be closely modified by microfluidic approaches. In addition, the generation frequency of microcapsules can be varied in a wide range (from ~ 100 to ~ 103 Hz), which enables hydrogel microencapsulation for both very few (i.e. pancreatic islets) and large quantity (i.e. stem cells) of biological samples. Furthermore, the quality of microcapsules, especially cell-laden hydrogel microcapsules, can be further improved by multiple on-chip manipulation strategies through incorporating various functional units, such as sorting and removal of carrier oil phase. Finally, with the advent and development of photolithography and soft lithography, most of current microfluidic device are made of PDMS. Its short fabrication period (can be less than one day) enables fast prototyping and quick proof-of-concept trials, and its optical transparency nature facilitates dynamic observation, administration, and operation.
However, there are still several daunting challenges before its widespread application and commercialization. First, cell-laden hydrogel microcapsules cannot simulate the in vivo microniches of all kinds of cells, tissues or organs. For instance, since the shell of microcapsule shields encapsulated cells from external flow and hydrodynamic shear stress, the 3D spheroids formed in microcapsules cannot resemble those highly vascularized organs, epithelial tissue for example, where the microvascular network and associated flow are essential for their behaviors and functions 217. Second, the microfluidic generation of hydrogel microcapsules has relatively low throughput compared to conventional electrospraying, especially when it is used for tissue engineering. The flow rates of the dispersed phase of cell suspension in droplet-based microfluidics are in the range of 10−1 ml/hr, while that of electrospray can easily go to 101 ml/hr or even higher 37, 79. Although this low-throughput can be mitigated by scale-up via parallelization, a network of many (> 100) droplet parallel generation units results in complex structure with multiple layers of channels which can be prohibitive to fabricate and operate 218. Last but not least, the operation of droplet-based microfluidics also demands multiple expensive micropumps, which impedes the integration, miniaturization, and transportation of the system.
In the future, these technical limitations and challenges of hydrogel microcapsules produced via droplet-based microfluidics should be resolved before this approach can prevail as the mainstream for controllable tissue engineering and high throughput drug screening 219. One strategy might utilize other manufacturing materials, such as poly(methyl methacrylate) (PMMA), glass, and silicon, rather than PDMS to improve the mechanical strength and reusability of devices, especially for mass production post proof-of-concept prototyping and experimentation. Another promising advance would be the emerging 3D printing technique, which allows fast microfabrication of microfluidic devices in regular lab settings with accelerating fabrication speed and uncompromising fidelity 44, 178. In addition, new types of hydrogel materials with higher biocompatibility, longer biostability and broader rangers of physical and chemical properties are required for the construction of more biomimetic ECM. Moreover, although many publications have explored the various impacts of hydrogel microcapsules on cell culture as summarized in this review, very few of them specifically illustrated the mechanisms and biochemical pathways of the cell-niches interactions on cell fate. This represents a major missing section in the investigation of hydrogel microencapsulation for mammalian cells and tissues, and certainly entails deeper insight and examination on this area in future studies.
6. Conclusions
Overall, droplet-based microfluidics is an extraordinary approach to generate cell-laden hydrogel microcapsules due to its versatile regulation of microcapsule’s size, morphology, and structure. In addition, multiple on-chip manipulation strategies, especially sorting and removal of carrier oil, can be readily integrated onto the microfluidic device to improve the quality of hydrogel microcapsules. As we have discussed in the preceding, these microcapsules can provide an excellent platform for 3D in vitro cell culture. Their miniaturized and homogeneous size, tunable properties, and controllable structures enable them for biomimetic or non-biomimetic in vitro cell culture. Various cellular activities, such as cell growth and proliferation, stem cell differentiation, tissue development, and cell interaction have been investigated in broad ranges via this approach. Although this technology is still in its infancy for 3D controllable cell culture, it holds the promise for widespread applications in drug screening and tissue engineering with improved accuracy and efficiency, under the assistance of ongoing advances in microfabrication and biomaterials.
Table 3.
Applications of cell 3D culture in hydrogel microcapsules
| Purpose | Cell | Microcapsule | Conclusion | References |
|---|---|---|---|---|
| Cell growth and proliferation | ESC; MSC; Fibroblast; Embryoid body | Plain hydrogel microcapsule, such as alginate, collagen, and PVA microbead | Diverse destinies of cell proliferation dependent on the interaction between cells and hydrogel ECM. | 78, 157–160 |
| Embryoid body; ESC; Islet; Cancer cell; Kidney cell | Liquid core-hydrogel shell microcapsule | Defined size and structure of cell aggregates; Prevention of cell leakage and protrusion. | 78, 80, 162–167 | |
| MSC; HEPM; Fibroblast; Cancer spheroid | RGD/nanofiber/groove modified hydrogel microcapsule | Enhanced cellular morphology and functionalities with those modifications. | 168–172 | |
| Multicellular spheroid | Liquid core-hydrogel shell microcapsule | Dramatic reorganization and improved motility for cells near the core-shell boundary. | 173 | |
| Stem cell differentiation | Neural stem cell; Cancer stem cell | Matrigel core-alginate shell; Alginate bead | Neuron differentiation. | 56, 178 |
| MSC | Hyaluronic acid; Collagen / PLGA core-alginate shell; Alginate bead; RGD/gelatin/methacrylate modified alginate bead | Adipogenic differentiation; Osteogenic differentiation; Chondrogenic differentiation; Myogenic differentiation. | 157, 179–184, 189 | |
| ESC | Alginate bead; Liquid core-alginate shell; PEG bead | Neuron differentiation; Hepatocyte differentiation; Cardiomyocyte differentiation; Endoderm differentiation. | 56, 79, 182, 185–187, 190 | |
| Tissue development | Preantral follicles | Collagen core-alginate shell microcapsule | Enhanced development and ovulation for preantral follicles. | 80, 195 |
| Epithelial cells | Aatrigel microcapsule | Lumen formation from single cell in single microcapsule. | 198 | |
| Madin Darby canine kidney cell | Geltrex microcapsule | 3D in vitro kidney epithelialization. | 201 | |
| Cell co-culture | Breast cancer cell MDA-MB-231 and breast cell MCF | Matrigel core-alginate shell microcapsule | Cell reorganization due to the minimization of overall interfacial energy. | 162 |
| Hepatocyte / HepG2 and fibroblasts/ endothelial progenitor cell | Liquid / matrigel core-alginate shell microcapsule; alginate-collagen beads | Improved liver function per microcapsule with co-culture of fibroblasts. | 162, 214, 215 | |
| MBA2 cell and M07e leukemia / umbilical cord blood cell | Agarose microcapsule | Cellular regulation via the paracrine secretion from MBA2 stromal cells. | 216 |
Acknowledgments
We would like to thank the NIH for funding this work through grants no. 5P41EB002503 (BioMEMS Resource Center) and 1R21EB020192-01.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Marx V. Nature. 2013;496:253–258. doi: 10.1038/496253a. [DOI] [PubMed] [Google Scholar]
- 2.Lee J, Cuddihy MJ, Kotov NA. Tissue engineering. Part B, Reviews. 2008;14:61–86. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- 3.Yamada KM, Cukierman E. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Prestwich GD. Journal of cellular biochemistry. 2007;101:1370–1383. doi: 10.1002/jcb.21386. [DOI] [PubMed] [Google Scholar]
- 5.Haycock JW. Methods in molecular biology. 2011;695:1–15. doi: 10.1007/978-1-60761-984-0_1. [DOI] [PubMed] [Google Scholar]
- 6.Howes AL, Richardson RD, Finlay D, Vuori K. PloS one. 2014;9:e108283. doi: 10.1371/journal.pone.0108283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pampaloni F, Reynaud EG, Stelzer EH. Nature reviews Molecular cell biology. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Nature protocols. 2009;4:309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 9.Rowley JA, Madlambayan G, Mooney DJ. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 10.Tibbitt MW, Anseth KS. Biotechnology and bioengineering. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrzesinski K, Magnone MC, Hansen LV, Kruse ME, Bergauer T, Bobadilla M, Gubler M, Mizrahi J, Zhang K, Andreasen CM. Toxicology Research. 2013;2:163–172. [Google Scholar]
- 12.Edmondson R, Broglie JJ, Adcock AF, Yang L. Assay and drug development technologies. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffith LG, Swartz MA. Nature reviews. Molecular cell biology. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 14.Suuronen EJ, Sheardown H, Newman KD, McLaughlin CR, Griffith M. International review of cytology. 2005;244:137–173. doi: 10.1016/S0074-7696(05)44004-8. [DOI] [PubMed] [Google Scholar]
- 15.Fennema E, Rivron N, Rouwkema J, van Blitterswijk C, de Boer J. Trends in biotechnology. 2013;31:108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Fey SJ, Wrzesinski K. Toxicological sciences. 2012:kfs122. doi: 10.1093/toxsci/kfs122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messner S, Agarkova I, Moritz W, Kelm JM. Archives of toxicology. 2013;87:209–213. doi: 10.1007/s00204-012-0968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen J, Hyllner J, Björquist P. Journal of cellular physiology. 2009;219:513–519. doi: 10.1002/jcp.21732. [DOI] [PubMed] [Google Scholar]
- 19.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrila J, Radtke AL, Crabbe A, Sarker SF, Herbst-Kralovetz MM, Ott CM, Nickerson CA. Nature reviews. Microbiology. 2010;8:791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- 21.Foty R. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quist AP, Oscarsson S. Expert opinion on drug discovery. 2010;5:569–581. doi: 10.1517/17460441.2010.489606. [DOI] [PubMed] [Google Scholar]
- 23.Souza GR, Molina JR, Raphael RM, Ozawa MG, Stark DJ, Levin CS, Bronk LF, Ananta JS, Mandelin J, Georgescu MM, Bankson JA, Gelovani JG, Killian TC, Arap W, Pasqualini R. Nature nanotechnology. 2010;5:291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan BP, Leong KW. European spine journal : official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 2008;17(Suppl 4):467–479. doi: 10.1007/s00586-008-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolesky DB, Truby RL, Gladman A, Busbee TA, Homan KA, Lewis JA. Advanced materials. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 26.Caliari SR, Burdick JA. Nature methods. 2016;13:405–414. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan J. Polymers. 2012;4:1084–1108. [Google Scholar]
- 28.Nicodemus GD, Bryant SJ. Tissue engineering. Part B, Reviews. 2008;14:149–165. doi: 10.1089/ten.teb.2007.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son Y-R, Chung J-H, Ko S, Shim S-M. Journal of microencapsulation. 2016;33:183–190. doi: 10.3109/02652048.2016.1144816. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Zhang X, Wang X, Wang C, Tang B. Journal of bioscience and bioengineering. 2012;114:1–8. doi: 10.1016/j.jbiosc.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Geckil H, Xu F, Zhang X, Moon S, Demirci U. Nanomedicine. 2010;5:469–484. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allazetta S, Hausherr TC, Lutolf MP. Biomacromolecules. 2013;14:1122–1131. doi: 10.1021/bm4000162. [DOI] [PubMed] [Google Scholar]
- 33.Cao Y, Lee BH, Peled HB, Venkatraman SS. Journal of biomedical materials research. Part A. 2016;104:2401–2411. doi: 10.1002/jbm.a.35779. [DOI] [PubMed] [Google Scholar]
- 34.Yamada M, Utoh R, Ohashi K, Tatsumi K, Yamato M, Okano T, Seki M. Biomaterials. 2012;33:8304–8315. doi: 10.1016/j.biomaterials.2012.07.068. [DOI] [PubMed] [Google Scholar]
- 35.Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, Matsunaga YT, Shimoyama Y, Takeuchi S. Nature materials. 2013;12:584–590. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 36.Yanagawa F, Sugiura S, Kanamori T. Regenerative Therapy. 2016;3:45–57. doi: 10.1016/j.reth.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stucky EC, Schloss RS, Yarmush ML, Shreiber DI. Cytotherapy. 2015;17:1353–1364. doi: 10.1016/j.jcyt.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Yang G, Zhang A, Xu LX, He X. Biomedical microdevices. 2010;12:89–96. doi: 10.1007/s10544-009-9363-z. [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann H, Ehrhart F, Zimmermann D, Müller K, Katsen-Globa A, Behringer M, Feilen P, Gessner P, Zimmermann G, Shirley S. Applied Physics A. 2007;89:909–922. [Google Scholar]
- 40.Bock N, Woodruff MA, Hutmacher DW, Dargaville TR. Polymers. 2011;3:131–149. [Google Scholar]
- 41.Tendulkar S, Mirmalek-Sani SH, Childers C, Saul J, Opara EC, Ramasubramanian MK. Biomedical microdevices. 2012;14:461–469. doi: 10.1007/s10544-011-9623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Wang J, Li C, Li Z, Liang R, Wang Q, Wang H, Zhu J, Yang Y. Science of Advanced Materials. 2015;7:902–909. [Google Scholar]
- 43.Wang Y, Li Y, Therien-Aubin H, Ma J, Zandstra PW, Kumacheva E. Biomicrofluidics. 2016;10:014110. doi: 10.1063/1.4940430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan AJ, Hidalgo San Jose L, Jamieson WD, Wymant JM, Song B, Stephens P, Barrow DA, Castell OK. PloS one. 2016;11:e0152023. doi: 10.1371/journal.pone.0152023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim C, Park J, Kang JY. Biomicrofluidics. 2014;8:066504. doi: 10.1063/1.4902943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou W-L, Lee P-Y, Yang C-L, Huang W-Y, Lin Y-S. Micromachines. 2015;6:1249–1271. [Google Scholar]
- 47.Seemann R, Brinkmann M, Pfohl T, Herminghaus S. Reports on progress in physics Physical Society. 2012;75:016601. doi: 10.1088/0034-4885/75/1/016601. [DOI] [PubMed] [Google Scholar]
- 48.Chen A, Byvank T, Chang WJ, Bharde A, Vieira G, Miller BL, Chalmers JJ, Bashir R, Sooryakumar R. Lab on a chip. 2013;13:1172–1181. doi: 10.1039/c2lc41201b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teh SY, Lin R, Hung LH, Lee AP. Lab on a chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 50.Selimovic S, Oh J, Bae H, Dokmeci M, Khademhosseini A. Polymers (Basel) 2012;4:1554. doi: 10.3390/polym4031554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu K, Deng Y, Zhang N, Li S, Ding H, Guo F, Liu W, Guo S, Zhao X-Z. Microfluidics and nanofluidics. 2012;13:761–767. [Google Scholar]
- 52.Pit AM, Duits MH, Mugele F. Micromachines. 2015;6:1768–1793. [Google Scholar]
- 53.Link DR, Anna SL, Weitz DA, Stone HA. Physical review letters. 2004;92:054503. doi: 10.1103/PhysRevLett.92.054503. [DOI] [PubMed] [Google Scholar]
- 54.Bremond N, Thiam AR, Bibette J. Physical review letters. 2008;100:024501. doi: 10.1103/PhysRevLett.100.024501. [DOI] [PubMed] [Google Scholar]
- 55.Park KJ, Lee KG, Seok S, Choi BG, Lee MK, Park TJ, Park JY, Kim DH, Lee SJ. Lab on a chip. 2014;14:1873–1879. doi: 10.1039/c4lc00070f. [DOI] [PubMed] [Google Scholar]
- 56.Kim C, Bang JH, Kim YE, Lee JH, Kang JY. Sensors and Actuators B: Chemical. 2012;166:859–869. [Google Scholar]
- 57.Boukellal H, Selimovic S, Jia Y, Cristobal G, Fraden S. Lab on a chip. 2009;9:331–338. doi: 10.1039/b808579j. [DOI] [PubMed] [Google Scholar]
- 58.Dai J, Kim HS, Guzman AR, Shim W-B, Han A. RSC Advances. 2016;6:20516–20519. [Google Scholar]
- 59.Murua A, Portero A, Orive G, Hernandez RM, de Castro M, Pedraz JL. Journal of controlled release : official journal of the Controlled Release Society. 2008;132:76–83. doi: 10.1016/j.jconrel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Orive G, Santos E, Pedraz JL, Hernandez RM. Advanced drug delivery reviews. 2014;67–68:3–14. doi: 10.1016/j.addr.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 61.Huang H, Choi JK, Rao W, Zhao S, Agarwal P, Zhao G, He X. Adv Funct Mater. 2015;25:6939–6850. doi: 10.1002/adfm.201503047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serra M, Correia C, Malpique R, Brito C, Jensen J, Bjorquist P, Carrondo MJ, Alves PM. PloS one. 2011;6:e23212. doi: 10.1371/journal.pone.0023212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen B, Wright B, Sahoo R, Connon CJ. Tissue engineering. Part C, Methods. 2013;19:568–576. doi: 10.1089/ten.TEC.2012.0489. [DOI] [PubMed] [Google Scholar]
- 64.DOLLÉ J-P, BARMINKO J, VERUVA S, MOURE C, SCHLOSS R, YARMUSH ML. Nano LIFE. 2013;3:1350004. doi: 10.1142/S1793984413500049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomei AA, Manzoli V, Fraker CA, Giraldo J, Velluto D, Najjar M, Pileggi A, Molano RD, Ricordi C, Stabler CL, Hubbell JA. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10514–10519. doi: 10.1073/pnas.1402216111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kauer TM, Figueiredo JL, Hingtgen S, Shah K. Nature neuroscience. 2011;15:197–204. doi: 10.1038/nn.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harting MT, Jimenez F, Xue H, Fischer UM, Baumgartner J, Dash PK, Cox CS. Journal of neurosurgery. 2009;110:1189–1197. doi: 10.3171/2008.9.JNS08158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chung BG, Lee KH, Khademhosseini A, Lee SH. Lab on a chip. 2012;12:45–59. doi: 10.1039/c1lc20859d. [DOI] [PubMed] [Google Scholar]
- 69.Cheng Y, Yu Y, Fu F, Wang J, Shang L, Gu Z, Zhao Y. ACS applied materials & interfaces. 2016;8:1080–1086. doi: 10.1021/acsami.5b11445. [DOI] [PubMed] [Google Scholar]
- 70.Baroud CN, Gallaire F, Dangla R. Lab on a chip. 2010;10:2032–2045. doi: 10.1039/c001191f. [DOI] [PubMed] [Google Scholar]
- 71.Gu H, Duits MH, Mugele F. International journal of molecular sciences. 2011;12:2572–2597. doi: 10.3390/ijms12042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hong J, deMello AJ, Jayasinghe SN. Biomedical materials. 2010;5:21001. doi: 10.1088/1748-6041/5/2/021001. [DOI] [PubMed] [Google Scholar]
- 73.Xu S, Nie Z, Seo M, Lewis P, Kumacheva E, Stone HA, Garstecki P, Weibel DB, Gitlin I, Whitesides GM. Angewandte Chemie. 2005;117:734–738. doi: 10.1002/anie.200462226. [DOI] [PubMed] [Google Scholar]
- 74.Nie Z, Li W, Seo M, Xu S, Kumacheva E. Journal of the American Chemical Society. 2006;128:9408–9412. doi: 10.1021/ja060882n. [DOI] [PubMed] [Google Scholar]
- 75.Nisisako T, Torii T. Advanced materials. 2007;19:1489–1493. [Google Scholar]
- 76.Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 77.Shah RK, Kim J-W, Agresti JJ, Weitz DA, Chu L-Y. Soft matter. 2008;4:2303–2309. [Google Scholar]
- 78.Kim C, Chung S, Kim YE, Lee KS, Lee SH, Oh KW, Kang JY. Lab on a chip. 2011;11:246–252. doi: 10.1039/c0lc00036a. [DOI] [PubMed] [Google Scholar]
- 79.Agarwal P, Zhao S, Bielecki P, Rao W, Choi JK, Zhao Y, Yu J, Zhang W, He X. Lab on a chip. 2013;13:4525–4533. doi: 10.1039/c3lc50678a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Agarwal P, Choi JK, Huang H, Zhao S, Dumbleton J, Li J, He X. Particle & particle systems characterization : measurement and description of particle properties and behavior in powders and other disperse systems. 2015;32:809–816. doi: 10.1002/ppsc.201500025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang H, He X. Lab on a chip. 2015;15:4197–4205. doi: 10.1039/c5lc00730e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang S, Guo F, Kiraly B, Mao X, Lu M, Leong KW, Huang TJ. Lab Chip. 2012;12:2097–2102. doi: 10.1039/c2lc90046g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cubaud T, Mason TG. Physics of Fluids (1994-present) 2008;20:053302. [Google Scholar]
- 84.Nabavi SA, Gu S, Vladisavljević GT, Ekanem EE. Journal of colloid and interface science. 2015;450:279–287. doi: 10.1016/j.jcis.2015.03.032. [DOI] [PubMed] [Google Scholar]
- 85.Zhou C, Yue P, Feng JJ. Physics of Fluids (1994-present) 2006;18:092105. [Google Scholar]
- 86.Hoang D, Portela L, Kleijn C, Kreutzer M, Van Steijn V. Journal of Fluid Mechanics. 2013;717:R4. [Google Scholar]
- 87.Utada AS, Fernandez-Nieves A, Stone HA, Weitz DA. Physical review letters. 2007;99:094502. doi: 10.1103/PhysRevLett.99.094502. [DOI] [PubMed] [Google Scholar]
- 88.Fu T, Wu Y, Ma Y, Li HZ. Chemical engineering science. 2012;84:207–217. [Google Scholar]
- 89.Samanipour R, Wang Z, Ahmadi A, Kim K. Journal of Applied Polymer Science. 2016:133. [Google Scholar]
- 90.Christopher GF, Noharuddin NN, Taylor JA, Anna SL. Physical review. E, Statistical, nonlinear, and soft matter physics. 2008;78:036317. doi: 10.1103/PhysRevE.78.036317. [DOI] [PubMed] [Google Scholar]
- 91.Utada A, Chu L-Y, Fernandez-Nieves A, Link D, Holtze C, Weitz D. Mrs Bulletin. 2007;32:702–708. [Google Scholar]
- 92.Nunes JK, Tsai SS, Wan J, Stone HA. Journal of physics D: Applied physics. 2013:46. doi: 10.1088/0022-3727/46/11/114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sauret A, Shum HC. International Journal of Nonlinear Sciences and Numerical Simulation. 2012;13:351–362. [Google Scholar]
- 94.Chaurasia AS, Sajjadi S. Chemical Engineering Science. 2015;129:260–270. [Google Scholar]
- 95.Yobas L, Martens S, Ong WL, Ranganathan N. Lab on a chip. 2006;6:1073–1079. doi: 10.1039/b602240e. [DOI] [PubMed] [Google Scholar]
- 96.Di Carlo D, Irimia D, Tompkins RG, Toner M. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang H, He X. Appl Phys Lett. 2014;105:143704. doi: 10.1063/1.4898040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moon S, Ceyhan E, Gurkan UA, Demirci U. PloS one. 2011;6:e21580. doi: 10.1371/journal.pone.0021580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bringer MR, Gerdts CJ, Song H, Tice JD, Ismagilov RF. Philosophical Transactions of the Royal Society of London A: Mathematical, Physical and Engineering Sciences. 2004;362:1087–1104. doi: 10.1098/rsta.2003.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Song H, Tice JD, Ismagilov RF. Angewandte Chemie. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 101.Tice JD, Song H, Lyon AD, Ismagilov RF. Langmuir. 2003;19:9127–9133. [Google Scholar]
- 102.Mũnoz Z, Shih H, Lin C-C. Biomaterials Science. 2014;2:1063–1072. doi: 10.1039/c4bm00070f. [DOI] [PubMed] [Google Scholar]
- 103.De Geest BG, Urbanski JP, Thorsen T, Demeester J, De Smedt SC. Langmuir. 2005;21:10275–10279. doi: 10.1021/la051527y. [DOI] [PubMed] [Google Scholar]
- 104.Xia B, Krutkramelis K, Oakey J. Biomacromolecules. 2016;17:2459–2465. doi: 10.1021/acs.biomac.6b00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma S, Natoli M, Liu X, Neubauer MP, Watt FM, Fery A, Huck WT. Journal of Materials Chemistry B. 2013;1:5128–5136. doi: 10.1039/c3tb20851f. [DOI] [PubMed] [Google Scholar]
- 106.Wang J, Cheng Y, Yu Y, Fu F, Chen Z, Zhao Y, Gu Z. ACS applied materials & interfaces. 2015;7:27035–27039. doi: 10.1021/acsami.5b10442. [DOI] [PubMed] [Google Scholar]
- 107.Tirella A, Liberto T, Ahluwalia A. Materials Letters. 2012;74:58–61. [Google Scholar]
- 108.Pfeifer GP, You YH, Besaratinia A. Mutation research. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 109.Pattison DI, Davies MJ. Exs. 2006:131–157. doi: 10.1007/3-7643-7378-4_6. [DOI] [PubMed] [Google Scholar]
- 110.Hoffman AS. Advanced drug delivery reviews. 2002;54:3–12. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 111.Choi C-H, Lee J-H, Shim H-W, Lee N-R, Jung J-H, Yoon T-H, Kim C-S, Lee D-P. 2006 [Google Scholar]
- 112.Wu F, Ju X-J, He X-H, Jiang M-Y, Wang W, Liu Z, Xie R, He B, Chu L-Y. Journal of Materials Chemistry B. 2016;4:2455–2465. doi: 10.1039/c6tb00209a. [DOI] [PubMed] [Google Scholar]
- 113.Quevedo E, Steinbacher J, McQuade DT. Journal of the American Chemical Society. 2005;127:10498–10499. doi: 10.1021/ja0529945. [DOI] [PubMed] [Google Scholar]
- 114.Zhang H, Tumarkin E, Peerani R, Nie Z, Sullan RM, Walker GC, Kumacheva E. Journal of the American Chemical Society. 2006;128:12205–12210. doi: 10.1021/ja0635682. [DOI] [PubMed] [Google Scholar]
- 115.Utech S, Prodanovic R, Mao AS, Ostafe R, Mooney DJ, Weitz DA. Advanced healthcare materials. 2015;4:1628–1633. doi: 10.1002/adhm.201500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hong S, Hsu HJ, Kaunas R, Kameoka J. Lab on a chip. 2012;12:3277–3280. doi: 10.1039/c2lc40558j. [DOI] [PubMed] [Google Scholar]
- 117.Sun Q, Deng Y. Langmuir. 2005;21:5812–5816. doi: 10.1021/la050403i. [DOI] [PubMed] [Google Scholar]
- 118.Huang SB, Chang YH, Lee HC, Tsai SW, Wu MH. Biomedical microdevices. 2014;16:345–354. doi: 10.1007/s10544-014-9837-5. [DOI] [PubMed] [Google Scholar]
- 119.Mazaheri H, Baghani M, Naghdabadi R. Journal of Intelligent Material Systems and Structures. 2016;27:324–336. [Google Scholar]
- 120.Gasperini L, Mano JF, Reis RL. Journal of the Royal Society, Interface. 2014;11:20140817. doi: 10.1098/rsif.2014.0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Olabisi RM. Journal of biomedical materials research. Part A. 2015;103:846–859. doi: 10.1002/jbm.a.35205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Headen DM, Aubry G, Lu H, Garcia AJ. Advanced materials. 2014;26:3003–3008. doi: 10.1002/adma.201304880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Collins DJ, Neild A, deMello A, Liu AQ, Ai Y. Lab on a chip. 2015;15:3439–3459. doi: 10.1039/c5lc00614g. [DOI] [PubMed] [Google Scholar]
- 124.Tan WH, Takeuchi S. Advanced materials. 2007;19:2696–2701. [Google Scholar]
- 125.Deng Y, Zhang N, Zhao L, Yu X, Ji X, Liu W, Guo S, Liu K, Zhao XZ. Lab on a chip. 2011;11:4117–4121. doi: 10.1039/c1lc20494g. [DOI] [PubMed] [Google Scholar]
- 126.Wen N, Zhao Z, Fan B, Chen D, Men D, Wang J, Chen J. Molecules. 2016:21. doi: 10.3390/molecules21070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sciambi A, Abate AR. Lab on a chip. 2015;15:47–51. doi: 10.1039/c4lc01194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sajeesh P, Sen AK. Microfluidics and nanofluidics. 2014;17:1–52. [Google Scholar]
- 129.Abate AR, Agresti JJ, Weitz DA. Applied Physics Letters. 2010;96:203509. [Google Scholar]
- 130.Baret JC, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD. Lab on a chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 131.Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hu H, Eustace D, Merten CA. Lab on a chip. 2015;15:3989–3993. doi: 10.1039/c5lc00686d. [DOI] [PubMed] [Google Scholar]
- 133.Schmid L, Weitz DA, Franke T. Lab on a chip. 2014;14:3710–3718. doi: 10.1039/c4lc00588k. [DOI] [PubMed] [Google Scholar]
- 134.Ding X, Li P, Lin SC, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F, Huang TJ. Lab on a chip. 2013;13:3626–3649. doi: 10.1039/c3lc50361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nam J, Lim H, Kim C, Kang JY, Shin S. Biomicrofluidics. 2012;6:024120. doi: 10.1063/1.4718719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Di Carlo D. Lab on a chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 137.Zhang J, Yan S, Yuan D, Alici G, Nguyen NT, Ebrahimi Warkiani M, Li W. Lab on a chip. 2016;16:10–34. doi: 10.1039/c5lc01159k. [DOI] [PubMed] [Google Scholar]
- 138.Di Carlo D, Edd JF, Humphry KJ, Stone HA, Toner M. Physical review letters. 2009;102:094503. doi: 10.1103/PhysRevLett.102.094503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gossett DR, Tse HT, Dudani JS, Goda K, Woods TA, Graves SW, Di Carlo D. Small. 2012;8:2757–2764. doi: 10.1002/smll.201200588. [DOI] [PubMed] [Google Scholar]
- 140.Sollier E, Amini H, Go DE, Sandoz PA, Owsley K, Di Carlo D. Microfluidics and Nanofluidics. 2015;19:53–65. [Google Scholar]
- 141.Nieuwstadt HA, Seda R, Li DS, Fowlkes JB, Bull JL. Biomedical microdevices. 2011;13:97–105. doi: 10.1007/s10544-010-9474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuntaegowdanahalli SS, Bhagat AA, Kumar G, Papautsky I. Lab on a chip. 2009;9:2973–2980. doi: 10.1039/b908271a. [DOI] [PubMed] [Google Scholar]
- 143.Morijiri T, Sunahiro S, Senaha M, Yamada M, Seki M. Microfluidics and nanofluidics. 2011;11:105–110. [Google Scholar]
- 144.Huh D, Bahng JH, Ling Y, Wei HH, Kripfgans OD, Fowlkes JB, Grotberg JB, Takayama S. Analytical chemistry. 2007;79:1369–1376. doi: 10.1021/ac061542n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McGrath J, Jimenez M, Bridle H. Lab on a chip. 2014;14:4139–4158. doi: 10.1039/c4lc00939h. [DOI] [PubMed] [Google Scholar]
- 146.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 147.Jing T, Ramji R, Warkiani ME, Han J, Lim CT, Chen CH. Biosensors & bioelectronics. 2015;66:19–23. doi: 10.1016/j.bios.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 148.Akbari S, Pirbodaghi T. Microfluidics and nanofluidics. 2014;16:773–777. [Google Scholar]
- 149.Wong EH, Rondeau E, Schuetz P, Cooper-White J. Lab on a chip. 2009;9:2582–2590. doi: 10.1039/b903774h. [DOI] [PubMed] [Google Scholar]
- 150.Zhang H, Tumarkin E, Sullan RMA, Walker GC, Kumacheva E. Macromolecular rapid communications. 2007;28:527–538. [Google Scholar]
- 151.Choi C-H, Wang H, Lee H, Kim JH, Zhang L, Mao A, Mooney DJ, Weitz DA. Lab on a chip. 2016;16:1549–1555. doi: 10.1039/c6lc00261g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang H, Sun M, Heisler-Taylor T, Kiourti A, Volakis J, Lafyatis G, He X. Small. 2015;11:5369–5374. doi: 10.1002/smll.201501388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Capretto L, Mazzitelli S, Luca G, Nastruzzi C. Acta biomaterialia. 2010;6:429–435. doi: 10.1016/j.actbio.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 154.Wieduwild R, Krishnan S, Chwalek K, Boden A, Nowak M, Drechsel D, Werner C, Zhang Y. Angewandte Chemie International Edition. 2015;54:3962–3966. doi: 10.1002/anie.201411400. [DOI] [PubMed] [Google Scholar]
- 155.Mendes AC, Baran ET, Lisboa P, Reis RL, Azevedo HS. Biomacromolecules. 2012;13:4039–4048. doi: 10.1021/bm301332z. [DOI] [PubMed] [Google Scholar]
- 156.Mayfield AE, Tilokee EL, Latham N, McNeill B, Lam B-K, Ruel M, Suuronen EJ, Courtman DW, Stewart DJ, Davis DR. Biomaterials. 2014;35:133–142. doi: 10.1016/j.biomaterials.2013.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chan HF, Zhang Y, Ho YP, Chiu YL, Jung Y, Leong KW. Scientific reports. 2013;3:3462. doi: 10.1038/srep03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wilson JL, McDevitt TC. Biotechnology and bioengineering. 2013;110:667–682. doi: 10.1002/bit.24802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barminko J, Kim JH, Otsuka S, Gray A, Schloss R, Grumet M, Yarmush ML. Biotechnology and bioengineering. 2011;108:2747–2758. doi: 10.1002/bit.23233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ishihara K, Xu Y, Konno T. Polymers in Nanomedicine. Springer; 2011. pp. 141–165. [Google Scholar]
- 161.Ravi M, Paramesh V, Kaviya SR, Anuradha E, Solomon FD. Journal of cellular physiology. 2015;230:16–26. doi: 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- 162.Lu Y-C, Song W, An D, Kim BJ, Schwartz R, Wu M, Ma M. Journal of Materials Chemistry B. 2015;3:353–360. doi: 10.1039/c4tb01735h. [DOI] [PubMed] [Google Scholar]
- 163.Rao W, Zhao S, Yu J, Lu X, Zynger DL, He X. Biomaterials. 2014;35:7762–7773. doi: 10.1016/j.biomaterials.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim C, Kang JY. BioChip Journal. 2014;8:199–208. [Google Scholar]
- 165.Bhujbal SV, de Haan B, Niclou SP, de Vos P. Scientific reports. 2014;4:6856. doi: 10.1038/srep06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ma M, Chiu A, Sahay G, Doloff JC, Dholakia N, Thakrar R, Cohen J, Vegas A, Chen D, Bratlie KM, Dang T, York RL, Hollister-Lock J, Weir GC, Anderson DG. Advanced healthcare materials. 2013;2:667–672. doi: 10.1002/adhm.201200341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Allazetta S, Kolb L, Zerbib S, Bardy J, Lutolf MP. Small. 2015;11:5647–5656. doi: 10.1002/smll.201501001. [DOI] [PubMed] [Google Scholar]
- 168.Dumbleton J, Agarwal P, Huang H, Hogrebe N, Han R, Gooch KJ, He X. Cellular and Molecular Bioengineering. 2016:1–12. doi: 10.1007/s12195-016-0428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shi X, Ostrovidov S, Zhao Y, Liang X, Kasuya M, Kurihara K, Nakajima K, Bae H, Wu H, Khademhosseini A. Advanced Functional Materials. 2015;25:2250–2259. [Google Scholar]
- 170.Lee HJ, Park YH, Koh WG. Advanced Functional Materials. 2013;23:591–597. [Google Scholar]
- 171.Chien HW, Tsai WB, Jiang S. Biomaterials. 2012;33:5706–5712. doi: 10.1016/j.biomaterials.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 172.Wang Y, Wang J. The Analyst. 2014;139:2449–2458. doi: 10.1039/c4an00015c. [DOI] [PubMed] [Google Scholar]
- 173.Alessandri K, Sarangi BR, Gurchenkov VV, Sinha B, Kießling TR, Fetler L, Rico F, Scheuring S, Lamaze C, Simon A. Proceedings of the National Academy of Sciences. 2013;110:14843–14848. doi: 10.1073/pnas.1309482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kwapiszewska K, Michalczuk A, Rybka M, Kwapiszewski R, Brzozka Z. Lab on a chip. 2014;14:2096–2104. doi: 10.1039/c4lc00291a. [DOI] [PubMed] [Google Scholar]
- 175.Yu L, Ni C, Grist SM, Bayly C, Cheung KC. Biomedical microdevices. 2015;17:33. doi: 10.1007/s10544-014-9918-5. [DOI] [PubMed] [Google Scholar]
- 176.Kumachev A, Greener J, Tumarkin E, Eiser E, Zandstra PW, Kumacheva E. Biomaterials. 2011;32:1477–1483. doi: 10.1016/j.biomaterials.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 177.Tsou Y-H, Khoneisser J, Huang P-C, Xu X. Bioactive Materials. 2016 doi: 10.1016/j.bioactmat.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Alessandri K, Feyeux M, Gurchenkov B, Delgado C, Trushko A, Krause KH, Vignjevic D, Nassoy P, Roux A. Lab on a chip. 2016;16:1593–1604. doi: 10.1039/c6lc00133e. [DOI] [PubMed] [Google Scholar]
- 179.Ma Y, Neubauer MP, Thiele J, Fery A, Huck W. Biomaterials Science. 2014;2:1661–1671. doi: 10.1039/c4bm00104d. [DOI] [PubMed] [Google Scholar]
- 180.Perez RA, Kim M, Kim TH, Kim JH, Lee JH, Park JH, Knowles JC, Kim HW. Tissue engineering. Part A. 2014;20:103–114. doi: 10.1089/ten.tea.2013.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Choi DH, Park CH, Kim IH, Chun HJ, Park K, Han DK. Journal of Controlled Release. 2010;147:193–201. doi: 10.1016/j.jconrel.2010.07.103. [DOI] [PubMed] [Google Scholar]
- 182.Tang M, Chen W, Weir MD, Thein-Han W, Xu HH. Acta biomaterialia. 2012;8:3436–3445. doi: 10.1016/j.actbio.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Coates EE, Riggin CN, Fisher JP. Journal of biomedical materials research. Part A. 2013;101:1962–1970. doi: 10.1002/jbm.a.34499. [DOI] [PubMed] [Google Scholar]
- 184.Liu J, Xu HH, Zhou H, Weir MD, Chen Q, Trotman CA. Acta biomaterialia. 2013;9:4688–4697. doi: 10.1016/j.actbio.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Maguire T, Novik E, Schloss R, Yarmush M. Biotechnology and bioengineering. 2006;93:581–591. doi: 10.1002/bit.20748. [DOI] [PubMed] [Google Scholar]
- 186.Richardson T, Kumta PN, Banerjee I. Tissue engineering. Part A. 2014;20:3198–3211. doi: 10.1089/ten.tea.2013.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhao S, Agarwal P, Rao W, Huang H, Zhang R, Liu Z, Yu J, Weisleder N, Zhang W, He X. Integrative biology : quantitative biosciences from nano to macro. 2014;6:874–884. doi: 10.1039/c4ib00100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chen Y, Dai X, Huang L, Sun Y, Chan HN, Shen B, Zeng X, Wu Z, Hsing I, Guo Z. Advanced Functional Materials. 2015;25:6189–6198. [Google Scholar]
- 189.Yao R, Zhang R, Luan J, Lin F. Biofabrication. 2012;4:025007. doi: 10.1088/1758-5082/4/2/025007. [DOI] [PubMed] [Google Scholar]
- 190.Siltanen C, Yaghoobi M, Haque A, You J, Lowen J, Soleimani M, Revzin A. Acta biomaterialia. 2016;34:125–132. doi: 10.1016/j.actbio.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ding S, Li L, Liu X, Yang G, Zhou G, Zhou S. Colloids and surfaces. B, Biointerfaces. 2015;133:286–295. doi: 10.1016/j.colsurfb.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 192.Khademhosseini A, Langer R. Biomaterials. 2007;28:5087–5092. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 193.Drury JL, Mooney DJ. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 194.Woodruff TK, Shea LD. Journal of assisted reproduction and genetics. 2011;28:3–6. doi: 10.1007/s10815-010-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Choi JK, Agarwal P, Huang H, Zhao S, He X. Biomaterials. 2014;35:5122–5128. doi: 10.1016/j.biomaterials.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Woodruff TK. Nature medicine. 2008;14:1190–1191. doi: 10.1038/nm1108-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jeruss JS, Woodruff TK. The New England journal of medicine. 2009;360:902–911. doi: 10.1056/NEJMra0801454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dolega ME, Abeille F, Picollet-D’hahan N, Gidrol X. Biomaterials. 2015;52:347–357. doi: 10.1016/j.biomaterials.2015.02.042. [DOI] [PubMed] [Google Scholar]
- 199.Mailleux AA, Overholtzer M, Brugge JS. Cell Cycle. 2008;7:57–62. doi: 10.4161/cc.7.1.5150. [DOI] [PubMed] [Google Scholar]
- 200.Zegers MM, O’Brien LE, Yu W, Datta A, Mostov KE. Trends in cell biology. 2003;13:169–176. doi: 10.1016/s0962-8924(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 201.Eydelnant IA, Betty Li B, Wheeler AR. Nature communications. 2014;5:3355. doi: 10.1038/ncomms4355. [DOI] [PubMed] [Google Scholar]
- 202.Li R, Albertini DF. Nature reviews. Molecular cell biology. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- 203.Chwalek K, Tsurkan MV, Freudenberg U, Werner C. Scientific reports. 2014;4:4414. doi: 10.1038/srep04414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Saini H, Navaei A, Van Putten A, Nikkhah M. Advanced healthcare materials. 2015;4:1961–1971. doi: 10.1002/adhm.201500331. [DOI] [PubMed] [Google Scholar]
- 205.Duguay D, Foty RA, Steinberg MS. Developmental biology. 2003;253:309–323. doi: 10.1016/s0012-1606(02)00016-7. [DOI] [PubMed] [Google Scholar]
- 206.Foty RA, Steinberg MS. The International journal of developmental biology. 2004;48:397–409. doi: 10.1387/ijdb.041810rf. [DOI] [PubMed] [Google Scholar]
- 207.Ferreira DS, Reis RL, Azevedo HS. Soft matter. 2013;9:9237–9248. [Google Scholar]
- 208.Li CY, Wood DK, Hsu CM, Bhatia SN. Lab on a chip. 2011;11:2967–2975. doi: 10.1039/c1lc20318e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liu Z, Shum HC. Biomicrofluidics. 2013;7:44117. doi: 10.1063/1.4817769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Goers L, Freemont P, Polizzi KM. Journal of the Royal Society, Interface. 2014:11. doi: 10.1098/rsif.2014.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nikkhah M, Strobl JS, Schmelz EM, Roberts PC, Zhou H, Agah M. Biomaterials. 2011;32:7625–7632. doi: 10.1016/j.biomaterials.2011.06.041. [DOI] [PubMed] [Google Scholar]
- 212.Li R, Lv X, Zhang X, Saeed O, Deng Y. Frontiers of Chemical Science and Engineering. 2016;10:90–98. [Google Scholar]
- 213.Usta OB, McCarty WJ, Bale S, Hegde M, Jindal R, Bhushan A, Golberg I, Yarmush ML. Technology (Singap World Sci) 2015;3:1–26. doi: 10.1142/S2339547815300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chan HF, Zhang Y, Leong KW. Small. 2016;12:2720–2730. doi: 10.1002/smll.201502932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen Q, Utech S, Chen D, Prodanovic R, Lin JM, Weitz DA. Lab on a chip. 2016;16:1346–1349. doi: 10.1039/c6lc00231e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tumarkin E, Tzadu L, Csaszar E, Seo M, Zhang H, Lee A, Peerani R, Purpura K, Zandstra PW, Kumacheva E. Integrative biology : quantitative biosciences from nano to macro. 2011;3:653–662. doi: 10.1039/c1ib00002k. [DOI] [PubMed] [Google Scholar]
- 217.Sudo R, Chung S, Zervantonakis IK, Vickerman V, Toshimitsu Y, Griffith LG, Kamm RD. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:2155–2164. doi: 10.1096/fj.08-122820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Jeong H-H, Issadore D, Lee D. Korean Journal of Chemical Engineering. 2016:1–10. [Google Scholar]
- 219.Erro E, Bundy J, Massie I, Chalmers SA, Gautier A, Gerontas S, Hoare M, Sharratt P, Choudhury S, Lubowiecki M, Llewellyn I, Legallais C, Fuller B, Hodgson H, Selden C. BioResearch open access. 2013;2:1–11. doi: 10.1089/biores.2012.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]