Abstract
Hereditary juvenile megaloblastic anemia due to vitamin B12 (cobalamin) deficiency is caused by intestinal malabsorption of cobalamin. In Imerslund–Gräsbeck syndrome (IGS), cobalamin absorption is completely abolished and not corrected by the administration of intrinsic factor (IF); if untreated, the disease is fatal. Biallelic mutations either in the cubilin (CUBN) or amnionless (AMN) gene cause IGS. In a series of families clinically diagnosed with likely IGS, at least six displayed no evidence of mutations in CUBN or AMN. A genome-wide search for linkage followed by mutational analysis of candidate genes was performed in five of these families. A region in chromosome 11 showed evidence of linkage in four families. The gastric IF (GIF) gene located in this region harbored homozygous nonsense and missense mutations in these four families and in three additional families. The disease in these cases therefore should be classified as hereditary IF deficiency. Clinically, these patients resembled those with typical IGS; radiocobalamin absorption tests had been inconclusive regarding the nature of the defect. In the diagnosis of juvenile cobalamin deficiency, mutational analysis of the CUBN, AMN, and GIF genes provides a molecular characterization of the underlying defect and may be the diagnostic method of choice.
Keywords: gastric intrinsic factor, Imerslund–Gräsbeck syndrome, intrinsic factor deficiency, vitamin B12 malabsorption, megaloblastic anemia
Both in adults and children, malabsorption of cobalamin (vitamin B12) is the most common cause of cobalamin deficiency, which usually manifests itself as megaloblastic anemia. The malabsorption may be general, such as in tropical sprue or celiac disease. In some cases, the malabsorption of cobalamin is relatively or very specific, as in genuine pernicious anemia, fish tapeworm infection, and in some instances of abnormal intestinal microbial flora associated with blind loops or diverticulosis (1). As in other nutritional deficiencies, a combination of causes, including hereditary and acquired conditions, are common (2, 3). Pediatric cases in developed countries appear mostly to be hereditary in nature. The disease entity or syndrome-selective vitamin B12 malabsorption with proteinuria was described independently by Imerslund (4) and Gräsbeck et al. (5) in 1960. The Imerslund–Gräsbeck syndrome (IGS) is characterized by recessive inheritance, relatively early onset, failure to thrive, infections, low serum cobalamin, mild and innocuous proteinuria, and variable neurological symptoms such as peripheral neuropathy, cognitive problems, and dementia (3). The diagnosis is reached by eliminating other causes of impaired cobalamin absorption and especially by measuring the absorption of radioactive cobalamin. Poor absorption of radiocobalamin given alone is first observed. Subsequently, the test is repeated by simultaneous administration of gastric intrinsic factor (IF), which in contrast to the situation in pernicious anemia (IF deficiency; IFD) fails to increase the absorption (“malabsorption response”). The Schilling test technique has been the most popular method of measuring the absorption of radioactive cobalamin. The absorbed radioactivity is measured indirectly after it has been flushed into the urine by intramuscular injection of a large quantity of nonradioactive cobalamin (6).
IGS has been thought to be rare, mostly occurring in Finland and Norway, because of genetic founder effects of rare mutations (7). It was recently shown that in Finland, all cases studied so far were due to mutations in the cubilin (CUBN) gene, and in Norway, all cases were due to mutations in the amnionless (AMN) gene (8, 9). Both proteins are involved in intestinal absorption and renal tubular reabsorption. These proteins form the heterodimer called cubam that accomplishes the internalization of the IF–cobalamin complex (10). Thus, the molecular mechanism of IGS has begun to be explained.
The gastric IF (GIF) gene had long been postulated to be implicated in classical IFD (11–15), and the defect was expected “to reflect the usual spectrum of nonsense and missense changes” (16). In 2004, the first and hitherto sole GIF mutation was described (17). The female IFD patient was diagnosed with a radiocobalamin absorption test that showed correction of absorption by coadministration of IF.
We previously demonstrated that patients and families with IGS of various ethnicities, mainly Mediterranean, had mutations in either CUBN or AMN; however, in a few families/patients, no mutations were found in these genes (9). To determine the molecular basis of the disease in these patients, we undertook the genetic studies reported in this work. We show that homozygous germ-line mutations in GIF, the gene encoding IF, cause a phenotype that for practical purposes had not been distinguishable from IGS. Although a properly performed radiocobalamin absorption test should have distinguished these patients from traditional IGS, in practice this distinction was difficult, mainly because the radiocobalamin absorption test is decisive only when cobalamin treatment has fully eliminated enterocyte malfunction and general malabsorption caused by the deficiency state (18). Unfortunately, the radiocobalamin tests are no longer available in many countries (3). We propose that patients with suspected hereditary juvenile cobalamin deficiency should be routinely diagnosed and characterized by their mutational status.
Materials and Methods
Patients. Turkish families AT, HT, and IT and Kuwaiti families 1 and 2 were described in refs. 9, 19, and 20. Newly diagnosed family LT is of Turkish origin. Family 8, now living in the United States, is from Guinea–Bissau, and no consanguinity was reported. French family 1 was of Caucasian origin and reportedly nonconsanguineous (Fig. 1). We obtained DNA samples extracted from venous blood after informed consent according to the Declaration of Helsinki. The study was covered by institutional review board approval.
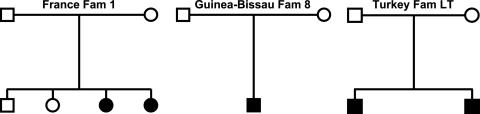
Fig. 1.
Pedigrees of three unrelated recently diagnosed putative IGS families with juvenile cobalamin deficiency (filled symbols). Molecular and genetic analysis excluded the CUBN and AMN genes as the cause of the disease. DNA sequence analysis identified mutations in the GIF gene that were homozygous in the affected children and heterozygous in the parents. Based on this finding, the patients' phenotypes could be reclassified as hereditary IFD. Open symbols signify unaffected individuals.
Linkage Analysis. A genome-wide search for linkage was performed at the Finnish Genome Center (Helsinki) by using blood-derived DNA samples from the 27 members of families AT, HT, IT, and Kuwait 1 and 2 (for pedigrees, see ref. 9). Data obtained from the 400-marker panel ABI Prism Linkage Mapping Set (Version 2.0, Applied Biosystems) were analyzed by using the programs linkage (http://linkage.rockefeller.edu/soft/linkage) and genehunter (www.fhcrc.org/labs/kruglyak/Downloads). We pursued the region on chromosome 11 near marker D11S4191 showing evidence of linkage because of its proximity to the GIF and TCN1 genes that are both located ≈400 kb centromeric to D11S4191.
Mutation Analysis. We amplified individual GIF and TCN1 exons from genomic DNA (GenBank database accession no. AP002347.3) by PCR and analyzed them by single-strand conformation polymorphism (21) and DNA sequencing. Sequencing was performed on ABI Prism 3730 DNA analyzers (Applied Biosystems). To study the common GIF c.68A>G; Q23R polymorphism in exon 1, we used a HpaII restriction enzyme assay to determine its frequency among 176 normal controls (40 grandparents of Centre d'Etude du Polymorphisme Humain families and 95 European and 41 Saudi volunteers). PCR primer sequences are available on request.
Results
To elucidate the genetic basis of a cohort of IGS patients in which involvement of AMN and CUBN had been excluded (9), we used samples from five families to perform a genome-wide search for linkage. As a result, we established linkage to the distal part of chromosome 11q12. Marker D11S4191 showed a nonparametric logarithm of odds score of 3.65. Inspection of the region revealed two potential candidate genes, i.e., GIF that encodes IF and TCN1 that encodes transcobalamin 1. These genes are located next to each other ≈400 kb centromeric to marker D11S4191. IF is a 417-aa protein secreted by gastric parietal cells that binds cobalamin in the proximal small intestine before binding to the cubam receptor for internalization in the distal third of the small bowel (16). Thus, it is involved as a cofactor in the intestinal uptake of cobalamin; however, inherited IFD should be distinguished easily from IGS by using the radiocobalamin absorption test with IF that corrects low cobalamin absorption (14). Transcobalamin 1 (haptocorrin) is not associated with metabolic cobalamin deficiency because this protein carries the major fraction of food-released cobalamin first into the stomach and later on binds it in the serum and does not deliver it to tissues (3, 16). Although neither gene was functionally a perfect candidate for the IGS phenotype, we proceeded to search both genes for mutations by single-strand conformation polymorphism and DNA sequencing.
Potentially inactivating mutations in the GIF gene were detected in four of the five families studied by linkage. In Turkish family AT, we found a 1-bp insertion in exon 8 (c.1175_1176insT) that led to a frameshift at amino acid residue 393 (T393fs). In family IT, we found a missense change in exon 2, c.137C>T, replacing the conserved serine residue 46 with leucine (S46L). Both Kuwaiti families shared the same acceptor splice site mutation in intron 1, c.80–1G>A, that destroyed the consensus sequence, thereby affecting the proper expression of the GIF mRNA. Turkish family HT did not show any sequence change in GIF using exon-by-exon analysis.
The screening of three more IGS families (Fig. 1) that had no AMN or CUBN mutations resulted in detecting a 4-bp deletion (183_186delGAAT) in exon 2 of GIF leading to a frameshift at methionine 61 (M61fs) in one African sibship (Fam 8). In French family 1, we identified another intron-1 splice mutation, this time affecting the consensus donor splice sequence (c.79+1G>A). Finally, in Turkish family LT we found a 1-bp deletion in exon 2 (c.161delA) that resulted in a frameshift at amino acid residue 54 (N54fs). All patients in these seven families were homozygous, whereas their respective parents were heterozygous for these changes. All healthy sibs were either heterozygous carriers or homozygous wild type, in conformation with their phenotypes. We further identified some GIF polymorphisms that are summarized in Table 1. Except for a common polymorphism (c.846C>T; S282S), no mutations were found in the TCN1 gene in our patient cohort, and it was not further studied.
Table 1. Mutations and polymorphisms in the GIF gene.
| Location | DNA change* | Predicted consequence† | Classification |
|---|---|---|---|
| Exon 1 | c.68A>G | Q23R | Polymorphism |
| Intron 1 | c.79+1G>A | Donor splice site | LOF mutation |
| Intron 1 | c.80-1G>A | Acceptor splice site | LOF mutation |
| Exon 2 | c.137C>T | S46L | Missense mutation |
| Exon 2 | c.161delA | N54fs | LOF mutation |
| Exon 2 | c.183_186delGAAT | M61fs | LOF mutation |
| Exon 2 | c.246C>T | S82S | Polymorphism |
| Exon 6 | c.764A>G | N255S | Polymorphism |
| Exon 7 | c.990C>T | N330N | Polymorphism |
| Exon 8 | c.1175_1176insT | T393fs | LOF mutation |
| Intron 8 | c.1192+24G>A | None | Polymorphism |
LOF, loss of function.
Numbering relative to adenine in the ATG start codon of GIF (GenBank database accession no. AP002347.3).
Numbering relative to the first methionine deduced from the cDNA sequence.
Discussion
In a cohort of seven diagnosed IGS families, we detected homozygous GIF mutations that predicted a complete loss of function in six families and possibly only reduced function in one sibship (Fam IT). Does this result mean that IFD [Online Mendelian Inheritance in Man genetics database (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM) accession no. 261000; juvenile or congenital pernicious anemia] and IGS (Online Mendelian Inheritance in Man accession no. 261100; selective malabsorption of vitamin B12; also known as juvenile or hereditary megaloblastic anemia, MGA1, juvenile pernicious anemia with proteinuria, or enterocyte IF receptor deficiency) are the same entity? Genetically and biochemically, the answer is clearly no. We deliberately listed so many synonyms above because they illustrate that most clinical manifestations leading to these descriptions overlap considerably. Differential diagnosis of IGS and IFD can only be accomplished by the radiocobalamin absorption test. Nevertheless, the relatively high cost and difficult logistics of measuring radiocobalamin absorption have led to a sharp decrease in its use and even its availability. Other diagnostic means, such as IF binding assays in gastric juice, are even less accessible (3). Because cobalamin deficiency of most etiologies can be corrected easily by parenteral injection of vitamin B12, in clinical practice it is tempting to proceed to therapy without a precise diagnosis. However, based on our results, straightforward and accurate noninvasive testing by genetic means is available to differentiate these disorders at the molecular level. This finding provides important information for the clinical management because proper diagnosis may modify the onset and form of cobalamin therapy (3). Larger series of patients with juvenile cobalamin deficiency may need to be studied before one can assess the sensitivity of mutation status to distinguish between inherited and acquired forms of the disease. Our unpublished observations (S.M.T. and A.d.l.C., unpublished data) so far suggest that the sensitivity is high: in a total of 55 families or sporadic patients referred to us for mutation analysis based on suspected hereditary cobalamin deficiency, only in six have we not detected a mutation. We interpret this finding to suggest that when mutation analysis is negative, acquired forms of cobalamin deficiency should be seriously considered.
Assessment of the available clinical and laboratory data provided in Table 2 shows great variation in, for instance, age at diagnosis (range 1–17 years), occurrence of proteinuria, and serum cobalamin concentration (range 30–411 pg/ml). Overall, particularly in the familial cases, the data are compatible with both typical IGS and typical IFD. A distinction between the two diagnoses probably could have been made by a radiocobalamin absorption test; however, in some cases at the time of diagnosis this test was not available for practical or economic reasons and, occasionally, because of parental refusal. Moreover, under difficult conditions of health care, even those radiocobalamin absorption tests that could be performed were not fully informative because apparently some general malabsorption still occurred at the time of the test. Thus, retrospectively, all of the patients in whom we found biallelic mutations in GIF could be classified as cases of IFD rather than IGS. One main reason for their initial classification as IGS probably is the fact that IGS is believed to be far more common, particularly in the Mediterranean region. Awareness of IGS is high in this area (9). After the publication of the articles implicating CUBN (22) and AMN (8, 9) many requests for testing from around the world emerged. It is likely that both IGS and IFD will turn out to be more frequent than previously thought, notably in populations with a high incidence of consanguineous marriages. Although in some families no consanguinity was reported, all 17 patients in all 7 families turned out to be homozygous for the detected GIF mutations.
Table 2. Clinical and laboratory data in hereditary IFD with mutations in GIF.
| Case | Sex | Age at diagnosis, years | Schilling test | Proteinuria, mg/liter | Serum cobalamin, pg/ml | Serum folate, ng/ml | Hemoglobin, g/dl | Mean cellular volume, fl | Reticulocyte count, % |
|---|---|---|---|---|---|---|---|---|---|
| Kuwait 1 | |||||||||
| Case 1 | M | 1.5 | Mal, NC by IF | 50 | 247 | 2.8 | 6 | 97.9 | NA |
| Case 2 | M | 15 | Mal, NC by IF | None | 31 | 5 | 7.9 | 123.7 | NA |
| Case 3 | M | 17 | Mal, NC by IF | None | 245 | 10.5 | 4.2 | 101 | NA |
| Case 4 | M | 5 | Mal, NC by IF | 170 | 94 | 6.8 | 5.4 | 104 | NA |
| Kuwait 2 | |||||||||
| Case 5 | M | 2.5 | Mal, NC by IF | 800 | 112 | 7.4 | 5.5 | 109 | NA |
| Case 6 | F | 4.6 | Mal, NC by IF | 200 | 117 | 13.6 | 6.5 | 97.4 | NA |
| Case 7 | M | 10 | Mal, NC by IF | 80 | 411 | 5.6 | 6.9 | 109.6 | NA |
| Fam 8 | |||||||||
| 8-01 | M | 15 | 58Co, 0.92%; 57Co, 3.57% | 117 | 100 | >20.7 | 5.9 | 103.6 | 1 |
| Turk AT | |||||||||
| A001 | F | 1 | NA | Yes | 96 | 4 | 6 | 94 | 1.1 |
| Turk IT | |||||||||
| I028 | F | 10 | 58Co, 2%; 57Co, 7% | Yes | 30 | 11 | 4 | 93 | 0.4 |
| I030 | M | NA | NA | NA | NA | NA | NA | NA | NA |
| I031 | F | NA | NA | NA | NA | NA | NA | NA | NA |
| I032 | F | 1.5 | NA | NA | NA | NA | NA | NA | NA |
| Turk LT | |||||||||
| L043 | M | 2 | 58Co, 1%; 57Co, 1.3% | None | 96 | 9 | 4 | 96 | 0.4 |
| France 1 | |||||||||
| EN95 | F | 6 | NA | 70 (none) | 75 | 13 | 6.5 | 100 | 8.1 |
| PN99 | F | 1.5 | NA | 180 | 75 | 12 | 10 | 83 | 1.77 |
NA, not available; M, male, F, female; Mal, malabsorption; NC, not corrected.
It is interesting to note that the detected 4-bp deletion (c.183_186delGAAT) in family 8 was the same mutation as that described by Yassin et al. (17). The ethnicity of their patient was reported to be African-American, but homozygosity vs. hemizygosity could not be established because paternal DNA was unavailable. Our discovery of the same mutation in a second family of African ancestry supports the hypothesis that the mutation might be common in some African populations through a founder effect. Haplotyping in both families could solve this question.
All mutations but one (S46L) predicted premature termination of protein translation with corresponding effects on protein expression or stability, loss of important functional domains, or nonsense-mediated mRNA decay (23). Recessive inheritance of a distinct phenotype and loss-of-function mutations are usually seen in concert, and IFD/IGS is no exception. It is interesting to note that only one missense mutation was detected. Although this result simply could be because we studied only few patients so far, it is not inconceivable that many missense changes in IF lead to only partial loss of IF binding affinity to cobalamin or the cubam receptor complex. We hypothesize that such residual activity might be sufficient to prevent early onset disease but might become apparent as adult-onset pernicious anemia as a result of age-related or autoimmune atrophic changes in the gastric or intestinal mucosa.
In a recent work by Gordon et al. (24), a genetic polymorphism in GIF was reportedly associated with IFD. The polymorphism they designated g.68A>G, Q5R is in fact identical to the c.68A>G, Q23R change that we found in exon 1 (for details on mutation nomenclature, see ref. 25). While studying GIF, we analyzed a set of 176 unrelated adult controls for exon 1 and observed 140 homozygotes for the c.68A allele, 6 homozygotes for the c.68G allele, and 30 heterozygotes. This analysis resulted in an observed allele frequency of 11.9% for the c.68G allele compared with 6.7% among 30 German newborns and 3.8% among 131 Spanish newborns as reported in ref. 24. Although the observed differences in frequency are likely just variations between populations, our results suggest that homozygosity for the c.68G polymorphism has no clinical significance in IFD. With an overall c.68G allele frequency of >10%, we would expect to see many more cases of IFD than those reported in the literature. However, we cannot totally exclude that the c.68G polymorphism might contribute to adult-onset pernicious anemia.
We were surprised to find that the “third” locus for IGS was indeed GIF, a gene postulated to cause IFD when mutated. Knowing that only a test involving ingested radioactivity in the patient can presently resolve the diagnosis between IGS and IFD, we conclude that in cases of juvenile cobalamin deficiency, genetic testing is warranted that should include the CUBN and AMN genes for IGS and the GIF gene for IFD. Prospective analysis of clinical and laboratory information eventually might permit a differential diagnosis when patients are correctly categorized. In addition, sequence changes in the GIF gene might contribute to late-onset cobalamin deficiency. This question remains to be studied in detail.
Acknowledgments
We thank Paivi Lahermo of the Finnish Genome Center for performing the genome-wide linkage analysis and Gérard Couillault, Hisham Abdel-Azim, and Candace Cressman Peterson for important clinical and laboratory details. This work was supported by the Liv och Hälsa Foundation (Finland), the Magnus Ehrnrooth Foundation (Finland), and the National Cancer Institute (United States).
S.M.T. and A.d.l.C. designed, coordinated, and supervised the study, performed research, interpreted the results, and codrafted the report; Z.L., J.D.P., and Z.Y. performed DNA isolations and linkage and mutational studies; C.O., M.C., C.A., K.L.D., L.F., and E.A.I. contributed samples and clinical laboratory results; R.G. reviewed clinical data and interpretation; and S.M.T., Z.L., J.D.P., C.O., M.C., C.A., Z.Y., K.L.D., L.F., E.A.I., R.G., and A.d.l.C. participated in discussing and writing up the final research work.
Abbreviations: IGS, Imerslund-Gräsbeck syndrome; IF, intrinsic factor; IFD, IF deficiency.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AP002347.3).
References
- 1.Chanarin, I. (1969) The Megaloblastic Anaemias (Blackwell Scientific, Oxford).
- 2.Gräsbeck, R. & Salonen, E. M. (1976) Prog. Food Nutr. Sci. 2, 193–231. [PubMed] [Google Scholar]
- 3.Carmel, R., Green, R., Rosenblatt, D. S. & Watkins, D. (2003) Hematology (Am. Soc. Hematol. Education Program Book), 62–81. [DOI] [PubMed]
- 4.Imerslund, O. (1960) Acta Paediatr. Scand. 49, Suppl. 119, 1–115. [PubMed] [Google Scholar]
- 5.Gräsbeck, R., Gordin, R, Kantero, I & Kuhlbäck, B. (1960) Acta Med. Scand. 167, 289–296. [DOI] [PubMed] [Google Scholar]
- 6.Schilling, R. F. (1953) J. Lab. Clin. Med. 42, 860–866. [PubMed] [Google Scholar]
- 7.Norio, R. (2003) Hum. Genet. 112, 470–526. [DOI] [PubMed] [Google Scholar]
- 8.Tanner, S. M., Aminoff, M., Wright, F. A., Liyanarachchi, S., Kuronen, M., Saarinen, A., Massika, O., Mandel, H., Broch, H. & de la Chapelle, A. (2003) Nat. Genet. 33, 426–429. [DOI] [PubMed] [Google Scholar]
- 9.Tanner, S. M., Li, Z., Bisson, R., Acar, C., Oner, C., Oner, R., Cetin, M., Abdelaal, M. A., Ismail, E. A., Lissens, W., et al. (2004) Hum. Mutat. 23, 327–333. [DOI] [PubMed] [Google Scholar]
- 10.Fyfe, J. C., Madsen, M., Hojrup, P., Christensen, E. I., Tanner, S. M., de la Chapelle, A., He, Q. & Moestrup, S. K. (2004) Blood 103, 1573–1579. [DOI] [PubMed] [Google Scholar]
- 11.Katz, M., Lee, S. K. & Cooper, B. A. (1972) N. Engl. J. Med. 287, 425–429. [DOI] [PubMed] [Google Scholar]
- 12.Katz, M., Mehlman, C. S. & Allen, R. H. (1974) J. Clin. Invest. 53, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmel, R. (1983) Am. J. Hum. Genet. 35, 67–77. [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, Y. M., Ducos, R., Rosenberg, A. J., Catrou, P. G., Levine, J. S., Podell, E. R. & Allen, R. H. (1985) J. Clin. Invest. 76, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remacha, A. F., Sambeat, M. A., Barcelo, M. J., Mones, J., Garcia-Die, J. & Gimferrer, E. (1992) Ann. Hematol. 64, 202–204. [DOI] [PubMed] [Google Scholar]
- 16.Rosenblatt, D. S. & Fenton, W. A. (1999) in Chemistry and Biochemistry of B12, ed. Banerjee, R. (Wiley, New York), pp. 367–384.
- 17.Yassin, F., Rothenberg, S. P., Rao, S., Gordon, M. M., Alpers, D. H. & Quadros, E. V. (2004) Blood 103, 1515–1517. [DOI] [PubMed] [Google Scholar]
- 18.Gräsbeck, R. & Kvist, G. (1967) Cahiers Coll. Med. Hop. Paris 8, 935–944. [PubMed] [Google Scholar]
- 19.Altay, C., Cetin, M., Gumruk, F., Irken, G. & Yetgin, S. (1995) Pediatr. Hematol. Oncol. 12, 19–28. [DOI] [PubMed] [Google Scholar]
- 20.Ismail, E. A., Al Saleh, Q., Sabry, M. A., Al Ghanim, M. & Zaki, M. (1997) Acta Paediatr. 86, 424–425. [DOI] [PubMed] [Google Scholar]
- 21.Liechti-Gallati, S., Schneider, V., Neeser, D. & Kraemer, R. (1999) Eur. J. Hum. Genet. 7, 590–598. [DOI] [PubMed] [Google Scholar]
- 22.Aminoff, M., Carter, J. E., Chadwick, R. B., Johnson, C., Gräsbeck, R., Abdelaal, M. A., Broch, H., Jenner, L. B., Verroust, P. J., Moestrup, S. K., et al. (1999) Nat. Genet. 21, 309–313. [DOI] [PubMed] [Google Scholar]
- 23.Wagner, E. & Lykke-Andersen, J. (2002) J. Cell Sci. 115, 3033–3038. [DOI] [PubMed] [Google Scholar]
- 24.Gordon, M. M., Brada, N., Remacha, A., Badell, I., del Rio, E., Baiget, M., Santer, R., Quadros, E. V., Rothenberg, S. P. & Alpers, D. H. (2004) Hum. Mutat. 23, 85–91. [DOI] [PubMed] [Google Scholar]
- 25.den Dunnen, J. T. & Antonarakis, S. E. (2001) Hum. Genet. 109, 121–124. [DOI] [PubMed] [Google Scholar]