Abstract
Background
Variable domains of camelid heavy-chain antibodies, commonly named nanobodies, have high biotechnological potential. In view of their broad range of applications in research, diagnostics and therapy, engineering their stability is of particular interest. One important aspect is the improvement of thermostability, because it can have immediate effects on conformational stability, protease resistance and aggregation propensity of the protein.
Methods
We analyzed the sequences and thermostabilities of 78 purified nanobody binders. From this data, potentially stabilizing amino acid variations were identified and studied experimentally.
Results
Some mutations improved the stability of nanobodies by up to 6.1 °C, with an average of 2.3 °C across eight modified nanobodies. The stabilizing mechanism involves an improvement of both conformational stability and aggregation behavior, explaining the variable degree of stabilization in individual molecules. In some instances, variations predicted to be stabilizing actually led to thermal destabilization of the proteins. The reasons for this contradiction between prediction and experiment were investigated.
Conclusions
The results reveal a mutational strategy to improve the biophysical behavior of nanobody binders and indicate a species-specificity of nanobody architecture.
General significance
This study illustrates the potential and limitations of engineering nanobody thermostability by merging sequence information with stability data, an aspect that is becoming increasingly important with the recent development of high-throughput biophysical methods.
Keywords: Single-domain antibody (sdAb, nanobody); Protein engineering; Protein stability; Protein aggregation; Protein design
1. Introduction
VHH domains derived from heavy-chain antibodies of camelids, commonly named nanobodies, are single-domain antigen-binding fragments with a large potential for numerous applications in research, biotechnology and medicine [1–4]. They exhibit several superior properties compared to conventional antibody scaffolds, such as enhanced solubility, low immunogenicity and the unique ability to bind cryptic epitopes; the last is due to their small size and a third complementarity determining region (CDR3) of unusual length [5]. The most interesting epitopes for which specific nanobodies have been generated are enzyme active sites, constant regions of virus particles, transient states of membrane proteins and non-amyloidogenic regions of human lysozyme [6–9]. Nanobodies are expected to contribute substantially to meeting the enormous demand for versatile, robust and stable binders.
Their structural simplicity and ease of production make recombinant nanobodies an ideal and easily accessible system for protein engineering approaches. Numerous benefits would arise particularly from engineering their thermostability. Nanobody-based therapeutics need to resist proteolytic degradation, a characteristic which was strengthened for various proteins by increasing their conformational stability [10–12]. Protein aggregation represents another challenge [13,14]. Its relevance becomes more and more apparent for therapeutic strategies, during which antibodies are usually administered at high concentrations [15,16]. While not detectable for some nanobodies [17], aggregation has been observed for molecules of this binder class [9,18]. Partial or full protein unfolding is usually a prerequisite for aggregation, making the increase of conformational stability a reasonable way to prevent this deleterious side reaction. Furthermore, application of nanobodies as in vivo tools would greatly benefit from stability-engineered binders. For example, in vivo expression in mammalian cells [19] requires nanobody folding in absence of a conserved disulfide bond in the nanobody framework, as the bond remains reduced under cytosolic conditions. Folding that is independent of a disulfide bond is expected to be more robust with stronger non-covalent interactions in the nanobody fold. Finally, recent attempts aimed at the engineering of stabilized nanobodies to construct biosensors and drugs for particularly harsh conditions [20–22], potentially increasing the application range of biological reagents to unaccustomed fields.
Protein thermostability is governed by diverse factors. Successful protein stabilization has been achieved through rigidifying flexible sites [23], optimizing surface charge [24], improving hydrophobic packing [25] and introducing disulfide bonds or salt bridges [26,27]. Interestingly, substantial stabilization was achieved both by the sum of several, marginally stabilizing mutations and by selecting single key positions [28,29]. Accordingly, it remains challenging to identify efficiently stabilizing positions in a protein fold, although several strategies have emerged that could meet this task [30]. Directed evolution methods, for example, explore the vast sequence space by high-throughput experiments [31]. Rational design exploits structural knowledge and general stabilizing features like salt bridges and disulfide bonds [26,27]. Sequence-based strategies identify stabilizing residues from a comparison of mesophilic and thermophilic homologues [32], or rely on the consensus-based strategy that attributes a high probability of stabilization to the most frequent residue at a given sequence position of a protein family [33].
The developments in high-throughput biophysical methods open up new avenues for protein engineering as they amend sequence data with quantitative information on conformation and stability [34–36]. This allows a comprehensive survey of the sequence space of a protein class with respect to thermodynamic parameters, as has been applied to antibody variable domains, for example [37,38]. We studied similar data on nanobody thermostabilities. The merger of sequence and quantitative protein stability information revealed sequence features that are characteristic for highly stable nanobodies. At a global level, our analysis identified species-specific subclasses of nanobody architecture, an insight important for nanobody engineering. With respect to particular sites, we identified stabilizing amino acid variations. In an experimental validation, some residue exchanges improved both conformational stability and aggregation behavior of several nanobodies, which were modified accordingly. Since we were working with recombinant nanobodies, such changes could be introduced easily in a systematic manner, yielding nanobody binders of improved thermostability. However, we also found positions in the nanobody framework that did not act as enhancers of stability if modified as predicted, apparently due to species-dependent interactions in the protein framework. A more complex combination of variations is needed in such cases for improving thermostability.
2. Materials and methods
2.1. Nanobody cloning, expression and purification
The nanobody data set comprised 57 dromedary, 4 alpaca and 17 llama nanobodies. Dromedary and alpaca nanobodies were obtained from phage-display screenings, representing high-affinity binders against different protein targets. They are cloned in the pMECS vector with a C-terminal HA- and His6-tag [39]. The llama nanobodies were obtained from a subtractive phage-display library against tumor lysates [40], cloned into the pHEN2 plasmid with a C-terminal Myc- and His6-tag [41]. N-terminal variants of the dromedary nanobodies were obtained by PCR using mutated primers (biomers.net, Ulm, Germany), while all other variants were purchased from Gen-9 (Cambridge, USA). All variants were cloned into the pMECS vector using NcoI and NotI restriction sites and verified by sequencing. Nanobody constructs present in pMECS and pHEN2 plasmids were expressed in the periplasm of E. coli cells WK6 or TG1, respectively, and purified as described [6]. Using sequence-based extinction coefficients [42], the nanobody concentration was assessed at 280 nm using a Nanodrop ND-1000 instrument (Peqlab Biotechnologie, Erlangen, Germany), measuring absorption at least in triplicate. The molecular weight of the N-terminal variants was verified by MALDI mass spectrometry. N- and C-terminal sequences were checked by in-source-decay [43].
2.2. Stability measurements
Melting temperatures were assessed in triplicate by the Thermofluor assays using Sypro Orange (Thermo Fisher Scientific, Waltham, USA) at a nanobody concentration of 0.5 mg/ml. Fluorescence was detected every 45 s using a LightCycler 480 (Roche Diagnostics, Mannheim, Germany) at a heating rate of 0.5 °C/min. Fluorescence traces were evaluated using a Matlab script, determining the transition point by finding the maximum of the numerical derivation of the smoothed trace data. Smoothing was performed by a linear box filter of 200 data points width. N-terminal variants were measured in triplicate by DSF using a Prometheus NT.48 instrument with back-reflection optics (NanoTemper Technologies, Munich, Germany) after dialysis against a PBS solution at pH 7.4 (Slide-A-Lyzer MINI, 3.5 kDa MWCO; Thermo Fisher Scientific, Waltham, USA). The molar protein concentration was adjusted to 32.7 μM (corresponding to 0.5 mg/ml), if not otherwise stated. The heating rate was 0.5 °C/min. Tm values were determined using the product software package based on first derivative analysis. Gibbs free energy of unfolding was measured by equilibrium denaturation on the same Prometheus NT.48 instrument using guanidinium chloride as denaturant and fitting the data to a two-state model of equilibrium unfolding [44]. Guanidinium chloride concentrations were determined using refractive indices according to Santoro and Bolen [45]. Since protein stickiness had been observed in the fluorescence cuvette filled with nanobody NbD1 or its N-terminal variant at a concentration of 2 μM, the protein concentration was adjusted to 30 μM to avoid artifacts resulting from surface binding. For both the Thermofluor and the DSF assays, the differences measured for mutations Q1E and Q5V were substantiated statistically by applying a paired t-test and a Wilcoxon signed-rank test.
2.3. Nanobody structure determination
Nanobody NbD2 (PDB ID: 5M7Q) was crystallized in 100 mM MES, pH 6.5, 1.6 M magnesium sulfate heptahydrate and was cryo-preserved in 100 mM MES pH 6.5, 1.375 M magnesium sulfate heptahydrate and 25% glycerol. Data were collected on beamline PROXIMA 1 at the SOLEIL synchrotron and processed with iMOSFLM [46]. Space group determination, scaling and merging was performed by means of the Pointless/Aimless routines of the CCP4 suite [47]. PHASER [48] provided a good molecular replacement solution using RCSB protein database entry 4W6X as a model. Refmac [49] was used for structure refinement while model building of the two copies contained in the asymmetric unit was performed with coot [50]. Data collection and refinement statistics are given in Supplementary Table 1.
2.4. Global Sequence Signature analysis
The GSS analysis [51] is based on the concept that any property of a protein is reflected in a signature of sequence positions, contributing differently to stability, for example. Here, a signature was statistically assessed using an MSA of nanobody sequences and the respective Tm values of the nanobodies by generating amino acid scoring tables for each individual position. Amino acids predominantly occurring in stable nanobodies will receive a high score, whereas residues predominantly present in low stability binders will receive a low score. This results in a ranking of residues per position with respect to protein stability. One could extract residues of the highest score at each position, basically corresponding to a conventional statistical analysis as applied to high-throughput experiments in protein engineering [37,38]. However, this would mask different “types” of signatures, which represent distinct solutions to the same problem like protein stability of the nanobody fold. Distinct solutions can be based on residue co-evolution [52], functional constraints [51] or species differences.
The GSS analysis separates possible subgroups in three steps. First, it quantifies the presence of a signature for each sequence by summing up individual, residue-specific scores, previously determined in the scoring tables. This sum is called GSS score. Second, distinct signatures share different residue compositions, most likely reflected in physicochemical properties like pI, aliphatic index, charge and others. Therefore, the GSS scoring procedure is performed again for such an additional property, adding a second dimension to the analysis. While a relationship between property raw values is expected to be devoid of any clear patterns (Fig. 1B), the related GSS scores yielded clear sequence clustering, revealing protein subgroups based on altered amino acid compositions (Fig. 1C). This allows comparing clusters in terms of sequence composition (contingency table chi2-test) and identifying those differences that are linked to significantly increased protein stability (t-test).
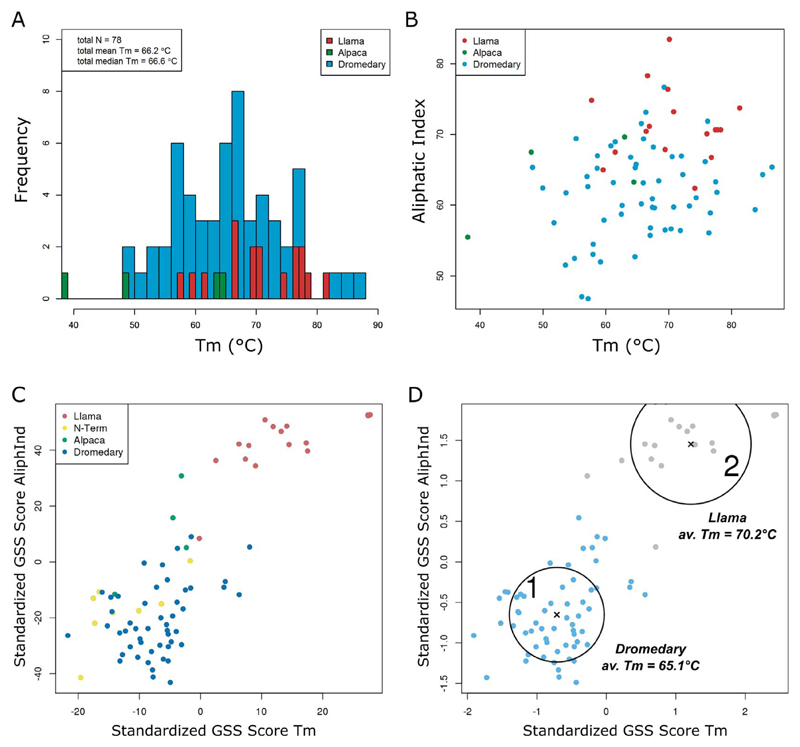
Fig. 1.
GSS analysis of nanobody stability and aliphatic indices. (A) Tm distribution of 78 nanobodies that bind to different antigens. (B) Plot of raw Tm values and aliphatic indices. (C) Plot of GSS scores for Tm and aliphatic indices. The GSS scores quantify the mutational signatures that are characteristic for stability, or the aliphatic index, respectively. Colors mark the nanobody origin as indicated in the legend; yellow data points indicate dromedary nanobodies chosen for N-terminal mutations. (D) K-means clustering of the data of panel C, yielding two clusters (blue and gray). The mean Tm values of both clusters are shown; the mean sequence identity is 0.68 and 0.77 for cluster 1 and 2, respectively. Circles around the cluster centers indicate one standard deviation. The correlation between GSS scores and respective Tm values and aliphatic indices is shown in Supplementary Fig. 4.
A general feature of nanobody sequences is their preferential use of more polar residues at four, so-called hallmark positions, which establish the binding to the light chain variable domain (VL) in conventional binders [5] (MSA numbering: positions 40, 47, 49 and 51; Kabat numbering: positions 37, 44, 45 and 47). However, mixtures of nanobody and conventional hallmark residues are observed only rarely. For this study, only nanobodies with all four hallmark positions in VHH configuration were included; VHH signifies the variable domain of the heavy chain of heavy-chain antibodies. This resulted in a set of 78 sequences, consisting of 4 alpaca, 17 llama and 57 dromedary sequences. They were analyzed with respect to pI, aliphatic index, GRAVY score [53], number of positive and negative charges, charge ratio and sequence length using the ExPASy server [42]. Only the nanobody domain was considered, irrespective of the type of C-terminal tag attached (HA- and His6-tag for dromedary and alpaca nanobodies, myc- and His6-tag for llama nanobodies). An MSA was generated with multAlin (multalin.toulouse.inra.fr) using default parameters, followed by CDR stacking towards N- and C-terminal loop ends by centering the gaps within each loop using JalView [54,55]. In case of odd-numbered loop length, the central residue was stacked towards the N-terminus. GSS analysis was performed using its web interface (gss.mrc-lmb.cam.ac. uk). GSS scoring calculation included the full MSA, as well as the CDR3 region but avoided C-terminal tags.
2.5. Sequence harmony analysis
The nanobody MSA for the GSS analysis was modified by removing binders from alpaca and separating llama- and dromedary-derived sequences. Subtype-specific positions were determined using the web-server www.ibi.vu.nl/programs/seqharmwww with a sequence harmony cut off value of 0.7. For Fig. 2C, sequence harmony values were inverted.
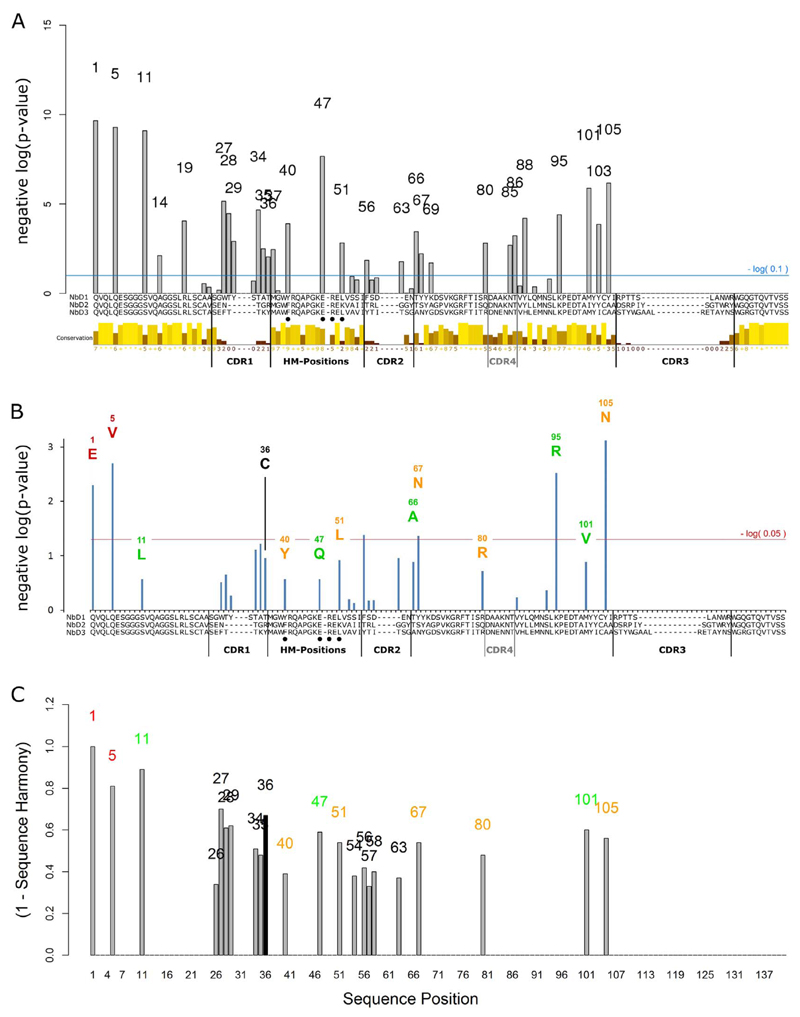
Fig. 2.
GSS stability signature. (A) Sequence differences between the nanobodies in clusters 1 and 2 of Fig. 1D, calculated by a contingency table chi-squared test. Plotted are respective negative log10(p) values, drawn above the nanobody MSA. The numbers represent the MSA-specific position. For CDR3, no signal is observed due to its high variability in both clusters. “CDR4” represents a fourth loop that is facing the antigen but commonly without making contacts. The bars below the nanobody MSA show the degree of conservation of physico-chemical properties for each column; the color and bar height encode the degree of conservation; yellow: high conservation, brown: low conservation. Nanobody hallmark positions are indicated by filled circles (HM-Positions). (B) Influence of sequence differences on nanobody thermostability. At each position, sequences bearing the most frequent residue were grouped within each cluster. The influence of two different residues in cluster 1 and 2 were evaluated by a t-test, considering the respective mean Tm values of the grouped sequences. The respective negative log10(p) values are plotted. Sequence numbering and labeling are identical to panel A. Position 36 indicates a cysteine that is attributed to the additional disulfide bond present in several dromedary nanobodies but not in llama. Red letters: vector-encoded N-terminal residues, specific for llama nanobodies; green letters: residues that do not make any contacts to CDRs according to the crystal structure 1F2X of a dromedary nanobody; orange letters: residues that make contacts to CDRs in structure 1F2X. (C) Subtype-specific sites from a comparison of dromedary- and llama-derived nanobodies using the sequence harmony server [62]. A value of 1 indicates a non-overlapping residue distribution while a value of 0 signifies an identical distribution between both groups. The black bar indicates position 36 which is occupied by a cysteine in some dromedary-derived nanobodies and is linked to enhanced stability, an aspect that cannot be revealed from this kind of analysis. The sequence numbering and the color code are as in Fig. 2B. Sequence Harmony analysis was performed with a cutoff value of 0.7. Afterwards, the sequence harmony score was inverted and values for the CDR3 region omitted.
2.6. Difference contact matrices
Crystallographic data and coordinate files were retrieved from the RCSB protein database (PDB; June 2016). Antigen-free nanobody structures were used that originate from dromedary (PDB ID: 1YC7, 1F2X, 5E7B, 3EAK) and llama (PDB ID: 1HCV, 1I3V, 1SHM, 2X1O, 3R0M, 4IDL, 4KRN, 4QGY, 4QLR, 5H8D), avoiding structures from mutational studies. Residue contact networks for each single protein chain and the consensus networks (structurally conserved residue contacts in a set of structures) for all dromedary and llama nanobodies were calculated as previously described [56]. To identify corresponding residue positions in different nanobody structures and sequences, we created a manually curated profile-to-profile alignment of a structural alignment of all nanobody chains (determined using Mustang [57]) with the nanobody MSA used for GSS analysis. Only one chain per entry was used for the consensus network calculation to avoid a bias towards entries with multiple chains. For multiple nanobody chains in a PDB entry, all chains were checked for consistency of residue contacts. Each consensus contact network was visualized in 3D using customized R scripts and PyMOL (PyMOL Molecular Graphics System, Version 1.5.0.4; Schrödinger, https://www.pymol.org) as previously described [56]. To identify contacts that are specifically conserved in llama or dromedary, the difference of consensus contacts between the two sets of PDB entries was calculated. The data were visualized as difference contact maps to identify structural differences that are conserved in nanobodies of the same organism.
2.7. Consensus approach
Amino acid frequencies for positions 1 and 5 in dromedary heavy-chain domains were obtained from the abYsis database (www.bioinf.org.uk/abysis2.7), assigning relative frequencies of Q and E for position 1 of 0.40 and 0.22, respectively, and of Q and V for position 5 of 0.11 and 0.83, respectively. The prediction of stabilization was calculated using the proposed equation of Steipe [58]: ΔΔG = − RT (lnfconsensus − ln foriginal). R represents the universal gas constant, T the temperature (298.15 K), foriginal the relative frequency of the residue prior mutation and fconsensus the relative frequency of the residue introduced after mutation, respectively.
3. Results
3.1. Identification of residues that potentially increase nanobody thermostability
We collected thermostability data by measuring apparent Tm values in a Thermofluor assay for 78 purified nanobodies, which originated from various phage-display libraries. The broad distribution of the individual Tm values, ranging from 38 °C to 86 °C (Fig. 1A), indicated a substantial potential for stability engineering. The respective Tm values of the nanobodies were related to physicochemical properties, such as the aliphatic index. The aliphatic index is defined as the relative volume of a protein occupied by aliphatic side chains (alanine, valine, isoleucine, and leucine) and considered to be a positive descriptor of thermostability [59]. The analysis resulted in a distribution of data points devoid of clearly separated subgroups but indicated a slight trend of increased aliphatic indices and melting temperatures for llama-derived nanobodies (Fig. 1B); To reveal common signatures of enhanced protein stability within the nanobody fold, we performed a Global Sequence Signature (GSS) analysis [51] on a multiple sequence alignment (MSA) constructed from the amino acid sequences of the 78 nanobodies. The GSS process indicates sequence signatures that reflect particular protein properties of interest, such as stability differences in our case. The analysis yielded a grouping of data points (Fig. 1C), separating nanobody sequences into two major clusters (Fig. 1D).
Notably, the clustering represented the species origin of the nanobodies, with cluster 1 containing predominantly dromedary molecules and cluster 2 only llama-derived nanobodies. This separation could be due to structural differences, such as an increased average CDR3 length in dromedary nanobodies, and a species-dependent amino acid distribution that is known for position 11 [60,61]. Interestingly, the mean Tm of the llama cluster is 5.1 °C higher than that of the dromedary cluster. Again, we used GSS analysis to compare the sequence composition between the clusters for each MSA position so as to find out how llama nanobodies differ in sequence from molecules in the dromedary cluster (Fig. 2A) and evaluated the influence of detected differences on Tm (see the method section for details). This resulted in a stability signature with a negative log10(p) value as a measure for the influence on Tm (Fig. 2B). The obtained signature only marginally varied when the GSS analysis was performed with physicochemical properties other than the aliphatic index (data not shown). For comparison, we also used the sequence harmony server [62] to compare dromedary- and llama-derived sequences. It allows identifying sites that are specific for protein subfamilies (Fig. 2C). Most positions that were identified in the GSS analysis were confirmed.
The large majority of detected residue changes are specific for the llama cluster, which is consistent with its increased mean Tm value. However, also few dromedary-specific features were found, particularly position 36 of the MSA, revealing the additional disulfide bond between CDR3 and CDR1 (Fig. 2B). It is commonly linked to increased stability [63] and occurs in 21 out of 57 dromedary nanobodies but not in any llama sequence of our data set. In the sequence harmony analysis, position 36 is indicated as a subtype-specific site. By combining sequence and stability data, the GSS analysis additionally predicts that a cysteine residue would be beneficial for stability. A comparison among dromedary nanobodies showed that the mean Tm value of the 21 nanobodies bearing the additional disulfide bond was on average 5.2 °C higher, which is in line with earlier results [64]. Despite the comparatively low negative log10(p) value for C36, its detection indicates the relevance of the obtained signature.
To verify that the observed stability differences were not caused by the myc-tags attached to the llama nanobodies or the HA-tags of the dromedary nanobodies, we checked the influence of C-terminal tags on nanobody stability (Supplementary Fig. 1). Comparing six llama nanobodies in both HA- and myc-tagged versions suggested a slightly stabilizing effect of the HA-tag. This shows that the observed stability difference was not due to the tag-molecules. On the contrary, they probably reduced the stability difference between the two clusters to some extent by increasing the stability of some dromedary nanobodies.
3.2. Testing stabilizing signatures
A suitable way to test the relevance of the identified signature experimentally was provided by positions 1 and 5 in the MSA. These positions are independent of the species origin but determined by the phage-display process (dromedary: pMECS vector encoding for 1Q, 5Q; llama: pHEN2 vector encoding for 1E, 5V). Therefore, they were advantageous for an experimental evaluation, since they are lacking any possible species bias resulting from the basic architecture of llama and dromedary nanobodies. As both positions had nevertheless been implicated in having an impact on protein stability, the actual effect of residue variations could be experimentally validated in an unbiased manner.
According to the stability signature, nanobodies should be stabilized by mutations Q1E and Q5V (Fig. 2B). These mutations were introduced into eight dromedary nanobodies, which were chosen so as to exhibit a high diversity in sequence and Tm (yellow dots in Fig. 1C; for MSA see Supplementary Fig. 2A). The locations of the two amino acids in the nanobody fold are indicated in the crystal structure of nanobody NbD2 (Fig. 3A; PDB ID: 5M7Q). Comparing the mutated variants to the original proteins in a Thermofluor assay showed that in all cases the mutations were either beneficial or neutral to Tm, resulting in an average ΔTm of 2.3 °C ( ± 2.7 °C); the strongest increase was 7.9 °C observed for nanobody NbD1 (Fig. 3B). In circular dichroism (CD) spectroscopy measurements with nanobodies NbD1 and NbD3 substantial aggregation was observed (data not shown). Aggregation is dependent on protein concentration and often influenced by the ionic strength of the solvent [65,66]. To exclude that the ΔTm values were biased by these factors, the original binders and the nanobody variants were dialyzed against a common PBS buffer and the molar concentrations were adjusted. Furthermore, the measurements were repeated with another, independent method, differential scanning fluorimetry (DSF), which is based on intrinsic tryptophan fluorescence (Fig. 3C). Again, the Q1E and Q5V variants were clearly stabilized compared to the original binders. In absolute numbers, there were slight differences to the Thermofluor assay: nanobody NbD1 exhibited a ΔTm of 5.0 °C, for example, while the value increased for others, for instance to 6.1 °C for NbD2. This could be explained by slightly varying buffer conditions in the Thermofluor assay affecting the aggregation behavior of some nanobodies and altering their apparent Tm values, or by artifacts introduced by the extrinsic dye used in the Thermofluor assay. With 2.3 °C ( ± 2.3 °C), the average increase in Tm for all eight nanobodies, however, was identical between the two methods.
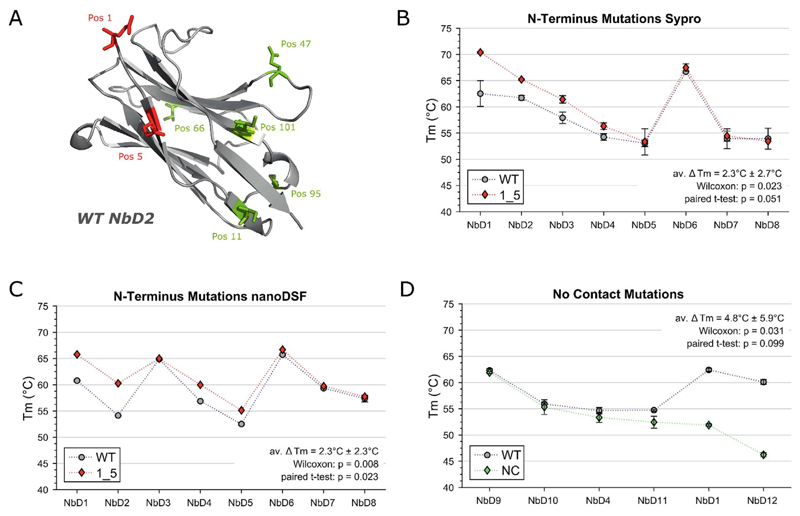
Fig. 3.
Stabilization of dromedary nanobodies by N-terminal mutations Q1E and Q5V. (A) Crystal structure of nanobody NbD2 (PDB ID: 5M7Q, for data collection and refinement statistics see Supplementary Table 1). The locations of N-terminal (red) and framework mutations (green) are indicated. (B) Effect of N-terminal mutations tested in a diverse set of eight dromedary nanobodies (mean sequence identity = 0.67) using the Thermofluor assay. The statistical significance of stabilization was calculated by a paired t-test and a Wilcoxon signed-rank test; the respective p-values are shown. Also, the mean amplitude of stabilization is indicated. Protein concentration: 0.5 mg/ml; heating rate: 0.5 °C/min. (C) Effect of N-terminal mutations Q1E and Q5V in the same dromedary nanobodies as listed in panel B, measured by differential scanning fluorimetry (DSF; based on intrinsic tryptophan fluorescence). Statistical evaluation and heating rate were as in panel B. (D) Destabilization of dromedary nanobodies by framework mutations 11L, 47Q, 66A, 95R and 101V measured in a Thermofluor assay. This group of mutations does not form contacts with CDRs in the crystal structure 1F2X of a dromedary nanobody. Statistical evaluation and assay conditions were as in panel B. All measurements were done in triplicate.
3.3. The mechanism of stabilization
To investigate the mechanism of nanobody stabilization in more detail, changes in thermodynamic stability were assessed by equilibrium unfolding transitions for NbD1 (Fig. 4A), the nanobody with the strongest stabilization in the Thermofluor assay (Fig. 3B). It shows a stability increase of ΔΔG = 6.6 kJ/mol upon mutations Q1E and Q5V, confirming their stabilizing effect. Furthermore, by exploiting the concentration dependence of protein aggregation, DSF and turbidity measurements revealed an improved aggregation behavior of NbD1 upon mutation (Fig. 4C–F). In this experiment, standard assay conditions at a concentration of 32.7 μM yielded a stabilization of 3.7 °C. Tm values were affected by aggregation as indicated by back-reflection signals and altered amplitudes of fluorescence transitions. At a protein concentration of 13.1 μM, aggregation ceased almost completely for both the original nanobodies and the N-terminal variants (Fig. 4D, F). This led to a reduction in the difference between original and variant protein of ΔTm = 2.7 °C. Accordingly, 1 °C (27%) of Tm stabilization measured at standard assay conditions could be attributed to a better aggregation behavior of the mutated variant, whereas 2.7 °C (73%) reflected the improved Gibbs free energy of unfolding measured in equilibrium unfolding experiments (Fig. 4A). Finally, both mutations seemed to contribute to thermostabilization, when looking at single mutation variants that we prepared for nanobody NbD4 (Fig. 4B). The stabilizing effects of the two single mutations Q1E (2.3 °C) and Q5V (0.9 °C) add up to 3.2 °C, nicely reflecting the combined effect in the double mutant of 3.1 °C.
Fig. 4.
Mechanism of stabilization by N-terminal mutations Q1E and Q5V. (A) Thermodynamic stability of nanobody NbD1 and its N-terminally mutated variant in guanidinium chloride dependent equilibrium unfolding experiments. The fraction of unfolded protein was measured by intrinsic tryptophan fluorescence and fitted according to Santoro and Bolen [45]. Red lines represent fitted curves. (B) Additive effect of thermostabilization by single and double mutations in nanobody NbD4, measured by differential scanning fluorimetry in triplicate using conditions as in Fig. 3C. Improvements by mutations Q5V (0.9 °C) and Q1E (2.3 °C) match the stabilization in the double mutant (3.1 °C). (C, D) Tryptophan fluorescence ratio (350 nm/330 nm) for melting nanobody NbD1 and its N-terminally mutated variant; in panel C, the standard assay concentration of 32.7 μM was used; in panel D, the concentration was reduced to 13.1 μM. Aggregation is indicated by a reduced amplitude of the unfolding transitions in the fluorescence traces and can be quantified by comparing Tm values of both concentration sets. Heating rate: 0.5 °C/min. (E, F) Back-scattered light obtained from a parallel turbidity measurement indicates aggregation onset temperatures.
3.4. Investigating stability signatures within the structural framework of nanobodies
Encouraged by the stabilizing effects of the Q1E and Q5V mutations, we explored the possibility of nanobody stabilization by mutating other positions identified by the GSS analysis. CDR regions were avoided to preserve antigen specificity. Potential variations in the nanobody framework were first examined in the crystal structure of a dromedary-derived nanobody (RCSB protein database entry 1F2X) to exclude major structural clashes. Also, residues with a negative log10(p) value slightly below a threshold of −log10(0.05) were considered. This way, we chose a set of five positions for mutation within the GSS-derived stability signature (indicated by green letters in Fig. 2B), comprising mutations 11L, 47Q, 66A, 95R and 101V. All five residues originate from the llama cluster, suggesting that stabilization should occur when introduced into dromedary nanobodies. Also, they do not make intramolecular contacts to CDRs in the crystal structure 1F2X of a dromedary nanobody. Their locations in the nanobody fold are indicated in Fig. 3A (green positions), a crystal structure of nanobody NbD2 that we solved successfully (PDB ID: 5M7Q), but which was not considered during the mutation selection process.
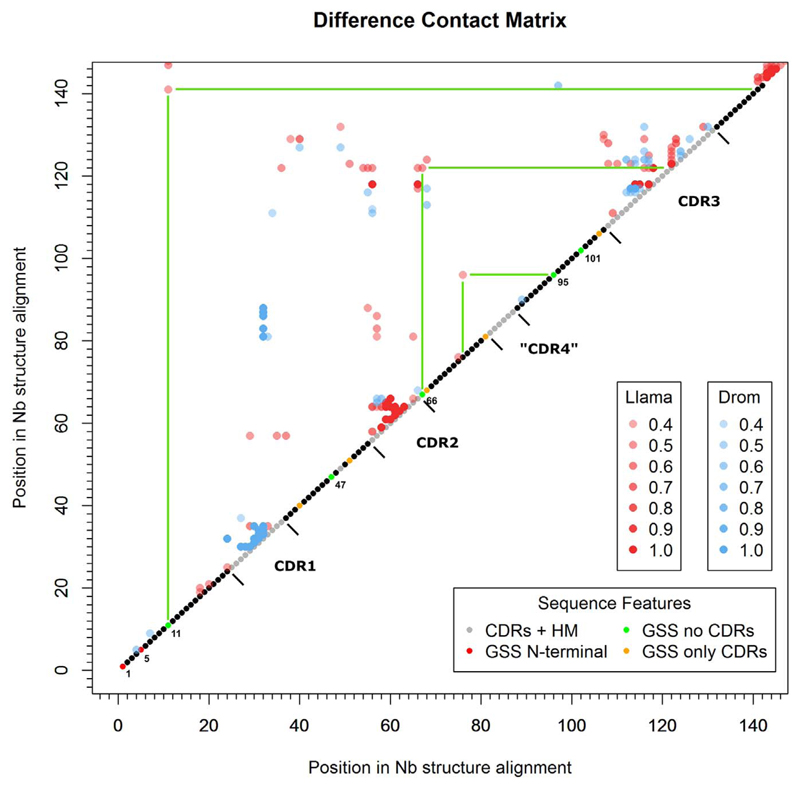
Surprisingly, introduction of the variations 11L, 47Q, 66A, 95R and 101V into six dromedary-derived nanobodies (for MSA see Supplementary Fig. 2B) led to markedly reduced purification yields (data not shown) and tended to be destabilizing (Fig. 3D). One explanation for this could be missing mutations that are necessary for synergistic effects, which are detected by GSS but not labeled as such. Alternatively, the species-dependent nanobody architecture may not tolerate the transfer of some llama-specific residues into the dromedary framework without compromising stability. Therefore, we investigated a possible species bias in nanobody architecture. Difference residue-contact maps [56] were calculated from crystal structures of antigen-free nanobodies so as to reveal species-dependent residue interactions in llama and dromedary nanobody structures (Fig. 5). Being under-represented in our data set, crystal structures from alpaca-derived nanobodies were excluded from this analysis. Interestingly, position 11, which is known to have a species-dependent residue distribution [60], and position 95 in the MSA share species-dependent contacts to other framework residues. Additionally, position 66 makes contacts to CDR3 residues predominantly in llama, which was not observed in the dromedary nanobody crystal structure 1F2X. Since all three positions were included in the mutational experiments (Fig. 3D), this supports the conclusion that transferring stabilizing mutations into these regions can be problematic, since residue contacts represent a species-dependent feature.
Fig. 5.
Species-dependent difference contact map of the nano-body fold. Connectivity networks of non-covalent contacts between amino acid residues were calculated on the basis of 10 llama and 4 dromedary monomeric crystal structures, respectively. The frequencies of a contact in each set of structures from a species provide a measure of the “conservation” of a residue contact in the respective species. Accordingly, subtracting species-specific frequencies allows the visualization of architectural differences between different species. The extent of contact differences between llama and dromedary nanobodies is indicated by spot intensity and color. For example, a red dot with frequency of 1.0 indicates a contact that occurs in every llama nanobody, and not in any dromedary nanobody. Green bars indicate contact differences for positions tested in mutational experiments (Fig. 3D). Sequence numbering is as in the MSA of Fig. 2.
Interestingly, the difference contact map revealed additional structural features specific for llama and dromedary nanobodies. The CDR loops 1 and 2, for example, exhibit a highly species-dependent contact pattern. This is particularly interesting, as generally nanobodies contain a much larger variety of loop conformations compared to the limited set of canonical loop structures in conventional antibodies [67,68]. The result suggests a species-specific mechanism of antigen-recognition, which would fit to the notion of species-specific differences between llama and dromedary frameworks. Furthermore, the species variance in both CDRs is reflected in their interactions to the fourth antigen facing loop, called CDR4 in our study, which is also more variable in nanobody sequences compared to heavy-chain variable domains (VH) of conventional antibodies [5].
4. Discussion
In this study, we looked for sequence variations in the nanobody fold that could increase their thermostability. Several tools exist to identify important residues, such as consensus-based engineering [33], structure-guided recombination [69] or methods of co-evolution [52,70]. We applied the GSS process [51] as it enables a merger of quantitative information about biophysical protein properties with sequence data from high-throughput experiments. This ability gains increasing importance considering the growing number of methods for large scale biophysical characterization of proteins [34–36]. GSS scoring of two properties allows distinguishing protein subgroups and relating them to stability. The two N-terminal sites in the nanobody sequences offered a means to study the influence of residue modifications that are not part of the species-specific framework of the analyzed nanobodies.
The degree of stabilization achieved by introducing two amino acid variations was significant in a set of eight nanobodies that were of high sequence diversity (mean sequence identity = 0.67; see MSA in Supplemental Fig. 2A). Interestingly, both positions have recently been tested in two nanobodies, however, in combination with additional mutations, leading to improved thermostability and solubility of the variants [22]. This further supports the general beneficial effect of both N-terminal mutations. Furthermore, the stabilization in Gibbs free energy for nanobody NbD1 was in the range of a predicted net-stabilization of 3.44 kJ/mol using the consensus approach of Steipe et al. [33], which uses naturally occurring amino acid frequencies to predict mutational effects in proteins. Nanobody thermostabilization was additive as predicted by the GSS analysis, while the consensus approach [33] predicted position 1 to be slightly destabilizing upon mutation Q1E (1.5 kJ/mol) and attributes the major stabilization of 4.9 kJ/mol to position 5 upon mutation Q5V. Accordingly, we hypothesize that mutation Q1E is mainly responsible for improved aggregation behavior by introducing a negative charge [24] and contributes indirectly to improved thermostability. Furthermore, as the NbD2 crystal structure shows, mutation Q5V sacrifices a hydrogen bond by introducing a more hydrophobic and much smaller valine at position 5 (Supplementary Fig. 3). This seems counter-intuitive but led to improved stability on its own and in combination with Q1E in nanobody NbD4. Therefore, we suggest considering both positions for a general modification in phage-display vectors of dromedary nanobodies.
In contrast to the N-terminal mutations, the other positions of the GSS stability signature reside in the natural amino acid framework of the nanobodies. Consequently, it was interesting to see whether a transfer of amino acids derived from the llama cluster into dromedary nanobodies led to stabilization, as the difference in average Tm between the llama and dromedary clusters and the GSS analysis suggested. The actually observed reduction in stability upon mutation of five modifications was somewhat surprising. The effect could have several reasons. For once, we are not claiming that llama nanobodies are per se more stable than binders from dromedary. The observed difference might rest upon the reduced sequence diversity within the llama cluster (mean sequence identity: llama cluster, 0.77; dromedary cluster, 0.68). Our dromedary nanobodies represent fully affinity-maturated binders against well-defined protein targets, while the llama nanobodies originate from a single library against complex cell lysates [40]. It is unclear if the latter binders are fully affinity-maturated, since their antigens are yet unknown. Accordingly, their sequences might lack final somatic mutations, which would be necessary for antigen specificity but possibly reduce stability to the same level as in the dromedary cluster. However, if this is the major difference between dromedary and llama nanobodies, it should be possible to reverse somatic mutations in the dromedary cluster and enhance their stability by introducing the residues suggested by the GSS analysis.
Destabilization could also be due to missing mutations that are required for synergistic effects. In a GSS analysis, synergistic variations would show up as increases for the individual sites that are responsible for stabilization when applied in combination. They would not be distinguishable from positions that exhibit a stabilizing effect on their own. Finally, species-dependent differences in the overall nanobody fold may limit the transferability of llama-specific residues into the dromedary framework, as indicated by the results of the difference contact map. The non-covalent residue contacts of three tested framework positions represent a species-dependent feature. In other words, the species-specific environment of these residues seems to be important for their stabilizing effect. Grafting them into the dromedary framework does not result in thermostabilization.
In conclusion, predictions can be performed successfully about the position and kind of amino acid exchanges that could lead to enhanced stability of nanobody binders by a combination of sequence information and corresponding thermostability of individual nanobodies. However, the analysis also demonstrated that the extent of stabilizing effects differs among individual binders due to the sequence diversity between the investigated nanobodies. Accordingly, proposed positions have to be tested experimentally. In addition, structural aspects have to be taken into account, as documented by the destabilization of the dromedary framework by the transfer of amino acids intended to stabilize the proteins and proposed on the basis of llama nanobody sequences. Furthermore, intramolecular interactions of introduced residues could compromise or contribute to binding specificity.
Last, as an alternative, one should not ignore the option of stabilizing binders by tag-sequences, a process that does not touch upon the framework sequence at all but exhibited effects that were similar to variations within the protein sequence.
Supplementary Material
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2017.06.014.
Acknowledgements
We thank NanoTemper Technologies in Munich for its generous support with free DSF measurements. Funding by the European Union (grant number: Health-F4-2010-241481) as part of the Affinomics consortium is gratefully acknowledged.
Abbreviations
- VHH
variable domain of the heavy chain of heavy-chain antibodies
- GSS algorithm
Global Sequence Signature algorithm
- CDR
complementarity determining region
- MSA
multiple sequence alignment
- CD
circular dichroism
- DSF
differential scanning fluorimetry
- VH
heavy chain variable domain
- VL
light chain variable domain
- HM
hallmark position
- GdmCl
guanidinium chloride
Footnotes
Transparency Document
The Transparency document associated with this article can be found, in online version.
References
- [1].Wesolowski J, Alzogaray V, Reyelt J, Unger M, et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol. 2009;198:157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Huang L, Muyldermans S, Saerens D. Nanobodies®: proficient tools in diagnostics. Expert Rev Mol Diagn. 2010;10:777–785. doi: 10.1586/erm.10.62. [DOI] [PubMed] [Google Scholar]
- [3].Siontorou CG. Nanobodies as novel agents for disease diagnosis and therapy. Int J Nanomedicine. 2013;8:4215–4227. doi: 10.2147/IJN.S39428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Helma J, Cardoso MC, Muyldermans S, Leonhardt H. Nanobodies and recombinant binders in cell biology. J Cell Biol. 2015;209:633–644. doi: 10.1083/jcb.201409074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- [6].Conrath KE, Lauwereys M, Galleni M, Frère J, et al. β-Lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Koromyslova AD, Hansman GS. Nanobody binding to a conserved epitope promotes norovirus particle disassembly. J Virol. 2015;89:2718–2730. doi: 10.1128/JVI.03176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].De Genst E, Chan P-H, Pardon E, Hsu S-TD, et al. A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation. J Phys Chem B. 2013;117:13245–13258. doi: 10.1021/jp403425z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hussack G, Hirama T, Ding W, Mackenzie R, Tanha J. Engineered single-domain antibodies with high protease resistance and thermal stability. PLoS One. 2011;6:e28218. doi: 10.1371/journal.pone.0028218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sieber V, Plückthun A, Schmid FX. Selecting proteins with improved stability by a phage-based method. Nat Biotechnol. 1998;16:955–960. doi: 10.1038/nbt1098-955. [DOI] [PubMed] [Google Scholar]
- [12].Wunderlich M, Martin A, Schmid FX. Stabilization of the cold shock protein CspB from Bacillus subtilis by evolutionary optimization of coulombic interactions. J Mol Biol. 2005;347:1063–1076. doi: 10.1016/j.jmb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- [13].Rosenberg AS. Effeects of protein aggregates: an immunologic perspective. AAPS J. 2006;8:E501–E507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Roberts CJ. Therapeutic protein aggregation: mechanisms, design, and control. Trends Biotechnol. 2014;32:372–380. doi: 10.1016/j.tibtech.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Frokjaer S, Otzen DE. Protein drug stability: a formulation challenge. Nat Rev Drug Discov. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- [16].Mitragotri S, Burke PA, Langer R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat Rev Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Akazawa-Ogawa Y, Takashima M, Lee Y, Ikegami T, et al. Heat-induced irreversible denaturation of the camelid single domain VHH antibody is governed by chemical modifications. J Biol Chem. 2014;289:15666–15679. doi: 10.1074/jbc.M113.534222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dumoulin M, Conrath K, Meirhaeghe AVAN, Meersman F, et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vercruysse T, Pardon E, Vanstreels E, Steyaert J, Daelemans D. An intrabody based on a llama single-domain antibody targeting the N-terminal alpha-helical multimerization domain of HIV-1 rev prevents viral production. J Biol Chem. 2010;285:21768–21780. doi: 10.1074/jbc.M110.112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Turner KB, Zabetakis D, Goldman ER, Anderson GP. Enhanced stabilization of a stable single domain antibody for SEB toxin by random mutagenesis and stringent selection. Protein Eng Des Sel. 2014;27:89–95. doi: 10.1093/protein/gzu001. [DOI] [PubMed] [Google Scholar]
- [21].Turner KB, Liu JL, Zabetakis D, Lee AB, et al. Improving the biophysical properties of anti-ricin single-domain antibodies. Biotechnol Rep. 2015;6:27–35. doi: 10.1016/j.btre.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Anderson GP, Liu JH, Zabetakis D, Liu JL, Goldman ER. Thermal stabilization of anti-α-cobratoxin single domain antibodies. Toxicon. 2017;129:68–73. doi: 10.1016/j.toxicon.2017.02.008. [DOI] [PubMed] [Google Scholar]
- [23].Wijma HJ, Floor RJ, Janssen DB. Structure- and sequence-analysis inspired engineering of proteins for enhanced thermostability. Curr Opin Struct Biol. 2013;23:588–594. doi: 10.1016/j.sbi.2013.04.008. [DOI] [PubMed] [Google Scholar]
- [24].Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Borgo B, Havranek JJ. Automated selection of stabilizing mutations in designed and natural proteins. Proc Natl Acad Sci U S A. 2012;109:1494–1499. doi: 10.1073/pnas.1115172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dombkowski AA, Sultana KZ, Craig DB. Protein disulfide engineering. FEBS Lett. 2014;588:206–212. doi: 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- [27].Makhatadze GI, Loladze VV, Ermolenko DN, Chen X, Thomas ST. Contribution of surface salt bridges to protein stability: guidelines for protein engineering. J Mol Biol. 2003;327:1135–1148. doi: 10.1016/s0022-2836(03)00233-x. [DOI] [PubMed] [Google Scholar]
- [28].Saab-Rincon G, Michelle M, Carbone M, Landwher M, Arnold FH. Protein Eng Handb. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim: 2008. Protein engineering by structure guided SCHEMA recombination; pp. 481–492. [Google Scholar]
- [29].Heinzelman P, Snow CD, Smith MA, Yu X, Kannan A, et al. SCHEMA recombination of a fungal cellulase uncovers a single mutation that contributes markedly to stability. J Biol Chem. 2009;284:26229–26233. doi: 10.1074/jbc.C109.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Magliery TJ. Protein stability: computation, sequence statistics, and new experimental methods. Curr Opin Struct Biol. 2015;33:161–168. doi: 10.1016/j.sbi.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- [32].Perl D, Schmid FX. Some like it hot: the molecular determinants of protein thermostability. Chembiochem. 2002;3:39–44. doi: 10.1002/1439-7633(20020104)3:1<39::AID-CBIC39>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- [33].Steipe B, Schiller B, Plückthun A, Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J Mol Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- [34].Razinkov VI, Treuheit MJ, Becker GW. Methods of high throughput biophysical characterization in biopharmaceutical development. Curr Drug Discov Technol. 2013;10:59–70. doi: 10.2174/157016313804998915. [DOI] [PubMed] [Google Scholar]
- [35].Fiedler S, Cole L, Keller S. Automated circular dichroism spectroscopy for medium-throughput analysis of protein conformation. Anal Chem. 2013;85:1868–1872. doi: 10.1021/ac303244g. [DOI] [PubMed] [Google Scholar]
- [36].Alexander CG, Wanner R, Johnson CM, Breitsprecher D, et al. Novel microscale approaches for easy, rapid determination of protein stability in academic and commercial settings. Biochim Biophys Acta. 2014;1844:2241–2250. doi: 10.1016/j.bbapap.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hsu H-J, Lee KH, Jian J-W, Chang H-J, et al. Antibody variable domain interface and framework sequence requirements for stability and function by high-throughput experiments. Structure. 2014;22:22–34. doi: 10.1016/j.str.2013.10.006. [DOI] [PubMed] [Google Scholar]
- [38].Chang H-J, Jain J-W, Hsu H-J, Lee Y-C, Chen H-S, et al. Loop-sequence features and stability determinants in antibody variable domains by high-throughput experiments. Structure. 2014;22:9–21. doi: 10.1016/j.str.2013.10.005. [DOI] [PubMed] [Google Scholar]
- [39].Vincke C, Gutiérrez C, Wernery U, Devoogdt N, et al. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol Biol. 2012;907:145–176. doi: 10.1007/978-1-61779-974-7_8. [DOI] [PubMed] [Google Scholar]
- [40].Kastelic D, Baty D, Truan G, Komel R, Pompon D. A single-step procedure of recombinant library construction for the selection of efficiently produced llama VH binders directed against cancer markers. J Immunol Methods. 2009;350:54–62. doi: 10.1016/j.jim.2009.08.016. [DOI] [PubMed] [Google Scholar]
- [41].Goffinet M, Chinestra P, Lajoie-Mazenc I, Medale-Giamarchi C, et al. Identification of a GTP-bound Rho specific scFv molecular sensor by phage display selection. BMC Biotechnol. 2008;8:34. doi: 10.1186/1472-6750-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hardouin J. Protein sequence information by matrix-assisted laser desorption/ionization in-source decay mass spectrometry. Mass Spectrom Rev. 2007;26:672–682. doi: 10.1002/mas.20142. [DOI] [PubMed] [Google Scholar]
- [44].Pace CN. Measuring and increasing protein stability. Trends Biotechnol. 1990;8:93–98. doi: 10.1016/0167-7799(90)90146-o. [DOI] [PubMed] [Google Scholar]
- [45].Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- [46].Powell HR, Johnson O, Leslie AGW. Autoindexing diffraction images with iMosflm. Acta Crystallogr D Biol Crystallogr. 2013;69:1195–1203. doi: 10.1107/S0907444912048524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Murshudov GN, Skubák P, Lebedev AA, Pannu NS, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kastelic D, Soler N, Komel R, Pompon D. The Global Sequence Signature algorithm unveils a structural network surrounding heavy chain CDR3 loop in camelidae variable domains. Biochim Biophys Acta. 2013;1830:3373–3381. doi: 10.1016/j.bbagen.2013.02.014. [DOI] [PubMed] [Google Scholar]
- [52].de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14:249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- [53].Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- [54].Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Flock T, Ravarani CNJ, Sun D, Venkatakrishnan AJ, et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- [58].Steipe B. Consensus-based engineering of protein stability: from intrabodies to thermostable enzymes. Methods Enzymol. 2004;388:176–186. doi: 10.1016/S0076-6879(04)88016-9. [DOI] [PubMed] [Google Scholar]
- [59].Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem. 1980;88:1895–1898. [PubMed] [Google Scholar]
- [60].Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol. 1997;34:1121–1131. doi: 10.1016/s0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- [61].Harmsen MM, Ruuls RC, Frenken LGJ, De Geus B. Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol Immunol. 2000;37:579–590. doi: 10.1016/s0161-5890(00)00081-x. [DOI] [PubMed] [Google Scholar]
- [62].Feenstra KA, Pirovano W, Krab K, Heringa J. Sequence harmony: detecting functional specificity from alignments. Nucleic Acids Res. 2007;35:W495–W498. doi: 10.1093/nar/gkm406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hagihara Y, Saerens D. Engineering disulfide bonds within an antibody. Biochim Biophys Acta. 2014;1844:2016–2023. doi: 10.1016/j.bbapap.2014.07.005. [DOI] [PubMed] [Google Scholar]
- [64].Govaert J, Pellis M, Deschacht N, Vincke C, et al. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J Biol Chem. 2012;287:1970–1979. doi: 10.1074/jbc.M111.242818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chi EY, Krishnan S, Kendrick BS, Chang BS, et al. Roles of conformational stability and colloidal stability in the aggregation of recombinant human granulocyte colony-stimulating factor. Protein Sci. 2003;12:903–913. doi: 10.1110/ps.0235703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang T, Kumru OS, Yi L, Wang YJ, et al. Effect of ionic strength and pH on the physical and chemical stability of a monoclonal antibody antigen-binding fragment. J Pharm Sci. 2013;102:2520–2537. doi: 10.1002/jps.23645. [DOI] [PubMed] [Google Scholar]
- [67].Decanniere K, Muyldermans S, Wyns L. Canonical antigen-binding loop structures in immunoglobulins: more structures, more canonical classes? J Mol Biol. 2000;300:83–91. doi: 10.1006/jmbi.2000.3839. [DOI] [PubMed] [Google Scholar]
- [68].Mahajan SP, Velez-vega C, Escobedo FA. Tilting the balance between canonical and noncanonical conformations for the H1 hypervariable loop of a llama VHH through point mutations. J Phys Chem B. 2013;117:13–24. doi: 10.1021/jp3075496. [DOI] [PubMed] [Google Scholar]
- [69].Trudeau DL, Smith MA, Arnold FH. Innovation by homologous recombination. Curr Opin Chem Biol. 2013;17:902–909. doi: 10.1016/j.cbpa.2013.10.007. [DOI] [PubMed] [Google Scholar]
- [70].Ochoa D, Pazos F. Practical aspects of protein co-evolution. Front Cell Dev Biol. 2014;2:1–14. doi: 10.3389/fcell.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.