Abstract
The only endogenous protein inhibitor known for metallocarboxypeptidases (MCPs) is latexin, a 25-kDa protein discovered in the rat brain. Latexin, alias endogenous carboxypeptidase inhibitor, inhibits human CPA4 (hCPA4), whose expression is induced in prostate cancer cells after treatment with histone deacetylase inhibitors. hCPA4 is a member of the A/B subfamily of MCPs and displays the characteristic α/β-hydrolase fold. Human latexin consists of two topologically equivalent subdomains, reminiscent of cystatins, consisting of an α-helix enveloped by a curved β-sheet. These subdomains are packed against each other through the helices and linked by a connecting segment encompassing a third α-helix. The enzyme is bound at the interface of these subdomains. The complex occludes a large contact surface but makes rather few contacts, despite a nanomolar inhibition constant. This low specificity explains the flexibility of latexin in inhibiting all vertebrate A/B MCPs tested, even across species barriers. In contrast, modeling studies reveal why the N/E subfamily of MCPs and invertebrate A/B MCPs are not inhibited. Major differences in the loop segments shaping the border of the funnel-like access to the protease active site impede complex formation with latexin. Several sequences ascribable to diverse tissues and organs have been identified in vertebrate genomes as being highly similar to latexin. They are proposed to constitute the latexin family of potential inhibitors. Because they are ubiquitous, latexins could represent for vertebrate A/B MCPs the counterparts of tissue inhibitors of metalloproteases for matrix metalloproteinases.
Keywords: metallocarboxypeptidase, metalloprotease, x-ray crystal structure
Latexin, also known as tissue or endogenous carboxypeptidase inhibitor (ECI), is a 222-residue protein in humans, and the only endogenous specific inhibitor of zinc-dependent metallocarboxypeptidases (MCPs) present in mammalians. It is sequentially unrelated to any structurally characterized protein and was originally found expressed in the lateral neocortex of rats. It is a marker of regionality and development in both central and peripheral rodent nervous systems and is down-regulated in the presenilin-1-deficient mouse brain, thus putatively playing a role in Alzheimer's disease (1, 2). The identification of latexin as an inhibitor of carboxypeptidase (CP) A (CPA) in a series of non-pancreatic tissues led to its isolation from the rat brain (3, 4). An experimental model of rat acute pancreatitis revealed that latexin expression is also induced in this condition, showing that tissue distribution of CPA and latexin correlate well in the rat (5). Latexin is also widespread in humans, although with a different distribution. In humans, expression of the protein is high in heart, prostate, ovary, kidney, pancreas, and colon but only moderate in brain (3).
MCPs can be classified into two subfamilies, the A/B (M14A according to the MEROPS database at http://merops.sanger.ac.uk.) and the N/E forms (M14B), previously referred to as pancreatic and regulatory CPs, respectively (6). A/B MCPs were among the first proteases studied as digestive enzymes synthesized in the pancreas of mammals (7). Molecular prototypes of the A/B MCPs are pancreatic bovine CPA (bCPA) and bovine CPB (bCPB) that excise C-terminal hydrophobic and basic amino acids, respectively. More recently, members of this subfamily have been found in archaea and bacteria, protozoa, fungi, nematodes, insects, and other invertebrates, plants, amphibians, birds, and mammals (8). In the last few years, functional and local ascription of A/B MCPs has moved away from the mere proteolysis of intake proteins in the digestive tract. In particular, they have been localized in brain, heart, stomach, colon, testis, and lung (4). They participate in peptide hormone activity and hormone-regulated tissue growth or differentiation, in fibrinolysis inhibition and bradykinin activation in blood serum, and in cellular response or complementing chymase in mast cells (9). One example is a gene product, human procarboxypeptidase A4 (hPCPA4), involved in prostate cancer (10). It is up-regulated via the histone hyperacetylation pathway as a downstream effect during sodium butyrate treatment of prostate cancer cell lines. The hPCPA4 gene is imprinted and may be responsible for prostate-cancer aggressiveness (11). Expression was detected in human hormone-regulated tissues; however, levels are very low in normal human adult tissues, including prostate, ovary, testis, and pancreas (10, 11).
A/B MCPs are secreted as inactive zymogens encompassing an N-terminal prodomain (PD) that blocks access to the activesite cleft of the enzyme. Activation occurs through limited proteolysis in a connecting segment at the end of the PD. This reaction releases the active CP from its PD, which acts as an autologous inhibitor (12). Heterologous MCP protein inhibitors have been reported from potato, tomato, the intestinal parasite Ascaris suum, medical leech, and the tick Rhipicephalus bursa (12, 13). A number of 3D structures are available for A/B MCPs, either in their active, inhibitor-complexed or zymogenic forms (see ref. 12 for a review), and for members of the N/E subfamily (14, 15). However, none of the former corresponds to a non-pancreatic protein. No structure of an endogenous human inhibitor for MCPs has been reported to date. We present the structure of hCPA4 in complex with the inhibitor latexin, and biochemical evidence for the role of the latter as a global inhibitor of vertebrate A/B MCPs.
Materials and Methods
Production and Purification of the Human CPA4 (hCPA4)/Latexin Complex. The cDNA for hPCPA4 was kindly provided by D. I. Smith and H. Huang (Mayo Clinic, Rochester, MN) and cloned into vector pPIC9. The protein was expressed and secreted to the extracellular medium by the methylotrophic yeast Pichia pastoris as described for other PCPs (16). Purification included hydrophobic interaction and anion exchange chromatography. The proenzyme was activated with trypsin and checked for functionality. The human latexin nucleotide sequence (GenBank accession no. NM 020169) was amplified from human brain cDNAs and cloned into the prokaryotic expression vector pGAT2 as a fusion construct with GST and a polyhistidine tag. Expression was achieved in BL21(DE3) Escherichia coli cells, and further processing included nickel Sepharose affinity chromatography. The hCPA4/latexin complex was produced by using fresh preparations of both proteins. Once obtained, the complex was incubated with thrombin to remove the fusion construct and subsequently purified by anion exchange chromatography. The complex was desalted and concentrated to ≈7 mg/ml.
Inhibition Assays of MCPs by Latexin. Inhibition constants were calculated by pre-steady-state analysis (Ki = koff/kon) (17). Kinetic association (kon) and dissociation (koff) constants were determined by a continuous photometric assay in which the inhibitor is added to a monitored progress curve obtained from an enzyme/substrate mixture. The following chromogenic substrates were used: N-(4-methoxyphenyl-azoformyl)-Phe-OH for hCPA1, hCPA2, bCPA, Helicoverpa armigera (ha) CPA, and hCPA4; N-(4-methoxyphenyl-azoformyl)-Arg-OH for hCPB and human thrombin-activatable fibrinolysis inhibitor; and N-(4-furylacryloyl)-Ala-Lys-OH for the hCPN assay. The assays were performed in 50 mM Tris·HCl, pH 7.5/0.1 M NaCl, with a substrate concentration of 100 μM and enzyme concentrations of 2 nM. Parallel steady-state kinetic measurements with various substrate and inhibitor concentrations were also carried out by using N-[3-(2-furyl) acryloyl]-Phe-Phe as a substrate.
Structure Analysis of the hCPA4/Latexin Complex. The latexin/hCPA4 complex was crystallized from hanging drops containing 1 μl of protein solution (7 mg/ml), 1 μl of reservoir solution (40% 2-methyl-2,4-pentanediol, 0.1 M bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane, pH 6.5), and 0.2 μl of 40% acetone at 20°C. The structure of the complex was solved by a combination of Patterson search and multiple-wavelength anomalous diffraction at the zinc absorption K-edge. To this end, three diffraction datasets were collected at 100 K from a single crystal on a marCCD 225 detector at beamline ID23–1 at the European Synchrotron Radiation Facility (Grenoble, France). A further high-resolution data set at 1.6-Å resolution was collected from the same crystal. Crystals contain two complexes per asymmetric unit. Data were processed, scaled, merged, and reduced with mosflm and scala from the ccp4 suite (18) (see Table 1). To calculate initial phases, a Patterson search was performed with the program amore (19) by using the coordinates corresponding to the active enzyme (excluding the catalytic zinc cation) of the structure of hPCPA4 (to be reported elsewhere) as a searching model. The rotated and translated coordinates were refined against the high-resolution data set, and the position of the two catalytic zinc cations was determined by difference Fourier synthesis. These positions were used to compute experimental phases by using the three data sets of the multiple-wavelength anomalous diffraction experiment and the program mlphare in ccp4. These phases were combined with those from the Patterson search solutions and subjected to a density modification step under twofold averaging and phase extension. Subsequently, manual model building on an SGI Graphics workstation with turbo-frodo alternated with crystallographic refinement with refmac5 within ccp4, initially applying noncrystallographic symmetry restraints, until the final model was obtained. It encompasses protein residues Ser-3 to Leu-308 for each of the two hCPA4 molecules (chains A and C) and residues Met-1 to Lys-217 of the two latexin molecules (chains B and D). Each protease chain bears one N-glycosylation attached to Asn-148 Nδ2. A free-standing valine residue was found in each of the hCPA4 active sites (Val998A and Val998C). The two latexin/hCPA4 complexes present in the crystal asymmetric unit are structurally equivalent (rms deviation of 0.27 Å). Accordingly, discussion will consider only the complex formed by molecule A (hCPA4) and molecule B (latexin).
Table 1. Crystallographic statistics on data collection and refinement.
| Data set | hCPA4/latexin |
| Space group | P21 |
| Cell constants (a, b, c in Å; β in °) | 78.9, 96.7, 92.4, 99.6 |
| Wavelength, Å | 1.0067 |
| No. of measurements | 678,157 |
| No. of unique reflections | 179,401 |
| Resolution range, Å (outermost shell) | 48.3-1.60 (1.69-1.60)* |
| Completeness, % | 99.8 (99.3) |
| Rmerge† | 0.068 (0.257) |
| Average intensity (<[</σ(>)]>) | 11.8 (4.8) |
| B factor (Wilson), Å2 | 13.5 |
| Average multiplicity | 3.8 (3.7) |
| Resolution range used for refinement, Å | 48.3-1.60 |
| No. of reflections used (test set) | 178,706 (678) |
| Crystallographic Rfactor (free Rfactor)‡ | 0.149 (0.176) |
| No. of solvent molecules/ions/other molecules | 1,153/2 (Zn2+)/1 (acetone), 12 (methylpentanediol) |
| rms deviation from target values | |
| Bonds, Å | 0.011 |
| Angles, ° | 1.35 |
| Average B factors for protein (Å2) | 16.0 |
Values in parentheses refer to the outermost resolution shell if not otherwise indicated.
Rmerge=ΣhklΣi|Ii(hkl) - <I(hkl)>|/ΣhklΣiIi(hkl), where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and <I(hkl)> is its average.
Rfactor = Σhkl∥Fobs|-k|Fcalc∥/Σhkl|Fobs|, with Fobs and Fcalc as the observed and calculated structure factor amplitudes; free Rfactor, same for a test set of reflections (> 500) not used during refinement.
Results and Discussion
Vertebrate A/B MCPs Are Inhibited by Latexin. Although the gene of hCPA4 was reported to code for a member of the MCP family (10), no direct studies at the protein level had been performed so far. The recombinant protein zymogen can be activated by trypsin. Inhibition studies with human latexin show that the mature form, hCPA4, is strongly inhibited in a noncompetitive manner, as are all of the vertebrate A/B-type MCPs tested (see Table 2). The kinetic inhibition constants (Ki), calculated by pre-steady-state analysis, are in the nanomolar range and are similar to those obtained with the inhibitor from leech. The Ki values were confirmed by parallel steady-state measurements that also indicated the noncompetitive nature of the inhibition. On the other hand, latexin does not inhibit members of the N/E class or an invertebrate A/B MCP from the cotton bollworm, H. armigera. The results of our inhibition studies compare well with those reported for rat latexin against various MCPs. The latter had also shown that rat latexin does not inhibit N/E MCPs, like mouse (m) CPH and hCPM, nor other metallopeptidases, like gluzincins, and serine proteases, like trypsin, chymotrypsin, elastase, and yeast CPY (3).
Table 2. Inhibition of MCPs by human latexin endogenous CP inhibitor.
| MCP | Inhibition constant Ki, nM |
|---|---|
| bCPA | 1.2 ± 0.2 |
| hCPA1 | 1.6 ± 0.2 |
| hCPA2 | 3.5 ± 0.3 |
| hCPB | 1.1 ± 0.1 |
| hCPA4 | 3.0 ± 0.3 |
| hTAFI | 1.8 ± 0.3 |
| haCP | No inhibition |
| hCPN | No inhibition |
TAFI, thrombin-activatable fibrinolysis inhibitor. Latexin does not inhibit dCPD2 and hCPE (L. D. Fricker, personal communication).
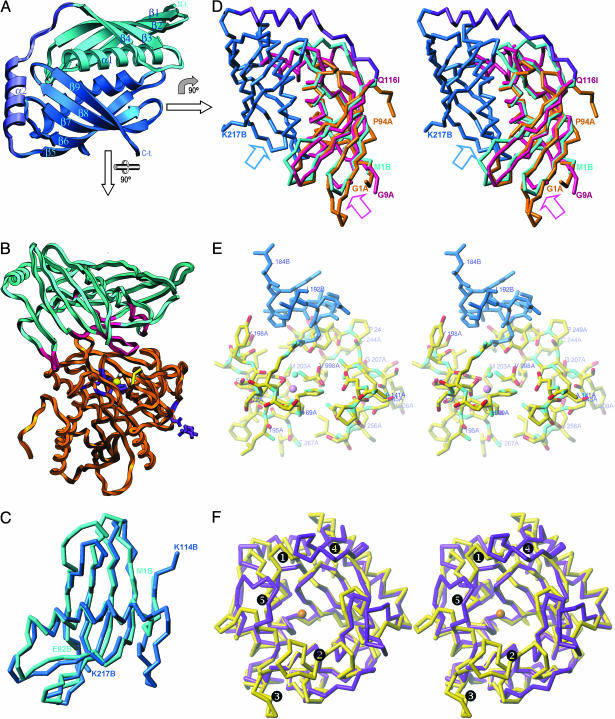
The Latexin Structure. Human latexin is elongated, with an α/β topology. It can be divided into two subdomains of same fold, an N-terminal subdomain (NTS; Met1B-Glu92B) and a C-terminal subdomain (CTS; Lys114B-Lys217B; see Figs. 1 and 2A). The structural similarity of the subdomains (Fig. 2C) means that they can be superimposed for 87 of their Cα atoms with an rms deviation of 2.1 Å, despite negligible sequence identity (14%). Each subdomain bears an extended α-helix (α1 in NTS and α3 in CTS) followed by a strongly twisted four-stranded antiparallel β-sheet of simple up-and-down connectivity (β1-β4 in NTS and β6-β9 in CTS) that embraces the helix establishing hydrophobic contacts. The subdomain topology is reminiscent of a left hand (Fig. 2C), with the helix resembling the thumb and the β-strands resembling the four fingers. The β-sheet of CTS contains an additional strand, β5, preceding the α-helix and running antiparallel to β6. The two subdomains are linked by a connecting segment (Gly93B-Met113B) that runs along the surface and contains helix α2. The overall molecular structure results from the packing of both subdomains against each other through the α-helices, running antiparallel to each other, with their axes ≈7.5 Å away and rotated ≈50° relative to each other. This arrangement positions both curved β-sheets on the molecular surface forming a flat, incomplete β-barrel. Two major surfaces can be distinguished on this barrel, an upper and a lower, each shaped by the central parts of helices α1 and α3 on opposite faces and the extremes of the β-strands (Fig. 2 A).
Fig. 1.
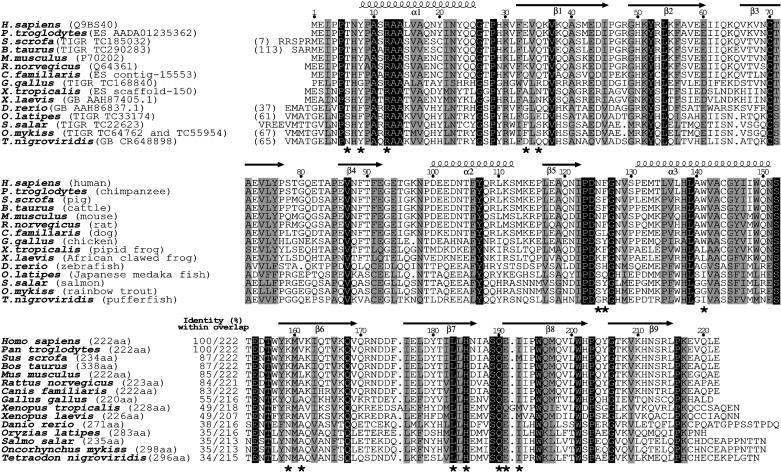
Sequence alignment of potential members (one per organism) of the latexin family of putative MCP inhibitors. Sequences were retrieved from Swiss-Prot/TrEMBL (www.expasy.ch), GenBank (GB; www.ncbi.nlm.nih.gov), TIGR (http://tigrblast.tigr.org/tgi), and ENSEMBL (ES; www.ensembl.org). The number of additional N-terminal protein residues is indicated in parentheses if appropriate. Some sequences are from unfinished genome projects and may be partial and still contain errors. They were compiled from EST libraries of diverse tissues and organs corresponding to different developmental stages (adult, fetal, and tailbud stages), among them from liver, kidney, gut, spleen, ovary, testis, brain, aorta, brain, cartilage, placenta, lung, thymus, and small intestine. Asterisks denote residues engaged in interactions with hCPA4. The length of each sequence, the identity to human latexin, and the number of overlapping residues are indicated. Absolutely conserved positions are shaded in black and conservatively substituted ones or those displaying two different residues are shaded in gray.
Fig. 2.
Structure of latexin and of its complex with hCPA4. (A) Ribbon plot of human latexin, view of the upper barrel surface. The N-terminal subdomain (cyan), the connecting segment (violet), and the C-terminal subdomain (blue) are shown. The constituting regular secondary structure elements are labeled (see also Fig. 1). (B) Ribbon plot of the inhibitor/enzyme complex showing hCPA4 in orange and latexin in cyan. Inhibitor segments contacting the protease are in magenta. The protein residues coordinating the catalytic zinc ion (yellow sphere) are violet, as is the glycosylation site (Asn148A). The catalytic solvent molecule is shown as a white sphere. (C) Superimposition of the NTS and CTS of latexin, with the same color coding as in A. (D) Stereo plot of the superimposition of human latexin (with the color coding as in A) with chicken egg-white cystatin (magenta; PDB ID code 1CEW) and sweet protein monellin (orange; PDB ID code 1MOL). The approximate position of a cysteine proteinase when inhibited by a cystatin [as inferred from the steffin/papain complex; PDB ID code 1STF (23)] is indicated by a magenta arrow, and the localization of hCPA4 in the complex with latexin is indicated by a blue arrow. (E) Closeup view in stereo of the section around the catalytic active-site cleft and the region of main peptidase/inhibitor interactions. Latexin is in blue and hCPA4 is colored according to atom type. The catalytic zinc ion (violet) and solvent molecule (cyan) are further shown as small spheres. (F) Stereo superimposition of the Cα backbone of hCPA4 (yellow), in its complex with latexin, and dCPD2 (violet; PDB ID code 1H81), as representatives of A/B- and N/E-type MCPs, respectively. The orientation corresponds to that in B rotated clockwise vertically 45° and then horizontally 90°. The regions shown comprise those shaping the rim of the funnel that leads to the active site of the enzymes, pinpointed by the catalytic zinc ion of hCPA4 (orange sphere). The five main insertions/deletions accounting for lack of interactions ( to
to  ) or potential steric clashes (
) or potential steric clashes ( to
to  ) with latexin on going from hCPA4 to dCPD2 are:
) with latexin on going from hCPA4 to dCPD2 are:  , Thr274A-Tyr277A;
, Thr274A-Tyr277A;  , Ser150A-Asn171A;
, Ser150A-Asn171A;  , Ser131A-Ile139A (all of hCPA4);
, Ser131A-Ile139A (all of hCPA4); 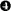 , Gln226-His241;
, Gln226-His241;  , Ser124-Val133 (both of dCPD2).
, Ser124-Val133 (both of dCPD2).
Structural Determinants for Latexin Inhibition. The mature hCPA4 enzyme shows the classical α/β-hydrolase fold of A/B MCPs, with a central mixed β-sheet flanked on both sides by several helices. This domain has a compact globular shape that has been hollowed out to render a funnel-like structure. The active-site cleft lies at the bottom of this funnel (Fig. 2 B, E, and F). The funnel rim is shaped by several loops that connect regular secondary structure elements and are responsible for interactions with the PD in hPCPA4 and protein inhibitors. The catalytic zinc ion is tetrahedrally coordinated by His69A, Glu72A, His196A, and a catalytic solvent molecule (Hoh501W), which in turn is further polarized by the side chain of the general base, Glu270A. Further residues traditionally identified as responsible for substrate binding and catalysis (8, 12) are Asn144A, Arg145A, Tyr248A, shaping active-site subsite S1′; Arg127A, and Glu270A for S1; Arg71A, Ser197A, Ty198A, and Ser199A for S2; and Phe279A for S3. The terminal carboxylate group of a substrate is fixed by Asn144A, Arg145A, and Tyr248A, whereas the scissile carbonyl group is near Glu270A, Arg127A, and the catalytic zinc. Typical CPA-like specificity toward hydrophobic side chains in substrates is accomplished by a hydrophobic S1′ pocket, shaped by the side chains of Met203A, Thr243A, Val247A, Ala250A, Ile255A, Thr268A, and Tyr248A.
In the latexin/hCPA4 complex, the inhibitor sits on top of the funnel rim, mainly clamping a loop encompassing residues Asp273A-Pro282A of the protease moiety through its lower barrel surface at the subdomain interface (Fig. 2B). The complex covers a surface of 2,340 Å2 at the protein interface, a higher value than typical protease-inhibitor interfaces that span ≈1,500 Å2 (20). However, complex formation implies rather few interactions. Forty-eight intermolecular contacts of <4 Å are observed, including 13 hydrogen bonds and 7 hydrophobic interactions. This finding is because of the shapes of the surfaces involved that are rather shallow in the present complex. In contrast to other protein inhibitors of metalloproteases, inhibition by latexin does not involve any of its termini. It is mainly caused by an inhibitory loop provided by CTS, shaped by the end of strand β7, the beginning of β8, and the connecting loop β7β8, which protrudes slightly into the protease moiety. In this respect, latexin is more reminiscent of cystatins that also inhibit via a β-ribbon structure (see below). Gln190B Nε2, at the tip of the inhibitory loop, is the part of latexin that approaches the active site most closely (up to 5.8 Å from the catalytic zinc ion; see Fig. 2E). In the latter, a free valine (Val998A), not found in the zymogen and possibly left behind after a proteolytic event during purification, fills the specificity pocket. Of particular importance for the complex stability is the interaction of Gln190B with Arg71A (Gln190B Oε1-Arg71A Nη2). This basic residue is present throughout A/B MCPs, and contributes to the maintenance of a pivotal salt bridge with the PDs of PCPs. It is, however, absent in N/E-forms that are not secreted as proenzymes. Gln190B also interacts through its Nε2 atom with atom Oη of Tyr248A. The latter residue side chain is in the down conformation as observed in other CPs and PCPs with an occupied specificity pocket (21, 22). The position of the former glutamine is maintained by an intramolecular interaction with the Nε2 atom of the preceding His185B. This histidine also contacts, edge to face, the side chain of Phe279A and Tyr198A Oη, the latter via its other hydrogen-bond donor, Nδ1. Further important interactions of the inhibitory loop are also the ones established by Glu191B Oε1, contacting the main-chain nitrogen of hCPA4 Glu163A, and by both Ile192B and Leu183B, which approach the side chain of Leu125A of the protease. The position of the inhibitory loop is fixed in latexin through intramolecular main-chain interactions with the neighboring strands β6 and β9, and loop α3β6. Here, Val161B forms a hydrophobic interaction with Leu125A of the protease, and Lys159B Nζ establishes a hydrogen bond with the main chain of Thr274A of hCPA4. Other intermolecular interactions stabilizing the complex encompass latexin loop β5α3 of the CTS, which approaches the funnel border of the protease. In particular, the tip of this loop, centered on Phe126B, contacts Arg124A, Trp73A, and, weakly, Ala283A. Two more regions, belonging to the NTS of latexin, are engaged in contacts, the beginning of helix α1 and a preceding residue, and the adjacent central part of strand β1. Here, we observe two hydrogen bonds and one hydrophobic interaction (Tyr8B N-Thr245A O, Arg12B Nη1-Val247A O, and Thr6B Cγ2-Gln239A Cβ) and three hydrogen bonds (Glu33B Oε2-Thr274A Oγ1, Glu33B Oε2-Thr276A Oγ1, and Gln35B Nε2-Glu237A Oε2), respectively. Finally, there is an additional isolated hydrophobic contact is encountered between Trp141B and the methyl group of Thr276A.
The interaction of latexin with hCPA4 may explain why A/B MCPs, secreted with the characteristic PD, can be inhibited by latexin whereas those that are not, N/E MCPs like hCPM and duck (d) CPD2, are not inhibited. There are a series of loops in the regions forming the funnel rim that are distinct for either the A/B or the N/E MCPs. Potential steric clashes and lack of interactions would be responsible for the selective inhibitory profile of latexin. The absence of a long insertion in hCPA4, Ser150A-Asn171A, reduced to Ala142-Ser149 in hCPM and Gln150-Pro157 in dCPD2 (see Fig. 2F), and the adjacent Ser131A-Ile139A, contributing in hCPA4 to back the previous loop and short-circuited to a single residue in hCPM (Asn-131) and dCPD2 (Asn-139), drastically reduce the possibility of interactions. The same holds for the loop region Thr274A-Tyr277A in hCPA4, absent in hCPM and dCPD2. However, the most important features are two characteristic loop insertions of N/E MCPs, Lys221-Asn233 (hCPM) or Gln226-His241 (dCPD2), and Val116-Ser124 (hCPM) or Ser124-Val133 (dCPD2), which would sterically clash with the inhibitor. In the invertebrate haCPA, which is not inhibited by latexin despite being an A/B MCP, the main structural difference observed with hCPA4 is an insertion in the former between the positions equivalent to Leu271A and Gly278A of the latter. This results in a loop that is four residues longer, which would collide with latexin helix α3.
Structural Homologues of Latexin. Latexin shows some functional similarities with cystatins in the way it inhibits its target, and bioinformatic searches have actually identified chicken egg-white cystatin [Protein Data Bank (PDB) ID code 1CEW] and monellin (PDB ID code 1MOL) as close structural homologues of latexin (Fig. 2D). Monellin is the main agent responsible for the sweet taste of plant berries and has the same fold as cystatins. The structural similarity of latexin with the latter encompasses one of the two subdomains (87 common Cα atoms, rms deviation of 2.2 Å, and 16% sequence identity). The motif comprising the α-helix and the wrapping β-sheets can be superimposed. The main difference is that cystatin has an additional α-helix, whereas monellin has shorter strands. Inhibition of cysteine proteinases through cystatins, as observed in the steffin/papain complex (23), occurs through the tips of the fingers, the loop connecting the strands equivalent to β1β2 and β3β4 of latexin NTS, or β6β7 and β8β9 considering CTS, and through the N terminus. In contrast, latexin inhibits through the interface between the two subdomains on the opposite face of the inhibitor (Fig. 2D). Furthermore, slight conformational differences, mainly because of deletions/insertions in both loop pairs of NTS and CTS, suggest that these should not inhibit cysteine proteinases. This finding was corroborated experimentally employing papain (data not shown).
The Latexin Family of Potential A/B MCP Inhibitors. Bioinformatic searches disclose a number of putative proteins with significant sequence similarity to human latexin, ranging from 34% to 100% identity (Fig. 1). Such sequences are found in mammalians and in other vertebrates like frogs, birds, and fish. The residues mainly engaged in inhibition are mostly conserved, conservatively substituted, in particular, around the inhibitory loop, or employ backbone atoms. Moreover, the tissue distribution analyses of mammalian latexins and the broad scattering in the EST libraries strongly suggest temporal and spatial ubiquity in the potential expression of these genes. In addition to the rodent orthologues of human latexin, a protein with similar sequence that has been studied at the biochemical level is ovocalyxin-32, a chicken eggshell matrix protein highly expressed and secreted by surface epithelial cells during eggshell formation (Swiss-Prot accession no. Q90YI1, 31% identity, ref. 24). Further similarity can be found to a protein whose expression is induced by retinoids in skin raft cultures, RARRES1 (Swiss-Prot accession no. P49788, 30% identity; ref. 25). In this case, RARRES1 could be a response to hyperproliferative and inflammatory processes like skin diseases, putatively acting against MCPs engaged in inflammation. We suggest including all these sequences within the latexin family of potential MCP inhibitors.
Conclusions
The strong similarity in structure of the latexin subdomains with cystatins suggests common ancestry. Genetic duplication of such a cystatin module, including the incorporation of a 41-residue linker and subsequent sequence divergence, may have given rise to the structure of latexin. However, different regions are involved in inhibition in cystatins and latexin. The action of the latter seems to be limited to vertebrate A/B MCPs, because invertebrate A/B MCPs and N/E subfamily MCPs are not inhibited. Accordingly, structural determinants simultaneously confer specificity for the A/B subfamily and pose a diversification between vertebrate and invertebrate forms but also sufficient variability to strongly inhibit all vertebrate forms tested, even across the species barrier.
There is a parallel evolution in the studies of MCPs and studies of matrix metalloproteinases (MMPs) that are zinc-dependent metalloendopeptidases. The latter were first identified because of their unspecific degrading activity toward extracellular matrix components in vertebrates. They were localized to the extracellular space. However, the potential roles of MMPs were reexamined when they were implicated in limited proteolysis events, as in ectodomain shedding. Furthermore, they were shown to be expressed in several organs and tissues and also in non-vertebrates, even possibly in bacteria and viruses (see ref. 26 and references therein). Likewise, the perceived roles of MCPs have evolved from indiscriminate protein digestive enzymes to sophisticated regulators associated with selective limited proteolysis. MMPs are inhibited by their endogenous tissue inhibitors, the tissue inhibitor of metalloproteinases (TIMPs) that display complementary spatial and temporal expression patterns, as do A/B MCPs and latexins. All these findings suggest that latexins could represent to A/B MCPs the same as TIMPs to MMPs, namely their endogenous inhibitors in vertebrates.
Acknowledgments
We thank Robin Rycroft for helpful contributions to the manuscript and Drs. Enghild and Valnickova (Aarhus University, Aarhus, Denmark) for a gift of human thrombin-activatable fibrinolysis inhibitor. This work was supported by Spanish Ministry for Science and Technology Grants BIO2003-00132, GEN2003-20642, and BIO2001-2046 and “FundacióLa Caixa” Grant ON03-7-0. Further support was provided by Commission of the European Communities Grant LSHC-CT-2003-503297 “CANCERDEGRADOME.” The assistance provided by European Molecular Biology Laboratory and European Synchrotron Radiation Facility synchrotron local contacts during data collection is further acknowledged. For these data collections, funding was provided by the European Synchrotron Radiation Facility and by European Union Grant RII3-CT-2004–506008.
Author contributions: F.X.A., J.V., and F.X.G.-R. designed research; I.P., R.B., R.G.-C., S.V., and F.X.G.-R. performed research; J.V., S.V., and F.X.G.-R. analyzed data; and F.X.A., J.V., and F.X.G.-R. wrote the paper.
Abbreviations: MCP, metallocarboxypeptidase; CP, carboxypeptidase; b, bovine; d, duck; h, human; m, mouse; ha, cotton bollworm Helicoverpa armigera; PCP, procarboxypeptidase; NTS, N-terminal subdomain; CTS, C-terminal subdomain; p, porcine; PD, prodomain; PDB, Protein Data Bank.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2BK7).
Note Added in Proof. At the proof stage of this publication, the structure analysis of mouse latexin was reported (27).
References
- 1.Mirnics, Z. K., Mirnics, K., Terrano, D., Lewis, D. A., Sisodia, S. S. & Schor, N. F. (2003) Mol. Psychiatry 8, 863–878. [DOI] [PubMed] [Google Scholar]
- 2.Hatanaka, Y., Uratani, Y., Takiguchi-Hayashi, K., Omori, A., Sato, K., Miyamoto, M. & Arimatsu, Y. (1994) Eur. J. Neurosci. 6, 973–982. [DOI] [PubMed] [Google Scholar]
- 3.Normant, E., Martres, M. P., Schwartz, J. C. & Gros, C. (1995) Proc. Natl. Acad. Sci. USA 92, 12225–12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Normant, E., Gros, C. & Schwartz, J. C. (1995) J. Biol. Chem. 270, 20543–20549. [DOI] [PubMed] [Google Scholar]
- 5.Ji, B., Chen, X. Q., Misek, D. E., Kuick, R., Hanash, S., Ernst, S., Najarian, R. & Logsdon, C. D. (2003) Physiol. Genomics 14, 59–72. [DOI] [PubMed] [Google Scholar]
- 6.Liu, Q., Yu, L., Gao, J., Fu, Q., Zhang, J., Zhang, P., Chen, J. & Zhao, S. (2000) Mol. Biol. Rep. 27, 241–246. [DOI] [PubMed] [Google Scholar]
- 7.Waldschmidt-Leitz, E. & Purr, A. (1929) Ber. Dt. Chem. Ges. 62, 2217–2226. [Google Scholar]
- 8.Auld, D. S. (2004) in Handbook of Proteolytic Enzymes., eds. Barrett, A. J., Rawlings, N. D. & Woessner, J. F., Jr. (Elsevier, London), Vol. 1, pp. 812–821. [Google Scholar]
- 9.Springman, E. B. (2004) in Handbook of Proteolytic Enzymes., eds. Barrett, A. J., Rawlings, N. D. & Woessner, J. F., Jr. (Elsevier, London), Vol. 1, pp. 828–830. [Google Scholar]
- 10.Huang, H., Reed, C. P., Zhang, J. S., Shridhar, V., Wang, L. & Smith, D. I. (1999) Cancer Res. 59, 2981–2988. [PubMed] [Google Scholar]
- 11.Kayashima, T., Yamasaki, K., Yamada, T., Sakai, H., Miwa, N., Ohta, T., Yoshiura, K., Matsumoto, N., Nakane, Y., Kanetake, H., et al. (2003) Hum. Genet. 112, 220–226. [DOI] [PubMed] [Google Scholar]
- 12.Vendrell, J., Querol, E. & Avilés, F. X. (2000) Biochim. Biophys. Acta 1477, 284–298. [DOI] [PubMed] [Google Scholar]
- 13.Arolas, J. L., Lorenzo, J., Rovira, A., Castella, J., Avilés, F. X. & Sommerhoff, C. P. (2004) J. Biol. Chem. 280, 3441–3448. [DOI] [PubMed] [Google Scholar]
- 14.Gomis-Rüth, F. X., Companys, V., Qian, Y., Fricker, L. D., Vendrell, J., Avilés, F. X. & Coll, M. (1999) EMBO J. 18, 5817–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reverter, D., Maskos, K., Tan, F., Skidgel, R. A. & Bode, W. (2004) J. Mol. Biol. 338, 257–269. [DOI] [PubMed] [Google Scholar]
- 16.Ventura, S., Villegas, V., Sterner, J., Larson, J., Vendrell, J., Hershberger, C. L. & Avilés, F. X. (1999) J. Biol. Chem. 28, 19925–19933. [DOI] [PubMed] [Google Scholar]
- 17.Morrison, J. F. (1982) Trends Biochem. Sci. 7, 102–105. [Google Scholar]
- 18.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–776.15299374 [Google Scholar]
- 19.Navaza, J. (1994) Acta. Crystallogr. A 50, 157–163. [Google Scholar]
- 20.Janin, J. & Chothia, C. (1990) J. Biol. Chem. 265, 16027–16030. [PubMed] [Google Scholar]
- 21.Guasch, A., Coll, M., Avilés, F. X. & Huber, R. (1992) J. Mol. Biol. 224, 141–157. [DOI] [PubMed] [Google Scholar]
- 22.García-Sáez, I., Reverter, D., Vendrell, J., Avilés, F. X. & Coll, M. (1997) EMBO J. 16, 6906–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stubbs, M. T., Laber, B., Bode, W., Huber, R., Jerala, R., Lenarcic, B. & Turk, V. (1990) EMBO J. 9, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautron, J., Hincke, M. T., Mann, K., Panheleux, M., Bain, M., McKee, M. D., Solomon, S. E. & Nys, Y. (2001) J. Biol. Chem. 276, 39243–39252. [DOI] [PubMed] [Google Scholar]
- 25.Nagpal, S., Patel, S., Asano, A. T., Johnson, A. T., Duvic, M. & Chandraratna, R. A. (1996) J. Invest. Dermatol. 106, 269–274. [DOI] [PubMed] [Google Scholar]
- 26.Gomis-Rüth, F. X. (2003) Mol. Biotechnol. 24, 157–202. [DOI] [PubMed] [Google Scholar]
- 27.Aagaard, A., Listwan, P., Cowieson, N., Huber, T., Ravasi, T., Wells, C. A., Flanagan, J. U., Kellie, S., Hume, D. A., Kobe, B. & Martin, J. L. (2005) Structure (London) 13, 309–317. [DOI] [PubMed] [Google Scholar]