Abstract
IL-22 is an IL-10 homologue that binds to and signals through the class II cytokine receptor heterodimer IL-22RA1/CRF2–4. IL-22 is produced by T cells and induces the production of acute-phase reactants in vitro and in vivo, suggesting its involvement in inflammation. Here we report the identification of a class II cytokine receptor designated IL-22RA2 (IL-22 receptor-α 2) that appears to be a naturally expressed soluble receptor. IL-22RA2 shares amino acid sequence homology with IL-22RA1 (also known as IL-22R, zcytor11, and CRF2–9) and is physically adjacent to IL-20Rα and IFN-γR1 on chromosome 6q23.3–24.2. We demonstrate that IL-22RA2 binds specifically to IL-22 and neutralizes IL-22-induced proliferation of BaF3 cells expressing IL-22 receptor subunits. IL-22RA2 mRNA is highly expressed in placenta and spleen by Northern blotting. PCR analysis using RNA from various tissues and cell lines showed that IL-22RA2 was expressed in a range of tissues, including those in the digestive, female reproductive, and immune systems. In situ hybridization revealed the dominant cell types expressing IL-22RA2 were mononuclear cells and epithelium. Because IL-22 induces the expression of acute phase reactants, IL-22RA2 may play an important role as an IL-22 antagonist in the regulation of inflammatory responses.
Cytokines play important roles in the regulation of hematopoiesis and immune responses and can influence lymphocyte development. The human class II cytokine family includes IFN-α, IFN-β, and IFN-γ, as well as IL-10 and IL-10-related molecules such as IL-19 (1), MDA-7 (2), IL-20 (2), IL-22 (3), and AK-155 (4). These class II cytokines typically bind and signal through class II cytokine receptors. Members of the human class II cytokine receptor family include IFN-αR1, IFN-αR2, IFN-γR1, IFN-γR2, IL-10R (5), CRF2–4 (6), IL-20Rα (7) [also known as zcytor7 (8) and CRF2–8 (9)], IL-20Rβ (7) [also known as DIRS1 (10)], IL-22RA1 (IL-22 receptor-α 1, submitted to The Human Genome Organization for approval) [also known as IL-22R (3), zcytor11 (11), and CRF2–9 (9)], and tissue factor.
Class II cytokine receptors are typically heterodimers composed of two distinct receptor chains, the α and β receptor subunits (12). In general, the α subunits are the primary cytokine binding proteins, and the β subunits are required for formation of high affinity binding sites, as well as for signal transduction. An exception is the IL-20 receptor in which both subunits are required for IL-20 binding (7).
The class II cytokine receptors are identified by a conserved cytokine-binding domain of about 200 aa (D200) in the extracellular portion of the receptor. This cytokine-binding domain is comprised of two fibronectin type III (FnIII) domains of ≈100 aa each (13, 14). Each FnIII domain contains conserved Cys, Pro, and Trp residues that determine a characteristic folding pattern of seven β-strands similar to the constant domain of immunoglobulins (15). The conserved structural elements of the class II cytokine receptor family make it possible to identify new members of this family on the basis of primary amino acid sequence homology. Previously we have successfully identified two new members of class II cytokine receptor family, zcytor7 (8) [also known as IL-20Rα (7)] and zcytor11 (11) [also known as IL-22R (3)], using this approach.
IL-22, or IL-TIF (IL-10-related T cell-derived inducible factor) (16), is a recently described IL-10 homologue. Mouse IL-22 was originally identified as a gene induced by IL-9 in T cells and mast cells in vitro (16). Acute phase reactant induction activity was observed in mouse liver upon IL-22 injection, and IL-22 expression was rapidly induced after lipopolysaccharide injection, suggesting that IL-22 contributes to the inflammatory response in vivo (17).
This report describes the cloning, chromosome localization, and expression pattern of a human class II cytokine receptor that binds and neutralizes IL-22 in a specific bioassay. This receptor, designated IL-22RA2 (IL-22 receptor-α 2, approved by The Human Genome Organization), appears to be expressed in a naturally occurring soluble form that represents a natural antagonist of IL-22 activity.
Materials and Methods
Radiation Hybrid Mapping of IL-22RA2.
Chromosome mapping was carried out by using a commercially available version of the Stanford G3 Human/Hamster Radiation Hybrid Mapping Panel (Research Genetics, Huntsville, AL) in conjunction with publicly available servers (http://shgc-www.stanford.edu/, http://www.ncbi.nlm.nih.gov/, and http://genome.UCSC.edu). The PCRs were carried out by using IL-22RA2-specific primers 5′-AGCTGCCTTCTTCACTTG-3′ and 5′-TTGCTCTGCCTCTTATTC-3′, which yield a 226-bp amplicon in the 3′ untranslated region. The PCR cycle conditions were as follows: an initial 1-cycle, 15-min denaturation at 95°C, followed by 35 cycles of a 1-min denaturation at 95°C, 1 min annealing at 56°C and 1 min and 15 sec extension at 72°C, and a final 1-cycle extension of 7 min at 72°C.
IL-22RA2-Fc4 Fusion Protein.
A PCR-generated IL-22RA2 DNA fragment encoding polypeptide Thr-22–Pro-231 was created by using PCR primers 5′-GCGGATCCACTCAGTCAACGCATGAGTCTCTG-3′ (BamHI site) and 5′-GCAGATCTTGGAATTTCCACACATCTCTCTTCA-3′ (BglII site). The excised DNA was subcloned into a mammalian expression vector Fc4/pzmp20 that is designed to attach a tPA leader to the N terminus of IL-22RA2 polypeptide and a Fc region derived from human IgG to the C terminus of IL-22RA2 polypeptide. The expression vector for IL-22RA2-Fc4 was transfected into BHK 570 cells (ATCC no. CRL-10314) by using Lipofectamine (GIBCO/BRL). Approximately 24 h posttransfection, the BHK cells were split into selection media with 1 μM methotrexate, and a stable BHK/IL-22RA2-Fc4 cell line was generated by repeat splitting. The IL-22RA2-Fc4 fusion protein was purified from serum-free conditioned media using Protein A affinity chromatography.
IL-22-CEE Fusion Protein.
The cDNA for human IL-22 coding region without its stop codon was amplified by PCR amplification using primers 5′-TTGGGTACCTCTGCAATGGCCGCCCTGCAGAAATCT-3′ (KpnI site) and 5′-TTGGGATCCAATGCAGGCATTTCTCAGAGACAT-3′ (BamHI site) and then inserted into the CEE/pHZ200 mammalian expression vector. CEE is a C-terminal Glu-Glu sequence tag with the sequence Glu-Tyr-Met-Pro-Met-Glu. The IL-22-CEE/pHZ200 vector was transfected into BHK cells and the stable BHK/IL-22-CEE was generated as described above. The IL-22-CEE fusion protein was purified from filtered BHK serum-free conditioned media using anti-EE Protein G-Sepharose and Sephadex S200 (Amersham Pharmacia) chromatography.
Cell Binding Assay.
A modified cell binding assay (18) procedure was performed on COS cells transfected with candidate class II cytokine cDNAs using IL-22RA2-Fc4 fusion protein as a probe. Briefly, COS cells were fixed with 1.8% formaldehyde, permeabilized with 0.1% Triton-X, then incubated with 1 μg/ml IL-22RA2-Fc4 for 1 h, washed, and incubated with goat-anti-human Ig-horseradish peroxidase (Jackson ImmunoResearch) for another hour. The positive signal was amplified with fluorescein tyramide reagent (NEN) and visualized by using a FITC filter on a fluorescent microscope.
BaF3 Stable Cell Line Expressing CRF2–4 and IL-22RA1.
An IL-3-dependent murine pre-B cell line BaF3 (19) was transfected by electroporation with expression vectors containing CRF2–4. The transfected BaF3 cells were split into selection media with 2 μg/ml puromycin to isolate the puromycin-resistant transfectants. Pools of the transfected BaF3 cells, hereinafter called BaF3/CRF2–4, were further transfected with IL-22RA1 expression vector using the same approach and selected in media containing 200 μg/ml zeocin. The Baf3/CRF2–4 cells expressing IL22RA1 were designated as BaF3/CRF2–4/IL-22RA1. The expression of both receptors was confirmed by reverse transcriptase–PCR.
BaF3 Proliferation Assay.
BaF3/CRF2–4/IL-22RA1 cells were washed, resuspended in IL-3-free media, and plated into a 96-well format at about 5,000 cells per well. Serial dilutions of IL-22-CEE protein were made in IL-3 free media and added to cells. The assay plates were incubated at 37°C, 5% CO2 for 3 days, then Alamar Blue (Accumed, Chicago) was added. After additional 24-h incubation, plates were read on a Fmax plate reader (Molecular Devices) by using the softmax pro program at 544 nm (excitation) and 590 nm (emission).
Northern Blot.
Northern blot analysis was performed by using human multiple tissue Northern blots I, II, and III (CLONTECH) and a monocyte U937 Northern blot (ZymoGenetics). A 364-bp cDNA probe for human IL-22RA2 was generated by PCR amplification using oligonucleotides 5′-AGTCAACGCATGAGTCTCTGAAG-3′ and 5′-ACCAACAAAGAGCCATTGACTTG-3′. Blots were hybridized overnight in ExpressHyb (CLONTECH) with a 32P random-labeled probe at 65°C, and then washed twice for 30 min in 0.1 × SSC, 0.1% SDS at 55°C, followed by exposure to x-ray film.
OriGene Assay.
Reverse transcriptase–PCR was performed on Rapid-Scan gene expression panels (OriGene Technologies, Rockville, MD). Rapid-Scan is a PCR-based system, using first-strand cDNAs derived from different tissues to generate an expression profile for the gene of interest. Individual first-strand cDNAs have been normalized by using human β-actin as an internal standard. Oligonucleotides 5′-TGGGAGGGCACTTACTGGCAACA-3′ and 5′-CTCTGTGAGCCCCTTCATAAACC-3′ were used to generate a 431-bp IL-22RA2 fragment. PCR conditions were 94°C for 2 min followed by 35 cycles of 94°C/15 sec, 65°C/30 sec, and 72°C/1 min, followed by 72°C for 2 min.
In Situ Hybridization.
In situ hybridization analysis was performed as described (7). A cDNA plasmid template, corresponding to nucleotides 237–930 of human IL-22RA2 (Fig. 1A), was constructed. A digoxygenin-labeled antisense RNA probe was generated by using T3 RNA polymerase and an in vitro transcription system (Promega). Hybridization was carried out at 60–65°C overnight in a 50% formamide solution, and slides were subsequently washed in 0.1 × SSC at 55°C. The signals were amplified with 2–3 rounds of tyramide signal amplification (TSA in situ indirect kit, NEN) and visualized with a Vector Red substrate kit (Vector Laboratories). Tissue slides were counterstained with hematoxylin (Vector Laboratories).
Figure 1.
IL-22RA2 encodes a soluble class II cytokine receptor. (A) Nucleotide and deduced amino acid sequence for human IL-22RA2. The predicted signal peptide is underlined. The upstream in-frame stop codon (TGA) is double-underlined. The potential N-linked glycosylation sites are underlined with dotted lines. The potential mRNA instability motif is underlined with a dashed line. (B) Alignment of deduced human IL-22RA2 and IL-22RA1 aa sequences using the Smith–Waterman algorithm (31). Colons and periods indicate identical amino acids and conservative substitutions, respectively. The FnIII domains are labeled.
Results and Discussion
Identification, Cloning, and Structural Analysis of IL-22RA2.
Scanning of a translated human genomic database resulted in the identification of a class II cytokine receptor that is highly related to IL22RA1 and IL-20Rα. Using the predicted cDNA sequence, a partial clone from a tonsil cDNA library (ZymoGenetics) was identified. Based on this confirmed sequence, nested oligonucleotides were designed, and 5′ and 3′ rapid amplification of cDNA ends (20) was performed on RNA isolated from human placenta and the monocytic cell line U937, yielding a 1.6-kb full-length cDNA sequence (Fig. 1A). The cDNA sequence, designated as IL-22RA2, revealed a 5′ untranslated region (nucleotides 1–236), an ORF encoding a 231-aa protein (nucleotides 237–932), and a 3′ untranslated region (nucleotides 933-1610) that contains a mRNA instability motif (21) and a poly(A) tail (Fig. 1A). There is an in-frame stop codon (TGA) 21-nt upstream of the first Met that is embedded in a consensus Kozak sequence typical of translation initiation in vertebrates (22).
IL-22RA2 contains a 21-aa signal peptide based on the signal peptidase cleavage rules (23), but the mature protein does not appear to have a transmembrane domain. IL-22RA2 is composed of a single characteristic cytokine-binding domain (D200) that contains two FnIII domains (13, 14). It shows sequence similarity with class II cytokine receptors, sharing the highest amino acid sequence identity with IL-22RA1, particularly in the first FnIII domain (Fig. 1B).
Several naturally occurring soluble forms of cytokine receptors have been described (24–26). They are generally truncated forms of the cellular receptor that retain the extracellular ligand-binding portion but lack the transmembrane and intracellular domains. Some soluble receptors arise from the proteolytic cleavage of the membrane-bound receptors, some are synthesized by alternatively spliced mRNAs, and others may be encoded by viruses and secreted after viral infection of cells. For example, the soluble isoform of IFN-αR2, designated as IFNAR 2a, is generated by alternative mRNA splicing of the IFN-αR2 gene (27). Most naturally occurring soluble receptors are within the class I cytokine receptor family, and are not the predominant form. However, it appears that IL-22RA2 is the first example in the class II cytokine receptor family for which the predominant and perhaps only naturally occurring form is a soluble receptor. We have tried by using 3′ rapid amplification of cDNA ends on various RNA sources to isolate an IL-22RA2 isoform containing a transmembrane domain, and thus far we have only obtained sequences encoding the soluble form (data not shown). That IL-22RA2 is predominantly a naturally occurring soluble receptor is also consistent with our observation that an orthologous murine IL-22RA2 full-length cDNA (unpublished data) also encodes a soluble receptor lacking an apparent transmembrane domain.
IL-22RA2 Maps to a Class II Cytokine Receptor Cluster.
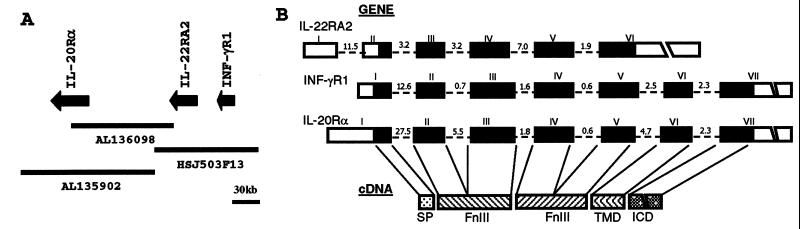
Radiation hybrid mapping was used to map IL-22RA2 to human chromosome 6q23.3–24.2, which is the same chromosomal region for IFN-γR1 (28) and IL-20Rα (8). This chromosome region is associated with various disorders, including neonatal, type II, and type I diabetes mellitus (29). More detailed analysis using publicly available genomic sequences allowed us to deduce the gene order of these cytokine receptors on chromosome 6q as centromere—IL-20Rα, IL-22RA2, IFN-γR1—telomere in the same transcriptional orientation (Fig. 2A). The distance between IL-22RA2 and IFN-γR1 is about 24 kb from the end of last exon of IFN-γR1 to the beginning of the first exon of IL-22RA2, and the distance between IL-22RA2 and IL-20Rα is about 100 kb. Interestingly, the ligands for IL-22RA2 and IFN-γR1 (IL-22 and IFN-γ) also are located within about 90 kb of each other in the same chromosomal region around 12q15 (17).
Figure 2.
IL-22RA2 shares the same chromosome location and exon-intron organization with IFN-γR1 and IL-20Rα. (A) Chromosome mapping of IL-22RA2. IL-22RA2 maps to human chromosome 6q23.3–24.2 and is tightly linked to IFN-γR1 and IL-20Rα. The genes for IL-22RA2, IFN-γR1, and IL-20Rα are indicated by arrows. (B) Exon-intron organization of the genes for IL-22RA2, IFN-γR1, and IL-20Rα. Exon and intron sequences are represented by boxes and dashed lines, respectively. Coding and untranslated regions are represented by filled boxes and open boxes, respectively. The exons are numbered and the sizes of introns are shown in kb. The common feature of all three cDNAs is schematically represented below. SP, signal peptide; TMD, transmembrane domain; ICD, intracellular domain.
Comparison of the IL-22RA2 cDNA with its genomic sequence revealed that the IL-22RA2 gene is organized into six exons and spans about 29 kb of genomic DNA. The intron insertion sites correlate with the junction between the signal peptide and the two FnIII domains. The exon-intron organization of IL-22RA2 gene resembles IFN-γR1 and IL-20Rα (Fig. 2B). The mapping studies indicate that at least three members of the class II cytokine receptor family comprise a cytokine receptor cluster. Clustering of family members may be the result of gene duplication with subsequent divergence of function and regulation. The known activities for the three class II cytokine receptor family members within this cluster reflect this functional divergence. IL-20Rα is a subunit for IL-20 receptor heterodimer, IFN-γR1 is the binding subunit for IFN-γ receptor heterodimer, whereas IL-22RA2 is a soluble receptor that binds IL-22 and acts as an IL-22 antagonist (see below). It will be interesting to see whether there are any additional class II cytokine receptor genes located within this chromosome region.
IL-22RA2 Binds to IL-22.
Because IL-22RA2 is a class II cytokine receptor, the binding of an IL-22RA2-Fc4 fusion protein to many of the known or orphan class II cytokines was tested. Expression vectors encoding human IFN-α, IFN-β, IFN-γ, IL-10, IL-19, IL-20, IL-22, MDA-7, AK-155, and mouse IL-22 were transiently transfected into COS cells, and the binding of IL-22RA2-Fc4 fusion protein to transfected COS cells was carried out by using a fluorescence-based binding assay. The expression of these cytokines in COS cells was confirmed by either (i) the binding of their soluble receptors (e.g., IL-10R soluble receptor Fc4 fusion protein binds COS cells transfected with IL-10 cDNA), or (ii) positively staining with mAb against EE-tag COS cells transfected with epitope-tagged cytokine cDNA (e.g., COS cells transfected with IL-22-CEE cDNA). Among the cytokines tested, only human and mouse IL-22 showed positive binding, indicating IL-22 and IL-22RA2 are a potential ligand-receptor pair. Because IL-22RA2 and IL-22RA1 share the highest homology within the first FnIII domain (Fig. 1B) and both bind to IL-22, it is likely that the first FnIII domain confers ligand-binding specificity.
IL-22RA2 Is an IL-22 Antagonist.
IL-22 is an IL-10 homologue that signals through a heterodimeric IL-22RA1/CRF2–4 receptor (3, 16, 17, 30). CRF2–4 is a shared signaling subunit for both IL-10 and IL-22 heterodimeric receptors (3), and IL-10 can induce a proliferative response in murine pre-B BaF3 cell line transfected with IL-10R1. Moreover, IL-22 can induce activation of STAT proteins (STAT1, STAT3, and STAT5) (16, 17). Therefore, it is reasonable to speculate that IL-22 might be able to deliver a proliferative signal in BaF3 cells that express the IL-22 receptors. To determine whether the binding of IL-22RA2 to IL-22 results in the neutralization of IL-22 activity, a BaF3 stable cell line that expresses IL-22 receptor (i.e., IL-22RA1 and CRF2–4) was generated as a functional assay for IL-22.
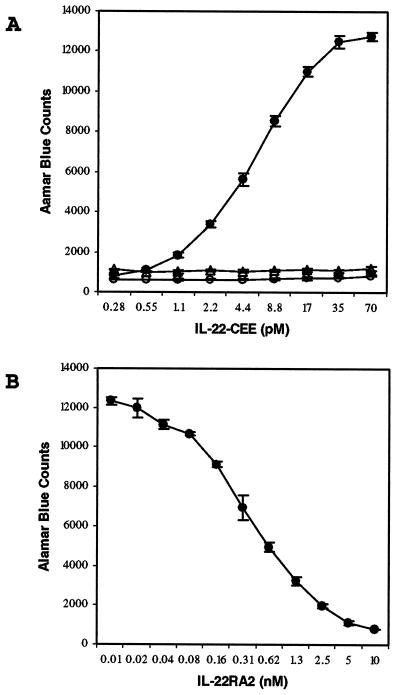
IL-22 protein was generated as a C-terminal Glu-Glu-tagged fusion protein (IL-22-CEE) in BHK cells. The purified protein migrated on a SDS/PAGE gel as four distinct bands in the region of about 25–35 kDa, in accordance with the three potential N-linked glycosylation sites in human IL-22 (3). When added to BaF3/CRF2–4/IL-22RA1 cells, IL-22-CEE induced a dose-dependent proliferative activity with half-maximal response at around 6 pM, whereas IL-22-CEE was inactive on control BaF3/CRF2–4 cells. The addition of about 10 nM IL-22RA2-Fc4 fusion protein completely blocked IL-22 activity (Fig. 3A), whereas it did not have any effect on BaF3 cell proliferation stimulated by IL-3, or BaF3/IL-20Rα/IL-20Rβ cell proliferation stimulated by IL-20 (data not shown). Therefore, IL-22RA2 specifically neutralized IL-22 activity. To test dose-dependent inhibition of IL-22 activity, serial dilutions of IL-22RA2-Fc4 were added into BaF3/CRF2–4/IL-22RA1 cell cultures containing 60 pM IL-22-CEE. The data revealed that IL-22RA2-Fc4 effectively inhibited IL-22-induced proliferation by 50% at a concentration of ≈0.4 nM (Fig. 3B). Thus IL-22RA2 not only binds to IL-22, but also antagonizes IL-22 activity in vitro. Although IL-22RA2 is clearly an IL-22 antagonist in vitro, its in vivo activity and physiological role in regulating IL-22 still needs further investigation, because some cytokine soluble receptor may actually potentiate the activity of their own cytokines in vivo.
Figure 3.
IL-22RA2 neutralizes the IL-22 activity in a BaF3 proliferation assay. (A) IL-22-CEE shows dose-dependent proliferative activity on BaF3 cells coexpressing IL-22RA1 and CRF2–4 (●), but not CRF2–4 alone (▵). The IL-22-CEE activity is completely blocked with the addition of 10 nM IL-22RA2-Fc4 fusion protein (○). (B) Dose-dependent inhibition of 60 pM IL-22-CEE activity by IL-22RA2-Fc4. Error bars indicate the standard deviation of triplicate samples.
Tissue Distribution of IL-22RA2 mRNA.
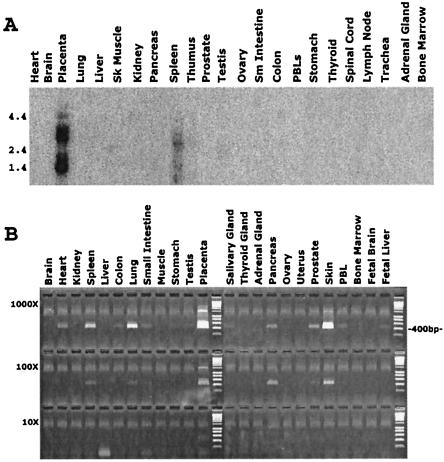
The expression pattern of human IL-22RA2 gene was examined by using Northern blot analysis. Transcripts of ≈1.6-kb and 3.0-kb size were detected in placenta and spleen but not in other tissues (Fig. 4A). The same size transcripts plus an additional ≈1.2-kb transcript were detected in the U937 cell line (data not shown). The different size of transcripts may be due to alternative mRNA splicing.
Figure 4.
Tissue distribution of human IL-22RA2. (A) Northern blot of human multiple tissue mRNA hybridized to an IL-22RA2 probe. Tissue sources are given at the top and the positions of size markers (in kb) are indicated on the left. The predominant hybridizing species correspond to transcripts of about 1.6 and 3.0 kb. (B) PCR analysis of IL-22RA2 mRNA level in human tissues. The first-strand cDNAs from each human tissue were subject to normalization, such that they all contain an equivalent concentration of β-actin cDNA. Each cDNA was diluted in water to a series of three concentrations (labeled 1,000×, 100×, 10×), with the lowest concentration (10×) being ≈10pg. The 400-bp marker is labeled at the right.
The relative level of IL-22RA2 mRNA accumulation in various tissues also was analyzed by semiquantitative PCR using a serially diluted cDNA array (OriGene Technologies). IL-22RA2 is most highly expressed in placenta, spleen, skin, and lung, with lower expression in a variety of other tissues such as heart, pancreas, and prostate (Fig. 4B). Additional cDNA and RNA panels (ZymoGenetics) from various normal and cancerous human tissues and cell lines also were analyzed. IL-22RA2 expression was detected in normal tissues such as digestive system (e.g., stomach, small intestine, esophagus, gastro-esophageal, pancreas, duodenum, ileum, colon, and small bowel), female reproductive system (e.g., mammary gland, endometrium, and breast), and other systems (e.g., lymph nodes, lung, skin, parotid, bladder, bronchus, heart ventricles, and kidney). Moreover, IL-22RA2 is expressed in certain tissue-specific tumors such as ovarian, uterine, and rectal cancers, but not in the corresponding normal tissues (data not shown).
IL-22RA2 Is Expressed in Monocytes, Activated B Cells, and Epithelium.
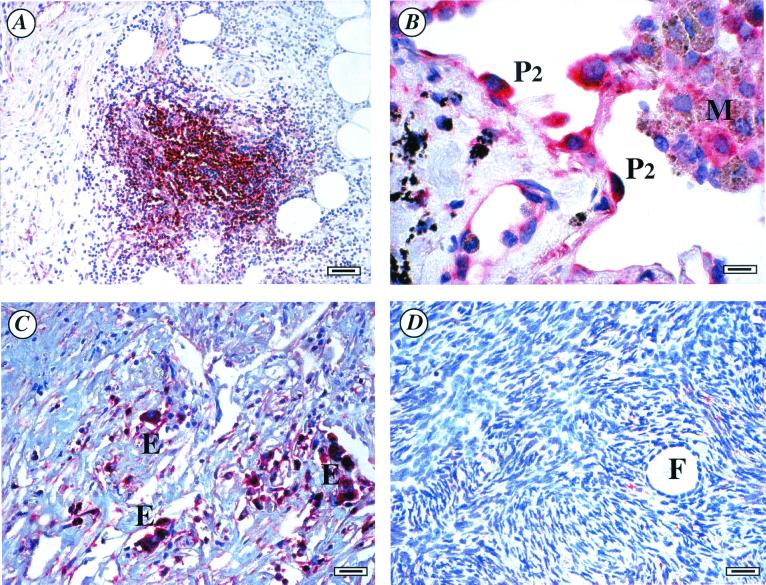
To identity potential cellular targets expressing IL-22RA2, in situ hybridization analysis was performed on various human tissues. Among tissues screened, strong signals were observed in the mononuclear cells of inflammatory infiltration sites, plasma cells, and a subset of epithelial cells. Tissues that gave positive signals included placenta, skin, inflamed appendix, lung, gastrointestinal tract, lymph node, thymus, and spleen (Fig. 5 A and B). In ovarian carcinoma, strong signals were detected in epithelial cells and some interstitial cells (most likely mononuclear cells), however, there was no signal observed in the normal ovarian tissue (Fig. 5 C and D). In summary, the in situ data for IL-22RA2 were consistent with the expression pattern generated by Northern blot and PCR analysis of cDNA/RNA panels. IL-22RA2 is predominantly expressed in mononuclear cells and epithelium.
Figure 5.
In situ hybridization of IL-22RA2 on human tissue sections. The positive signals were visualized with Vector Red, producing red signals in cells expressing IL-22RA2 messages. Results are representative of at least three different samples. (A) Inflamed appendix, strong signal is observed in the mononuclear cells (possibly lymphocytes) of inflammatory infiltration sites. (B) Lung, type II pneumocytes are positive, some macrophages may also be weakly positive. (C) Ovary carcinoma, carcinoma epithelial cells are positive. (D) Normal ovary is negative. P2, type II pneumocyte; M, macrophage; E, carcinoma epithelium; F, follicle. (Scale bars: A, 40 nm; B, 8 nm; C and D, 20 nm.)
To further determine which cell type(s) express IL-22RA2, a first-strand cDNA panel made from human blood fractions (CLONTECH) was analyzed for IL-22RA2 expression by PCR. The results indicated that activated CD4+ T cells and CD19+ B cells expressed IL-22RA2, whereas resting CD4+ and CD19+ cells did not (data not shown). Additional data indicated that IL-22RA2 was expressed in activated tonsillar B cells, but not resting B cells, in blood (data not shown). Furthermore, IL-22RA2 also is expressed in resting and activated U937 monocytic and HL60 promonocytic cell lines by reverse transcriptase–PCR analysis (data not shown). The data indicate that IL-22RA2 is predominantly expressed in monocytes and activated B cells. These results suggest IL-22RA2 may play a role in inflammatory, autoimmune diseases, and some cancers.
A crucial step in understanding the function of a novel cytokine receptor is the identification and characterization of its cognate ligand. We have successfully isolated a soluble class II cytokine receptor, IL-22RA2, and identified it as a naturally occurring IL-22 antagonist. It could represent a potentially important negative/positive regulatory mechanism of IL-22 signaling and expands our understanding of the human class II cytokine receptor family. One unique aspect of IL-22RA2 is that it is a naturally occurring soluble receptor, and no membrane-bound form has been identified yet. IL-22RA2 may function as a modulator of IL-22 activity in vivo, and it also may serve as a useful therapeutic agent for pathologies that may be associated with IL-22 overexpression. As therapeutic agents for the inhibition of cytokine activity, soluble receptors have several advantages such as specificity, high affinity, low immunogenicity, and reduced side effects. Because IL-22 induces the expression of acute-phase proteins, it is suggested IL-22RA2 may be important as an IL-22 antagonist in the regulation of inflammatory responses.
Acknowledgments
We thank our colleagues at ZymoGenetics, Inc. for their technical support and contributions to the work: Abdiazis Adan, Betty Haldeman, Cindy Sprecher, David Taft, Debra Leith, Dennis Dong, Diane Durnam, Emily Cooper, Frank Collins, Gary McKnight, Jane Gross, Jennifer Johnson, Janet Kramer, Jane Gross, Joe Kuijper, Katherine Henderson, Kristine Swiderek, Margaret Moore, Margo Rogers, Nels Hamacher, Patrick O'Hara, Sarah Pownder, Shannon Sexson, Steve Hughes, Tom Bukowski, and Will Lint.
Abbreviations
- IL-22RA2
IL-22 receptor-α 2
- FnIII
fibronectin type III
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY044429).
References
- 1.Rosen, C. A. & Kenny, J. J. (1999) U.S. Patent 5,985,614.
- 2.Jiang H, Lin J J, Su Z Z, Goldstein N I, Fisher P B. Oncogene. 1995;11:2477–2486. [PubMed] [Google Scholar]
- 3.Xie M H, Aggarwal S, Ho W H, Foster J, Zhang Z, Stinson J, Wood W I, Goddard A D, Gurney A L. J Biol Chem. 2000;275:31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 4.Knappe A, Hor S, Wittmann S, Fickenscher H. J Virol. 2000;74:3881–3887. doi: 10.1128/jvi.74.8.3881-3887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Wei S H, Ho A S, de Waal Malefyt R, Moore K W. J Immunol. 1994;152:1821–1829. [PubMed] [Google Scholar]
- 6.Lutfalla G, Gardiner K, Uze G. Genomics. 1993;16:366–373. doi: 10.1006/geno.1993.1199. [DOI] [PubMed] [Google Scholar]
- 7.Blumberg H, Conklin D, Xu W F, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman B A, Hammond A, et al. Cell. 2001;104:9–19. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 8.Lok, S., Kho, C. J., Jelmberg, A. C., Adams, R. L., Whitmore, T. E. & Farrah, T. M. (1999) U.S. Patent 5,945,511.
- 9.Kotenko S V, Pestka S. Oncogene. 2000;19:2557–2565. doi: 10.1038/sj.onc.1203524. [DOI] [PubMed] [Google Scholar]
- 10.Parham, C. L., Moore, K. W., Murgolo, N. J. & Bazan, J. F. (1999) International Patent Application Publication Number WO 99/46379.
- 11.Lok, S., Adams, R. L., Farrah, T. M., Jelmberg, A. C. & Whitemore, T. E. (1999) U.S. Patent 5,965,704.
- 12.Stahl N, Yancopoulos G D. Cell. 1993;74:587–590. doi: 10.1016/0092-8674(93)90506-l. [DOI] [PubMed] [Google Scholar]
- 13.Bazan J F. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thoreau E, Petridou B, Kely P A, Djiane J, Mornon J P. FEBS Lett. 1991;282:16–31. doi: 10.1016/0014-5793(91)80437-8. [DOI] [PubMed] [Google Scholar]
- 15.Uze G, Lutfalla G, Mogensen K E. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- 16.Dumoutier L, Louahed J, Renauld J C. J Immunol. 2000;164:1814–1819. doi: 10.4049/jimmunol.164.4.1814. [DOI] [PubMed] [Google Scholar]
- 17.Dumoutier L, Roost E V, Colau D, Renauld J C. Proc Natl Acad Sci USA. 2000;97:10144–10149. doi: 10.1073/pnas.170291697. . (First Published August 22, 2000; 10.1073/pnas.170291697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis S, Aldrich T H, Jones P F, Acheson A, Compton D L, Jain V, Ryan T E, Bruno J, Radziejewski C, Maisonpierre P C, Yancopoulos G D. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 19.Palacios R, Steinmetz M. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 20.Frohman M A. Methods Enzymol. 1993;218:340–362. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 21.Zubiaga A M, Belasco J G, Greenberg M E. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Heijine G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandes-Botran R. FASEB J. 1991;5:2526–2574. [Google Scholar]
- 25.Debets R, Savelkoul H F J. Immunol Today. 1994;15:455–457. doi: 10.1016/0167-5699(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 26.Upton C, Mossman K K, McFadden G. Science. 1992;258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- 27.Novick D, Cohen B, Tal N, Rubinstein M. J Leukocyte Biol. 1995;57:712–718. doi: 10.1002/jlb.57.5.712. [DOI] [PubMed] [Google Scholar]
- 28.Papanicolaou G J, Parsa N Z, Meltzer P S, Trent J M. Cytogenet Cell Genet. 1997;76:181–182. doi: 10.1159/000134542. [DOI] [PubMed] [Google Scholar]
- 29.Arthur E I, Zlotogora J, Lerer I, Dagan J, Marks K, Abeliovich D. Eur J Hum Genet. 1997;5:417–419. [PubMed] [Google Scholar]
- 30.Kotenko S V, Izotova L S, Mirochnitchenko O V, Esterova E, Dickensheets H, Donnelly R P, Pestka S. J Biol Chem. 2001;276:2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 31.Smith T F, Waterman M S. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]