Abstract
A high flux of reactive oxygen species during oxidative stress results in oxidative modification of cellular components including DNA. Oxidative DNA “damage” to the heterocyclic bases is considered deleterious because polymerases may incorrectly read the modifications causing mutations. A prominent member in this class is the oxidized guanine base 8-oxo-7,8-dihydroguanine (OG) that is moderately mutagenic effecting G→T transversion mutations. Recent reports have identified that formation of OG in G-rich regulatory elements in the promoters of the VEGF, TNFα, and SIRT1 genes can increase transcription via activation of the base excision repair (BER) pathway. Work in our laboratory with the G-rich sequence in the promoter of VEGF concluded that BER drives a shift in structure to a G-quadruplex conformation leading to gene activation in mammalian cells. More specifically, removal of OG from the duplex context by 8-oxoguanine glycosylase 1 (OGG1) produces an abasic site (AP) that destabilizes the duplex, shifting the equilibrium toward the G-quadruplex fold because of preferential extrusion of the AP into a loop. The AP is bound but inefficiently cleaved by apurinic/apyrimidinic endoDNase I (APE1) that likely allows recruitment of activating transcription factors for gene induction. The ability of OG to induce transcription ascribes a regulatory or epigenetic-like role for this oxidatively modified base. We compare OG to the 5-methylcytosine (5mC) epigenetic pathway including its oxidized derivatives, some of which poise genes for transcription while also being substrates for BER. The mutagenic potential of OG to induce only ~one-third the number of mutations (G→T) compared to deamination of 5mC producing C→T mutations is described. These comparisons blur the line between friendly epigenetic base modifications and those that are foes, i.e. DNA “damage,” causing genetic mutations.
Keywords: 8-Oxo-7,8-Dihydroguanine; Base Excision Repair; Epigenetics; G-Quadruplex; Oxidative Stress; Mutagenesis
1. Introduction
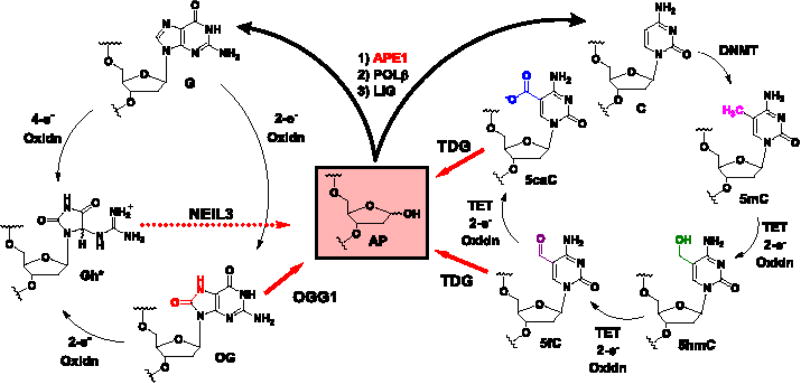
Reactive oxygen species formed during oxidative stress are electron deficient and readily oxidize proteins, lipids, RNA, and particularly DNA. Oxidative modification of the genomic DNA bases is well documented and can result in mutations responsible for initiation of a number of diseases [1]. The guanine (G) heterocycle is the most susceptible of the four DNA bases to oxidation leading to many products [2]. Chief among these oxidatively modified products is 8-oxo-7,8-dihydroguanine (OG; Fig. 1). Cellular levels of OG in the genome are routinely monitored as a biomarker to assess the extent of oxidative stress to which a cell has been exposed [3]. Moreover, OG is moderately mutagenic, if not repaired, causing G→T transversion mutations that are thought responsible for initiating and driving some cancers [1]. These mutations are a consequence of OG base pairing with A on the Hoogsteen face rather than C on the Watson-Crick face [4]. To counteract mutations from damaged DNA nucleotides, an elaborate DNA repair system has evolved to return modified sites back to the original canonical nucleotides [5]. Repair of OG is achieved by base excision repair (BER) that initiates removal of OG when base paired with C by the action of 8-oxoguanine glycosylase 1 (OGG1) in mammals (Fig. 1); in contrast, when OG is incorrectly base paired with A, MutY DNA glycosylase (MUTYH) removes the A allowing a second chance for a polymerase to insert C opposite OG for further action by OGG1 [5]. Following removal of OG by OGG1, an abasic site (AP) is formed that is a substrate for apurinic/apyrimidinic endoDNase I (APE1) to cleave the 5′-phosphodiester linkage yielding a nick in the DNA (Fig. 1) [5]. The repair process is completed by polymerase β (POLB) that removes the sugar fragment at the nick site followed by inserting the correct G nucleotide, and finally ligase (LIG) seals the nick to return the DNA back to its native state (Fig. 1) [5]. This dynamic process of G oxidation to OG followed by DNA repair has been estimated to occur up to 105 times per cell per day [6].
Fig. 1.
Comparison of the G oxidative modification cycle with the C methylation and oxidative modification cycle to illustrate the centrality of the abasic site (AP) to return the sequence back to the original active state. *For the sake of brevity, the 4-electron oxidation product of G, or 2-electron oxidation product of OG yielding 5-guanidinohydantoin (Gh) is shown; the other 4-electron product spiroiminodihydantoin (Sp) is not shown [7]. The yields of Gh and Sp show strong dependency on the reaction conditions and context, favoring Gh in duplex DNA oxidations or reactions at pH < 6 and favoring Sp in single-stranded and G-quadruplex DNA oxidations or reactions at pH > 7 [8, 9].
The long-standing view has been that OG is mutagenic and detrimental to cellular processes such as transcription. For instance, the presence of OG in template strands can stall the advancement of RNA pol II [10], and initiation of OG repair causes polymerases to stop [11], thus ascribing a role to OG as a transcriptional repressor. However, there are a few notable examples of oxidative stress leading to increased OG formation in the genome in tandem with increased gene expression. This has been documented in livers from mice with infection-induced colitis [12], and rat pulmonary artery endothelial cells exposed to hypoxic conditions [13]. Observations like these led our laboratory and others to inspect how the VEGF [14], TNFα [15], and SIRT1 [16] genes respond when G is oxidized to OG in their promoters. The most interesting finding in these cellular studies showed that OG can increase gene transcription via the BER pathway [14–16]. These results identify an intertwining of DNA repair with gene activation that is a phenomenon gaining appreciation [17]. Therefore, oxidative modification of G to OG may have regulatory and possibly epigenetic-like features in cells that are responding to oxidative stress. This perspective will discuss the discovery that OG can stimulate transcription via BER activity. These results provide the background for a comparative discussion between OG as a possible epigenetic-like DNA modification vs. the traditional 5-methylcytosine (5mC) epigenetic modification. Additionally, the ten-eleven translocation (TET) proteins oxidize 5mC in a stepwise fashion to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Fig. 1) in genes poised for activation by BER removal after being silenced by 5mC [18–21]. The observation of oxidative modification to DNA bases in the form of OG or oxidized 5mC highlights a possibility that base oxidation is a DNA-based mechanism for gene activation. Finally, the ability of OG to regulate gene expression vs. its ability to cause mutations will be discussed.
2. Initial reports that OG is epigenetic-like
A few initial reports proposed that OG, if present in key regions of the genome, could impact cellular processes. For instance, synthetic oligonucleotides with OG in protein transcription factor binding sequences found this modification negatively impacted factor binding affinity. This effect was demonstrated in the consensus sequences of specificity protein 1 (SP1) [22], nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) [23], and CAMP responsive element binding protein 1 (CREB) [24]. The work with the CREB transcription factor in the Strauss laboratory led them to propose that OG might be epigenetic by decreasing protein binding resulting in OG as a transcriptional repressor [24]. The Olinski laboratory quantified OG in heterochromatin vs. euchromatin from porcine thymus DNA to find that the transcriptionally active euchromatin DNA harbored more OG [25]. Their observation of OG concentrations varying throughout the genome led them to speculate that OG might be an epigenetic modification. Lastly, Park, et al. developed a method to demonstrate that G oxidation to OG could occur site specifically in vivo under oxidative stress conditions leading them to propose OG as an epigenetic modulator [26]. These observations of OG as a regulatory modification (i.e., epigenetic-like) were all lacking in cellular experiments demonstrating G oxidation to OG can form in critical regions of the genome and impact transcription.
3. OG activates mRNA synthesis by facilitating promoter G-quadruplex formation
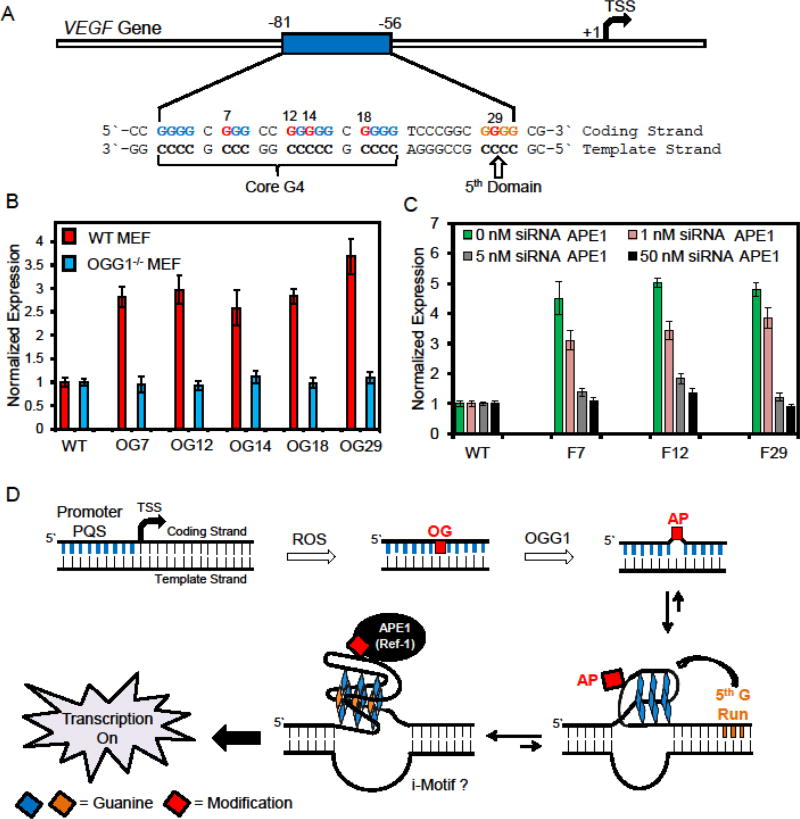
The vascular endothelial growth factor A (VEGF) gene harbors a G-rich promoter element critical for regulation of mRNA synthesis [27]. The G-rich element is located between positions −86 and −56 relative to the transcription start site (TSS) in the coding strand (Fig. 2A); further, this region is bound by three equivalents of the SP1 transcription factor [27]. Cellular regulation of VEGF by SP1 has been documented, and oxidative stress results in less SP1 binding followed by up-regulated transcription [27]. Moreover, this G-rich element is a potential G-quadruplex sequence (PQS) as demonstrated by Hurley and co-workers with the ability to adopt a G-quadruplex (G4) structure for regulation of transcription [28, 29] (Fig. 2A). Additionally, the Gillespie laboratory found hypoxia-induced oxidative stress increased transcription of the VEGF gene, in addition to OG formation in the vicinity of the gene promoter, likely near the PQS [13, 30]. These observations prompted us to inquire if oxidation of G to OG in the G-rich promoter element of VEGF could facilitate activation of transcription, whether BER is involved in the reactivation process, and whether there is a role for the G4 structure in gene induction.
Fig. 2.
Sequence for the PQS in the coding strand of the VEGF gene that upon oxidation of G to OG provides a substrate for BER that unmasks the G-quadruplex for gene induction. (A) The sequence of the G-rich element in the VEGF promoter. The Gs marked in red are sites in which OG was synthetically incorporated to demonstrate the proposed pathway in part D [14]. (B) Data illustrating the presence of OG in the VEGF promoter increased luciferase expression by >2.5 fold in MEF cells, and knocking out OGG1 results in the signal remaining unchanged relative to the wild type (WT) plasmid. (C) Utility of APE1-specific siRNAs in glioblastoma cells provided a dose response impact on luciferase expression. The data in panels B and C demonstrate OGG1 and APE1, respectively, are required for gene induction when OG is present in the VEGF promoter PQS. These data were adapted from the original publication [14]. (D) Proposed pathway for oxidation of the PQS to yield OG and guide the BER process by unmasking the G-quadruplex for gene activation, thus illustrating an intertwining of DNA repair and transcriptional induction.
To test our hypothesis, we developed a luciferase reporter plasmid with the VEGF PQS regulating the Renilla luciferase gene [14]. The system developed allowed synthetic incorporation of OG within the G-rich sequence with single-nucleotide precision. To guide selection of appropriate sites for modification, we first oxidized the VEGF PQS in a short oligonucleotide folded as a G4 [31] or in the duplex context with a reactive oxygen species (ROS) found in inflammation to identify G sites readily oxidized (Fig. 3) [32]. Armed with the knowledge of the most reactive Gs in the VEGF promoter sequence, we then synthetically incorporated OG into the plasmid at specific sites in the promoter (Fig. 2A red Gs) [14]. Next, transfection of the synthesized reporter plasmids into mouse embryonic fibroblasts (MEFs) revealed that the OG-containing plasmids produced >2.5-fold more luciferase protein than a wild-type (WT) plasmid without OG (Fig. 2B). Furthermore, luciferase expression was always increased regardless of where the OG was located (Fig. 2B). Gene induction with the reporter system was verified to produce a similar increase in luciferase expression in glioblastoma cells. Demonstration that BER of OG was essential in the initiation of gene activation was accomplished by repeating the studies in MEFs that had OGG1 knocked out (Fig. 2B OGG1−/− MEFs). When OGG1 was absent, the presence of OG in the reporter plasmid did not yield an increase in expression relative to the WT control. The null result in OGG1−/− MEFs highlights a critical role for OGG1 in the process of gene activation when OG is in the coding strand of the VEGF PQS element.
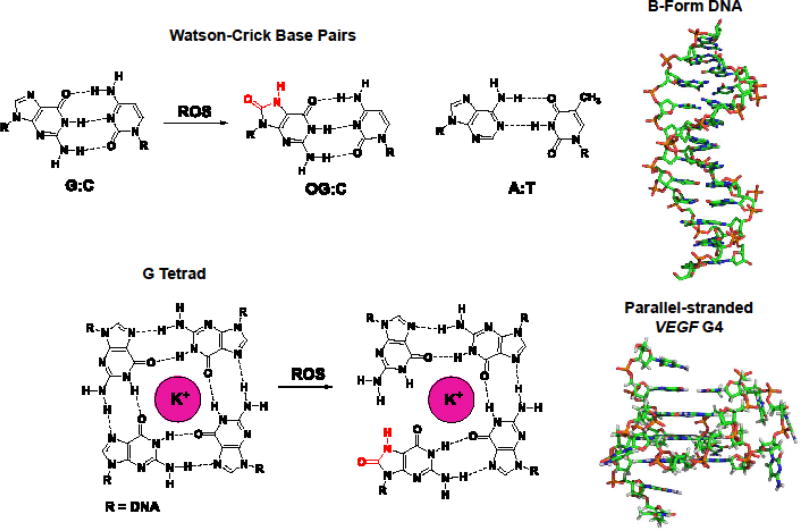
Fig. 3.
Structural comparison and base pairing properties of B-form and G4 DNA. The B-form DNA structure was derived from pdb 1BNA [40], and the VEGF G4 structure was derived from pdb 2M27 [31].
When OGG1 removes OG from the DNA in the cellular context, the product is an AP site that is subsequently the substrate for the next enzyme in the BER process, APE1 (Fig. 1) [11, 33]. Accordingly, we next synthesized catalytically competent, yet stable, AP analogs (tetrahydrofuran, F) in the reporter plasmids and then transfected them into MEF and glioblastoma cells. These AP-modified plasmids were studied to determine the importance of an AP and of APE1 in the gene activation process. First, when an AP analog was present in the reporter system, the luciferase gene was induced >4-fold, a value somewhat higher than that obtained for OG (Figs. 2B and 2C) [14]. Second, demonstration that APE1 was essential for gene induction was achieved by transfecting cells with AP analog-containing plasmids and knocking down APE1 with siRNAs. As siRNAs specific to APE1 were titrated into cells transfected with AP-analog containing plasmids, the level of luciferase expression decreased with a dose response (Fig. 2C). These experiments concluded that the AP site resulting from OG release from the DNA is critical for gene induction with APE1 playing a major role in the process. The activity of APE1 expands beyond it being a key player in BER to include interactions via its redox-effecter factor-1 (Ref-1) domain with protein factors such as HIF1-α, STAT3, and CBP/p300 that promote gene transcription [13, 34]. Future studies will begin to unravel more details of the activating transcription factors involved in the gene induction process when OG is processed by BER in the VEGF gene promoter region.
Beyond the importance of the BER process for activation of transcription, we found that the ability of the VEGF PQS to possibly adopt a G-quadruplex fold is also essential for gene activation. G-Quadruplexes are structures that can fold in DNA sequences with four or more contiguous runs of >3 Gs with small intervening sequences forming loops between the G runs [35]. These structures diverge from the native B-form of DNA because they fold around cellular K+ ions to G tetrads held together by four G:G Hoogsteen base pairs (Fig. 3). In DNA, generally three or more G tetrads stack to adopt unique four-stranded folds that are structurally different than B-form DNA (Fig. 3) [36, 37]. Protein interactions with Bform and G4 DNA differ resulting in redirecting the downstream signals [38]. The ability of G4 structures to regulate transcription was best demonstrated by the Hurley laboratory in the c-MYC gene [38]. Global cellular confirmation of PQS to adopt G-quadruplex folds and alter transcription was recently demonstrated by the Balasubramanian laboratory through a combination of G4 ChIP-Seq and RNA-Seq experiments in human cells [39]. The VEGF PQS was found to adopt a parallel-stranded G4 structure on the basis of NMR structural analysis (Fig. 3) [31]. These studies and many others not referenced provide a solid background for us to consider the possibility that a G4 structure may exist in the VEGF PQS when OG is present and aid in the activation process.
Comparisons were made between plasmids containing OG in either the PQS sequence, or one judiciously mutated to be incapable of G4 formation, while still retaining the ability to be bound by the SP1 transcription factor [14]. The comparative studies found that the G4 structure was essential to induce transcription; in contrast, the G4 negative sequence provided no signal enhancement relative to the control. These results support the importance of the G4 fold in gene induction. Our previous analyses found a significant number of promoter PQSs possess additional G tracks flanking the core G4 structure (Fig. 2A) [32]. We proposed when a core G4 sequence is damaged leading to the unraveling of a G tetrads and the loss of global structure, the 5th G run is recruited as a “spare tire” to maintain the G-quadruplex fold (Figs. 2D and 3). The recruitment of the 5th G run allows extrusion of the modification into a large loop to achieve G-quadruplex folding; furthermore, the plasticity of these five G-track sequences allow BER enzymes to bind the modifications in the large loop. The Burrows and Wallace laboratories found that without the 5th domain present, DNA repair initiation was abolished in the VEGF PQS (Fig. 2A and D) [32, 41]. As a final study, comparisons were made between transfected plasmids containing OG in the native sequence with all five G tracks (G5) present to a modified sequence with only the essential four core G tracks (G4). The results of these studies led to the conclusion that both sequence contexts yield induction of transcription when OG is present, but the expression was significantly greater for the G5 sequence [14]. This observation supports an important role for the 5th domain in achieving the maximal transcriptional increase when OG is present.
These results led to the following proposed mechanism for transcriptional activation when OG is present in the coding strand of the VEGF promoter PQS (Fig. 2D) [14]. The G-rich PQS element renders this site highly susceptible to oxidative modification of G to OG in the duplex context. This oxidation yields an OG base paired with C that has a negligible impact on the B-form DNA structure (Fig. 3). Oxidation of G to OG allows recruitment of OGG1 for removal of OG to yield an AP. On the basis of initial biophysical studies, the AP results in melting of the duplex to unmask the more thermodynamically favorable G-quadruplex structure (Fig. 2D) [14]. The G-quadruplex fold is favored because the 5th G track allows extrusion of the damaged G run into a large loop while maintaining the fold. The AP site is then presented to APE1 and bound by this protein; consequently, the reaction kinetics are highly attenuated by the G4 structure prolonging APE1 binding [42] and aiding in the possible recruitment of other activating transcription factors for gene induction (e.g., HIF1-α) [13]. These initial studies identify an intertwining of G oxidation in PQS elements to provide a structural switch for recruitment of BER proteins to activate transcription (Fig. 2D). Furthermore, we identify the AP site and APE1 as central players in the gene activation process initiated by G oxidation to OG in the PQS of the coding strand of the VEGF gene.
In our publication of these results [14], the concept of a PQS structural switch was also demonstrated in a PQS found in the coding strand of the NTHL1 DNA repair gene. The presence of OG in the NTHL1 PQS yielded a >4-fold transcriptional enhancement. This example expands the possibility that the mechanism proposed may be a more general phenomenon for gene activation under oxidative stress conditions. The VEGF and NTHL1 results set the stage for many future inquiries. For instance, G4 ChIP-Seq in human skin cells found ~10,000 PQSs responsible for gene induction [39]; are any of these genes also activated by oxidative modification of G to OG in the PQSs? The strand distribution of PQSs is nearly equal on the coding and template strands [43], and so far, the PQS structural switch leading to gene induction has only been reported for sequences in the coding strand. Will the gene output be different when the PQS is in the template strand vs. the coding strand? Structural studies have found G-quadruplex sequences adopt many different types of folded structures [36, 37]. How does G4 folding (parallel, anti-parallel, or hybrid) impact the structural switch? These interesting questions should be addressed in the near future.
The proposal of an AP being processed by APE1 as the key step for gene activation begs the question: do other oxidatively modified DNA bases that are processed to yield APs also impact transcription? There exist a few notable examples of related modifications to study. First, the 4-electron oxidation products of G (or 2-electron oxidation products of the highly redox sensitive OG) are 5-guanidinohydantoin and spiroiminodihydantoin (Gh and Sp; Fig. 1) [7–9], both of which are excellent substrates for the NEIL glycosylases [44]. Processing of Gh and Sp by the monofunctional NEIL3 glycosylase yields an AP (Fig. 1) [45]; in contrast, NEIL1 or NEIL2 are proposed to be bifunctional glycosylases leading directly to a strand break [44]. The details of hydantoin processing in PQSs and the subsequent impact on transcription will guide a better understanding of the substrate and glycosylase requirements to activate transcription. Another interesting modification for study is C deamination to uracil (U) because in mammals, U is removed by the monofunctional glycosylases UNG and SMUG1 to yield an AP for processing by APE1 [46]. Future inspection of these modifications and others will be fascinating for expanding our knowledge of the PQS switching mechanism activated by BER to induce transcription.
4. Other examples of OG as an epigenetic-like DNA base modification
Boldogh and co-workers recently provided an additional observation of possible OG formation in promoters followed by OGG1 recruitment to induce transcription [15]. Briefly, their studies in HEK 293 and MEF cells found tumor necrosis factor-alpha (TNFα) possibly induced G oxidation to OG upstream of NF-κB consensus sequences in pro-inflammatory genes (e.g., TNFα). The result of OG formation was recruitment of OGG1 followed by NF-κB protein to up-regulate transcription of these genes. These results provide strong additional support for the oxidatively modified G residue, OG, as a regulatory modification by facilitating BER and transcriptional regulation networks working together on promoter sequences. The Tell laboratory studied HeLa cells under oxidative stress conditions to propose G oxidation to OG in the negative calcium responsive elements (nCaRE) in the promoter of the sirtuin-1 (SIRT1) gene [16]. Their work led them to propose that OGG1 removed OG in the nCaRE sequence to yield an AP followed by APE1 binding to the site. Next, APE1 functions in tandem with Ku70 and RNA Pol II to increase SIRT1 transcription; more importantly, their work, like ours [14], supports both AP and APE1 as key elements for gene activation associated with OG formation in gene promoters. The studies in our laboratory with the VEGF promoter PQS are the only experiments to date demonstrating that OG, when site-specifically found in the coding strand of a regulatory element, can upregulate transcription.
Not all examples of OG formation in cellular DNA lead to enhanced transcription. The Hanawalt laboratory found OG in template strands slightly inhibited advancement of RNA pol II and recruits the transcription-coupled repair machinery slowing transcription [10, 47]; additionally, this work concluded that the AP site generated by OGG1 release of OG blocked transcription and initiated transcription-coupled repair. The Khobta laboratory identified that OG is a barrier to transcription when located in either the coding or template strand in a gene coding region [11, 48]. Further, OG is only a transcriptional barrier after conversion to an AP site by OGG1. The experiments in our laboratory and the others demonstrate that OG can modulate transcription, and the direction of the modulation is dependent on the context in which OG is located within the genome. Further investigation into sequence contexts, such as PQSs, outside of promoters is also warranted to advance our knowledge of the impact OG has during synthesis of an entire mRNA sequence.
5. How does OG fit into the epigenetic landscape?
Many of the studies described propose that OG is an epigenetic modification [14, 15, 24–26]. Can OG actually be classified as an epigenetic mark, and if so, how does it fit into a classification system that typically involves heritability? In the traditional 5mC epigenetic system, there has emerged a clear picture of the protein writers, readers, and erasers for modifications on the global genome [18–21]. A wealth of data has led to a better understanding of how these modifications change in the genomes of different cell types, how these modifications are heritable from mother to daughter generations, and how these modifications impact gene activity and expression. This knowledge forms the modern epigenetic definition [49]. Some of these features with respect to OG are clear while others need further inquiry before the mechanisms are understood.
For 5mC, there exist DNA methyltransferases (DNMT) to write the modifications site specifically into the genomes (Fig. 1) [18]. It is well established that OG is formed in genomic DNA by direct oxidation of G via ROS [2], or indirectly written by remote oxidation and electron transfer through the DNA π stack to induce G oxidation [50]. These mechanisms generally effect oxidation of a 5′ G in the sequence context 5′-GpG-3′ that provides a pathway to obtain OG in a specific dinucleotide context, just as 5mC occurs in the 5′-CpG-3′ context. The ability of ROS to selectively modify critical Gs for cellular regulation is challenging to envision. Work by Perillo, et al. found that chromatin remodeling could induce region-specific oxidation of G to OG in the BCL-2 promoter of MCF7 cells for gene activation [51]. They identified that the flavin-dependent lysine-specific demethylase 1A or 1B (LSD1 or LSD2) remodelers generated H2O2 in the vicinity of the genome for oxidation of G to OG, most likely by the Fenton reaction [52]. More interestingly, the Perillo, et al. work documented that OGG1 and BER were essential for BCL-2 gene induction [51]; in our studies [14], we note the region oxidized in BCL-2 is a PQS proposed to be involved in regulation of this gene [38], and therefore, the activation mechanism Perillo, et al. observed may have functioned through a mechanism similar to our proposal for VEGF (Fig. 2D) [14]. Thus, one possibility is that chromatin remodeling by LSD1/2 can induce region-specific G oxidation to OG for gene regulatory purposes, although the LSD1/2 mechanism relies on diffusion-controlled delivery of the H2O2 oxidant to the regulatory site of G oxidation. Evolution of this approach to write OG into the genome for gene activation remains less refined compared to a direct protein-catalyzed oxidation mechanism. Future exploration to better understand the details of writing OG into the genome are warranted; for example, are there protein writers to site-specifically install OG in the genome?
The presence of OG in genomic DNA is well established by nuclease and phosphatase digestion of cellular DNA followed by mass spectrometric quantification; however, this approach does not allow knowledge of the sequence or region in which OG is located. Thus, a major hurdle along the way to elucidating the role of OG in gene regulation will be the development and implementation of genome-level OG sequencing. Many laboratories have developed antibody-based OG sequencing that provides a low resolution sequence map of OG (~10–1000 kbp) [53, 54]. Recently, our laboratory developed an OG sequencing approach with single-nucleotide resolution implemented on plasmid DNA [55], in addition to an OG sequencing method with ~0.15-kbp resolution (i.e., OG-Seq) that was implemented on the mouse genome [56]. Expansion of sequencing studies, ideally at single-nucleotide resolution, to different cell types under a variety of conditions (i.e., different stressors or cell states) will be essential to the determination of the sequences in which OG is preferentially formed, whether these sequences and regions change based on cellular conditions, and whether they are gene regulatory regions.
The 5mC modification is read by methyl-CpG-binding proteins to silence transcription. Upon oxidation of 5mC to 5hmC, 5fC, or 5caC by TET enzymes in gene regulatory regions, the sites become poised for activation (Fig. 1) [18]. Oxidative modification of 5mC recruits a new set of reader proteins, some of which are involved in DNA repair, such as TDG, NEIL1, and NEIL3 [19]. These DNA repair readers also constitute the erasing mechanism for 5mC from the genome (Fig. 1, TDG) [19]. The recruitment of BER proteins to oxidized 5mC is similar to the recruitment of OGG1 by OG, providing a fascinating link between these two systems involved in gene activation. Both TDG binding and removal of 5fC or 5caC and binding and removal of OG by OGG1 yield an AP product (Fig. 1) [11, 57]. The AP product recruits APE1 to continue BER in both cases [57]. From our studies, an AP in a coding strand G-quadruplex provides the structural switch that stalls APE1, possibly recruiting additional factors and leading to activation of transcription. Are oxidized base modifications such as 5fC, 5caC, and OG gatekeepers for site-specific introduction of AP, in which the AP facilitates gene activation in both cases? In the case of OG, OGG1 is the best established reader protein; however, systematic studies to identify OG-specific readers have yet to be conducted. It is very possible that for OG, the reading and erasing mechanisms are coupled by overlapping DNA repair and gene activation functions. This coupling concept allows cells to complete two necessary tasks during oxidative stress: 1) Repair of oxidatively modified DNA bases, and 2) alteration of the cellular phenotype in response to oxidative stress.
Whether OG is truly epigenetic or a regulatory modification is not clear. For instance, heritability of OG from mother to daughter generations is not known and will require deep inquiry to answer this question. Methylation of C is well established to guide cellular differentiation during embryonic development for long-term cellular information storage [19]; in contrast, OG appears to be a modification allowing cells a rapid response pathway during oxidative stress. Beyond the toxicology of OG formation during oxidative stress, OG formation may be important during embryonic growth. Take the following example: VEGF is essential for vascularization; as embryos grow they become slightly hypoxic leading to the need to vascularize [58]. Does OG formation in the VEGF PQS drive this process via a mechanism such as we have outlined (Fig. 2D) [14]? If this is possible, it would provide a link between OG formation in the genome and cellular development. It is hard to envision that OG would possess the long-term information potential that 5mC has because OG is directly recognized by BER [5], while 5mC must be modified by TET proteins to become a substrate for BER [19]. Thus, OG may fit into the epigenetic landscape as a DNA modification allowing cells to adapt and respond to changes important on the short term, such as oxidative stress, while 5mC is relegated to longer term cellular information storage. Is this sufficient to define OG as epigenetic? Future studies and more discussion on this topic will craft how we best describe OG and other base modification in the future.
6. OG as a friend and foe
The experiments and discussion so far have acknowledged, against conventional wisdom, OG may be a cellular friend by facilitating gene activation via a DNA repair mechanism in response to oxidative stress [13–16]. However, the toxicological aspects of OG cannot be ignored. Cells deficient in BER of OG accumulate G→T transversion mutations highlighting the mutagenic potential of OG [59]. The VEGF example provides a case in which OG formation driving gene expression can be viewed as a foe, or unwanted outcome. For instance, as solid tumors grow, they become hypoxic inducing oxidative stress leading to VEGF activation and vascularization allowing tumor growth and metastasis [58]. This process is good for the tumor and bad for the organism. This underscores the importance of context for these modifications in regulating biological processes. This is even true for 5mC in which cancer cells hijack the methylation systems to change cellular phenotype to benefit the cancer at the expense of the organism [60].
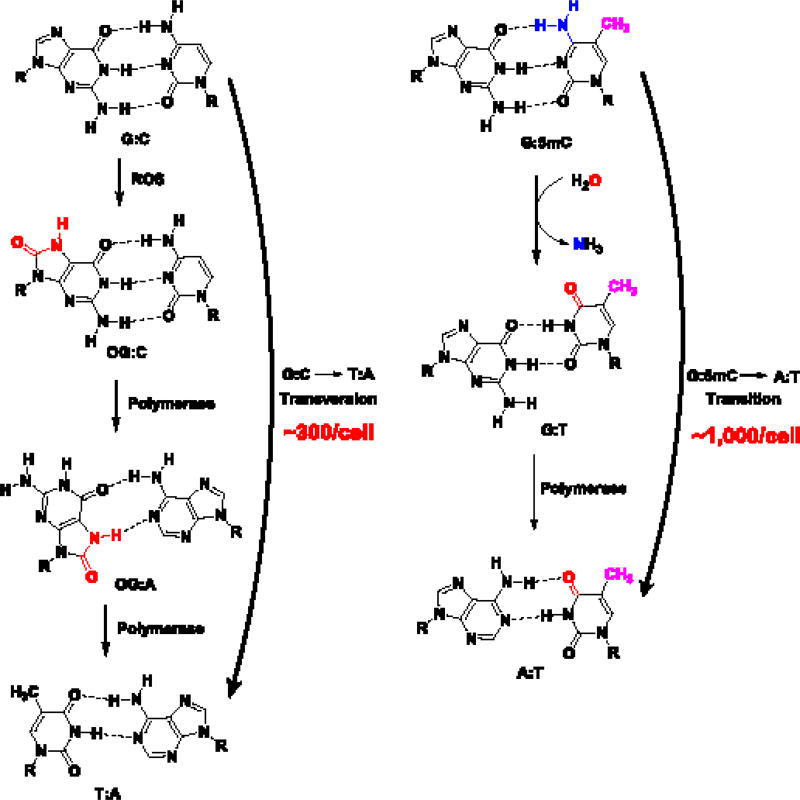
Lastly, we compare the mutagenicity of OG vs. 5mC. The mutagenic potential of OG to effect G→T transversion mutations is well established (Fig. 4) [4]. The transversion mutations result from the facile formation of OG:A base pairs by polymerase activity that upon a second polymerase bypass, inserts a T opposite A to occupy the original position of G (Fig. 4) [4]. The frequency for OG to cause this mutation signature is estimated at ~1% [61]. There exists a steady state level of ~30,000 OGs (mouse embryonic stem cell) [62] that could result in ~300 mutation events under replicating conditions (Fig. 4). The epigenetic mark 5mC can also cause C→T transition mutations upon deamination of 5mC to T (Fig. 4) [63]. The C→T transition rate for 5mC is estimated at ~2×10−3% [63]. There exist ~50 million 5mCs (in mouse embryonic stem cells) [62] that could lead to ~1,000 mutations (Fig. 4). Therefore, on the basis of this estimate, the epigenetic mark 5mC induces ~3-fold more mutations than OG. This is consistent with recent deep genome sequencing experiments that identified more C→T mutations than G→T [64]. The line that differentiates DNA modifications as mutagenic vs. epigenetic is blurred, and that should result in a reevaluation of modifications considered mutagenic. The former “DNA damage” bases 5-hydroxymethyluracil and 6-methyladenine have recently been reconsidered as possible epigenetic modifications [62, 65], and recent findings support OG joining this list [13–16]. The context in which these modifications exist, especially OG, is exceedingly important in defining whether they are a friend or foe to biological processes and the overall organism: coding vs. template strands, promoter vs. transcribed regions, G-quadruplexes vs. CpG islands—all appear to modulate the roles played by base modifications in cellular function and survival.
Fig. 4.
Pathways to generate genomic mutations for OG and 5mC.
Acknowledgments
The work from our laboratory was funded by a National Cancer Institute grant R01 CA090689. We are grateful to our wonderful collaborators Dr. Yun Ding (Univ. of Utah) as well as Dr. Susan Wallace and laboratory members (Univ. of Vermont) for their exceptional experimental work that provided the early backbone of our hypotheses.
Abbreviations
- AP
abasic site
- APE1
apurinic/apyrimidinic endoDNase 1
- BCL2
B-cell lymphoma 2
- BER
base excision repair
- CBP
CREB binding protein
- ChIP-Seq
chromatin immunoprecipitation assay with sequencing
- c-MYC
V-myc avian myelocytomatosis viral oncogene homolog gene
- CREB
CAMP responsive element binding protein 1
- 5caC
5-carboxylcytosine
- DNMT
DNA methyltransferase
- 5fC
5-formylcytosine
- F
tetrahydrofuran
- G4
G-quadruplex
- G
guanine
- Gh
5-guanidinohydantoin
- HIF1-α
hypoxia inducible factor 1 alpha
- 5hmC
5-hydroxymethylcytosine
- Ku70
protein encoded by the x-ray repair cross complementing 6 gene
- LSD1-2
lysine demethylase 1A and 2A
- LIG
ligase
- 5mC
5-methylcytosine
- MCF-7
Michigan Cancer Foundation-7 cell line
- MEF
mouse embryonic fibroblast
- MUTYH
MutY DNA glycosylase
- nCaRE
negative calcium response elements
- NEIL1-3
endonuclease VIII-like 1–3
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NTHL1
Nth-like DNA glycosylase 1 gene
- OG
8-oxo-7,8-dihydroguanine
- OGG1
8-oxoguanine glycosylase 1
- OGG1−/−-MEF
mouse embryonic fibroblast with OGG1 knocked out
- OG-Seq
8-oxo-7,8-dihydroguanine sequencing
- p300
E1A binding protein P300
- POLB
polymerase β
- PQS
potential G-quadruplex sequence
- Ref-1
redox effector factor 1
- RNA pol II
RNA polymerase II
- ROS
reactive oxygen species
- SIRT1
sirtuin 1 gene
- SMUG
single-stranded-selective monofunctional uracil-DNA glycosylase 1
- Sp
spiroiminodihydantoin
- SP1
specificity protein 1
- STAT3
signal transducer and activator of transcription 3
- TDG
thymine-DNA glycosylase
- TNFα
tumor necrosis factor α gene
- U
uracil
- UNG
uracil-DNA glycosylase
- VEGF
vascular endothelial growth factor A gene
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare that there are no conflicts of interest.
References
- 1.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadet J, Wagner JR, Shafirovich V, Geacintov NE. One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int. J. Radiat. Biol. 2014;90:423–432. doi: 10.3109/09553002.2013.877176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gedik CM, Collins A. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 4.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 5.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park EM, Shigenaga MK, Degan P, Korn TS, Kitzler JW, Wehr CM, Kolachana P, Ames BN. Assay of excised oxidative DNA lesions: isolation of 8-oxoguanine and its nucleoside derivatives from biological fluids with a monoclonal antibody column. Proc. Natl. Acad. Sc.i U. S. A. 1992;89:3375–3379. doi: 10.1073/pnas.89.8.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of 13C-, 15N-, and 18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J. Am. Chem. Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 8.Fleming AM, Burrows CJ. G-Quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol. 2013;26:593–607. doi: 10.1021/tx400028y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem. Res. Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 10.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Allgayer J, Kitsera N, Bartelt S, Epe B, Khobta A. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016;44:7267–7280. doi: 10.1093/nar/gkw473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, Gillespie MN. An oxidative DNA "damage" and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309:L1367–1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2016:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, Boldogh I. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J. Biol. Chem. 2016;291:25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antoniali G, Lirussi L, D'Ambrosio C, Dal Piaz F, Vascotto C, Casarano E, Marasco D, Scaloni A, Fogolari F, Tell G. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell. 2014;25:532–547. doi: 10.1091/mbc.E13-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol. Cell. 2013;52:291–302. doi: 10.1016/j.molcel.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, Zhao Boxuan S, He C. Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 2016;23:74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Booth MJ, Raiber E-A, Balasubramanian S. Chemical methods for decoding cytosine modifications in DNA. Chem. Rev. 2015;115:2240–2254. doi: 10.1021/cr5002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song C-X, Zhang K, He C, Xu G-L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramon O, Sauvaigo S, Gasparutto D, Faure P, Favier A, Cadet J. Effects of 8-oxo-7,8-dihydro-2`-deoxyguanosine on the binding of the transcription factor Sp1 to its cognate target DNA sequence (GC box) Free Radical Res. 1999;31:217–229. doi: 10.1080/10715769900300781. [DOI] [PubMed] [Google Scholar]
- 23.Hailer-Morrison MK, Kotler JM, Martin BD, Sugden KD. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2'-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry. 2003;42:9761–9770. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 24.Moore SP, Toomire KJ, Strauss PR. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst) 2013;12:1152–1158. doi: 10.1016/j.dnarep.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Zarakowska E, Gackowski D, Foksinski M, Olinski R. Are 8-oxoguanine (8-oxoGua) and 5-hydroxymethyluracil (5-hmUra) oxidatively damaged DNA bases or transcription (epigenetic) marks? Mutat. Res. 2014;764–765:58–63. doi: 10.1016/j.mrgentox.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Park J, Park JW, Oh H, Maria FS, Kang J, Tian X. Gene-specific assessment of guanine oxidation as an epigenetic modulator for cardiac specification of mouse embryonic stem cells. PLoS ONE. 2016;11:e0155792. doi: 10.1371/journal.pone.0155792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, Hocker M. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J. Biol. Chem. 2003;278:8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- 28.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo K, Gokhale V, Hurley LH, Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008;36:4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark DW, Phang T, Edwards MG, Geraci MW, Gillespie MN. Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radical Biol. Med. 2012;53:51–59. doi: 10.1016/j.freeradbiomed.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41:10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming AM, Zhou J, Wallace SS, Burrows CJ. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these "spare tires" have an evolved function? ACS Cent. Sci. 2015;1:226–233. doi: 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace SS. Base excision repair: A critical player in many games. DNA Repair. 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid. Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran PL, De Cian A, Gros J, Moriyama R, Mergny JL. Tetramolecular quadruplex stability and assembly. Top. Curr. Chem. 2013;330:243–273. doi: 10.1007/128_2012_334. [DOI] [PubMed] [Google Scholar]
- 36.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 40.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broxson C, Hayner JN, Beckett J, Bloom LB, Tornaletti S. Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res. 2014;42:7708–7719. doi: 10.1093/nar/gku417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013;12:1159–1164. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoguanine in human cells. Nucleic Acids Res. 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitsera N, Stathis D, Luhnsdorf B, Muller H, Carell T, Epe B, Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011;39:5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Genereux JC, Barton JK. Mechanisms for DNA charge transport. Chem. Rev. 2010;110:1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 52.Alshykhly OR, Fleming AM, Burrows CJ. 5-Carboxamido-5-formamido-2-iminohydantoin, in addition to 8-oxo-7,8-dihydroguanine, is the major product of the iron-Fenton or X-ray radiation-induced oxidation of guanine under aerobic reducing conditions in nucleoside and DNA contexts. J. Org. Chem. 2015;80:6996–7007. doi: 10.1021/acs.joc.5b00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohno M, Miura T, Furuichi M, Tominaga Y, Tsuchimoto D, Sakumi K, Nakabeppu Y. A genomewide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16:567–575. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshihara M, Jiang L, Akatsuka S, Suyama M, Toyokuni S. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 2014;21:603–612. doi: 10.1093/dnares/dsu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riedl J, Ding Y, Fleming AM, Burrows CJ. Identification of DNA lesions using a third base pair for amplification and nanopore sequencing. Nat. Commun. 2015;6:8807. doi: 10.1038/ncomms9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding Y, Fleming AM, Burrows CJ. Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017;139:2569–2572. doi: 10.1021/jacs.6b12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 58.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 60.Thienpont B, Steinbacher J, Zhao H, D'Anna F, Kuchnio A, Ploumakis A, Ghesquiere B, Van Dyck L, Boeckx B, Schoonjans L, Hermans E, Amant F, Kristensen VN, Koh KP, Mazzone M, Coleman ML, Carell T, Carmeliet P, Lambrechts D. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohno M, Sakumi K, Fukumura R, Furuichi M, Iwasaki Y, Hokama M, Ikemura T, Tsuzuki T, Gondo Y, Nakabeppu Y. 8-oxoguanine causes spontaneous de novo germline mutations in mice. Sci. Rep. 2014;4:4689. doi: 10.1038/srep04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, Schuermann D, Michalakis S, Kosmatchev O, Schiesser S, Steigenberger B, Raddaoui N, Kashiwazaki G, Muller U, Spruijt CG, Vermeulen M, Leonhardt H, Schar P, Muller M, Carell T. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 63.Cooper DN, Mort M, Stenson PD, Ball EV, Chuzhanova NA. Methylation-mediated deamination of 5-methylcytosine appears to give rise to mutations causing human inherited disease in CpNpG trinucleotides, as well as in CpG dinucleotides. Hum. Genomics. 2010;4:406–410. doi: 10.1186/1479-7364-4-6-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sahakyan AB, Balasubramanian S. Single genome retrieval of context-dependent variability in mutation rates for human germline. BMC Genomics. 2017;18:81. doi: 10.1186/s12864-016-3440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, Li C, Liu B, Luo Y, Zhu Y, Zhang N, He S, He C, Wang H, Chen D. N(6)-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]