Abstract
We reported that single oncosuppressor-mutated (SOM) cells turn malignant when exposed to cancer patients’ sera. We tested the possibility to incorporate this discovery into a biological platform able to detect cancer in healthy individuals and to predict metastases after tumor resection. Blood was drawn prior to tumor resection and within a year after surgery. Blood samples from healthy individuals or metastatic patients were used as negative and positive controls, respectively. Patients at risk for cancer were included in the screening cohort. Once treated, cells were injected into nonobese diabetic/severe combined immunodeficiency mice to monitor tumor growth. All samples of sera coming from metastatic patients transformed SOM cells into malignant cells. Four samples from screened patients transformed SOM cells. Further clinical tests done on these patients showed the presence of early cancerous lesions despite normal tumor markers. Based on the xenotransplants size, we were able to predict metastasis in three patients before diagnostic tests confirmed the presence of the metastatic lesions. These data show that this serum-based platform has potentials to be used for cancer screening and for identification of patients at risks to develop metastases regardless of the Tumor Node Metastasis (TNM) stage or tumor markers level.
Keywords: In vitro platform, transformation, metastasis, screening, prediction, patient serum, cell culture
Introduction
Despite progress in the understanding of the molecular and genetic basis of cancers, cure or even the 5-year survival rate has remained very low due to metastatic disease, which is recognized as the prominent cause of cancer-related death.1 Understanding the mechanisms underlying the metastatic process is the cornerstone to improve cancer patient survival, and such knowledge is needed to develop new prognostic and diagnostic tools.
Traditionally, metastasis is described as a multistage process initiated by cancer cells detachment from the primary tumor site, circulation of the cells in the blood flow, with subsequent homing in distant sites for the establishment of secondary foci of disease.2 In this context, research has mainly been focused on the determination of the identity of these circulating tumor cells (CTCs). Nowadays, the detection and molecular characterization of CTCs are one of the most active areas of translational cancer research.3 If on one hand, tremendous increase in the amount of research, examining the potential clinical utility of CTCs in the management of cancer has been accomplished, on the other hand, the analytical specificity and clinical utility of these detection methods have not been demonstrated unequivocally.4 Controversies have arisen since reports from different investigators have shown conflicting results regarding the prognostic relevance of CTCs, and their exploitation, as a prognostic marker, is still a subject of many ongoing investigations.5–9 Furthermore, the lack of correlation between the presence of CTCs and development of metastatic disease has triggered questions regarding the undisputed validity of the “seed and soil” theory.10
Recent studies have reported that human cancer cells could transfer signaling molecules to target cells predisposing them to malignant transformation.11–13 This suggests that metastases might occur via transfer of biologically active circulating factors, derived from the primary tumor, to susceptible target cells located in distant organs. This alternative theory has been strengthened by the discovery that blood-circulating factors (i.e. cell-free nucleic acids) or factors carried in circulating microvesicles (such as mRNA, micro-RNA, mutated and amplified oncogene sequences and retrotransposon elements) are indeed shed from several types of human tumors and have different biological effects on distinct types of cells.14–21
The oncogenic potential of circulating factors has been first described in murine immortalized fibroblasts and was called “genometastasis.”22–24 More recently, we observed that exposing immortalized or single oncosuppressor-mutated (SOM) human cells (i.e. human embryonic kidney 293 (HEK293) cells and breast cancer 1 (BRCA1)-deficient fibroblasts) to metastatic cancer patients’ serum induced their transformation into malignant cells, confirming the validity of the genometastasis theory in human cells. The above effect was not seen if these cells were exposed to sera coming from healthy patients.25,26 Cells transformed even when they were exposed to conditioned media obtained from colon cancer cell cultures proving that the oncotransforming factors were actually produced by cancer cells. Based on these observations, we hypothesize that the ability of SOM cells to incorporate cancer factors could constitute the basis of a novel in vitro serum-based platform that could function both as a cancer screening test for healthy patients and as a predictor of metastatic recurrence after primary tumor resection in cancer patients.
Materials and methods
Blood collection and serum preparation
Blood samples were collected in vacutainer tubes containing clot-activation additive and a barrier gel to isolate serum (Becton Dickinson, Franklin Lakes, New Jersey, USA). After 60 min clotting at room temperature, tubes were centrifugation at 1500 × g for 15 min. Serum was collected and subjected to a second centrifugation at 2000 × g for 10 min to clear it from contaminating cells. Serum was aliquoted and stored at −80°C until use.
Patient categorization
Recruitment for this study was conducted at the department of General Surgery at the Royal Victoria Hospital and St Mary’s Hospital (Montreal, Canada), in accordance to an approved ethics protocol by the Ethics Committee of our institution (SDR-10-057). Thirty-seven patients were studied (Table 1). The test was conducted using, as a detecting biological platform, HEK293 cells and BRCA1 knockout (BRCA1-KO) fibroblasts.26 In the HEK293 group, five patients who had undergone resection of primary cancer and readmitted for treatment of metastatic disease served as positive controls (cases M1–M5). Eight healthy individuals (cases C1–C8) were enrolled in the study as a negative control group. The inclusion criteria considered to be enrolled in the healthy cohort were (i) age (30–60-year old), (ii) no signs and symptoms or personal history of cancer, and (iii) family history negative for cancer. Eight patients with clinical suspicion for cancer were enrolled in the screening cohort (cases S1–S8). Sixteen patients admitted for resection of different cancers were recruited and monitored for metastatic recurrence (cases F1–F16). In the BRCA1-KO fibroblast group (Table 2), 3 healthy individuals were used as negative controls (cases C1–C3), 10 patients with metastatic disease served as positive controls (cases M6–M15), 2 patients (cases S8 and S9) were enrolled in the screening group, and 2 patients (cases F17 and F18) in the monitoring group. Patient medical statuses and follow-up durations (mean = 22 months; range 3–60 months) are summarized in Tables 1 and 2. Blood samples were obtained with written consent from all participants.
Table 1.
Clinical profiles of the patients enrolled (HEK293 cells group).
| Cases ID | Blood collection | Disease | TNM status | Transforming potential | Metastases | Follow-up (months) | CEA (ng/ml) |
|---|---|---|---|---|---|---|---|
| C1 | – | Healthy | – | No | – | – | – |
| C2 | – | Healthy | – | No | – | – | – |
| C3 | – | Healthy | – | No | – | – | – |
| C4 | – | Healthy | – | No | – | – | – |
| C5 | – | Healthy | – | No | – | – | – |
| C6 | – | Healthy | – | No | – | – | – |
| C7 | – | Healthy | – | No | – | – | – |
| C8 | – | Healthy | – | No | – | – | – |
| M1 | Post-op (visit 1) | CRC | LM | Yes | Yes | – | – |
| M1-1 | Post-op (visit 2) | CRC | LM | Yes | Yes | – | – |
| M2 | Post-op | PcC | LM | Yes | Yes | – | – |
| M3 | Post-op | CRC | LM | Yes | Yes | – | – |
| M4 | Post-op | BC | LM | Yes | Yes | – | – |
| M5 | Post-op | CRC | LM | Yes | Yes | – | – |
| S1 | – | Healthy | – | No | – | – | – |
| S2 | – | Healthy | – | No | – | – | – |
| S3 | – | Healthy | – | No | – | – | – |
| S4 | – | Thyroid cyst | Benign | No | No | 6 | 2.0 |
| S5 | Pre-op | Panc. Cyst. | Carcinoma in situ | Yes | No | 3 | 1.2 |
| S6 | Pre-op | CRC | T1N1 | Yes | No | 8 | 1.0 |
| S7 | Pre-op | Liver mass | Benign | No | No | 3 | 2.0 |
| S8 | Pre-op | Sigmoid cancer | T3N2 | Yes | No | 3 | 2 |
| F1* | Pre-op | CRC | T3N0 | Yes | No | 7 | 3 |
| F1 | Post-op | CRC | T3N0 | Yes | No | 7 | 3.9 |
| F2* | Pre-op | CRC | T3N0 | Yes | No | 7 | 12 |
| F2 | Post-op | CRC | T3N0 | Yes | Yes | 7 | 11 |
| F3* | Pre-op | CRC | T3N0 | Yes | No | 6 | 6.7 |
| F3 | Post-op | CRC | T3N0 | Yes | No | 6 | 1.4 |
| F4 | Post-op | CRC | T3N2 | Yes | Yes | 3 | 22 |
| F5 | Post-op | CRC | T3N0 | Yes | No | 4 | 0.4 |
| F6 | Post-op | CRC | T4N1C | Yes | No | 20 | 3.7 |
| F7 | Post-op | CRC | T4N0 | Yes | No | 16 | 1.6 |
| F8 | Post-op | CRC | T3N0 | Yes | No | 12 | 1.8 |
| F9 | Post-op | CRC | T2N1 | No | No | 48 | 1.5 |
| F10 | Post-op | CRC | T4N0 | Yes | No | 60 | 1.3 |
| F11 | Post-op | CRC | T3N0 | Yes | No | 36 | 0.7 |
| F12a | Post-op | CRC | T2N0 | Yes | No | 60 | 1.4 |
| F13 | Post-op | SBC | – | Yes | No | 60 | 1.6 |
| F14 | Post-op | CRC | T3N0 | Yes | Yes | 60 | 6.7 |
| F15 | Post-op | Lymphoma | – | Yes | No | 12 | – |
| F16 | Post-op | Lung cancer | T1N0 | Yes | No | 6 | 2.1 |
BC: breast cancer; CRC: colorectal cancer; LM: liver metastasis; PcC: pancreatic cancer; Panc. cyst.: pancreatic cystic cancer; SBC: small bowel cancer; HEK293: human embryonic kidney 293; CEA: carcinoembryonic antigen.
aPatient F12 was suspected to have a metastatic lesion, but the biopsy revealed a nonmalignant lesion.
*Blood collected pre-op.
Table 2.
Clinical profiles of the patients enrolled (BRCA 1-KO fibroblasts group).
| Cases ID | Blood collection | Disease | Site of metastasis | Transforming potential | Metastases | Follow-up (months) | CEA (ng/ml) |
|---|---|---|---|---|---|---|---|
| C1 | – | Healthy | – | No | – | – | – |
| C2 | – | Healthy | – | No | – | – | – |
| C3 | – | Healthy | – | No | – | – | – |
| M6 | Post-op | Adrenal cancer | Lung | Yes | Yes | – | – |
| M7 | Post-op | BC | Lung and liver | Yes | Yes | – | – |
| M8 | Post-op | NET | Liver | Yes | Yes | – | – |
| M9 | Post-op | BC | Liver | Yes | Yes | – | – |
| M10 | Post-op | CRC | Liver | Yes | Yes | – | – |
| M11 | Post-op | Anal SCC | Liver | Yes | Yes | – | – |
| M12 | Post-op | CRC | Liver | Yes | Yes | – | – |
| M13 | Post-op | CRC | Liver | Yes | Yes | – | – |
| M14 | Post-op | CRC | Liver | Yes | Yes | – | – |
| M15 | Post-op | CRC | Liver | Yes | Yes | – | – |
| S9 | Pre-op | PcC | Peritoneal carcinomatosis | Yes | Yes | 3 | 40 |
| S8 | Pre-op | CRC | – | Yes | No | 3 | 2 |
| F17 | Post-op | CRC | Liver | Yes | Yes | 5 | 6 |
| F18 | Post-op | Lung cancer | – | Yes | No | 3 | 2 |
BC: breast cancer; CRC: colorectal cancer; PcC: pancreatic cancer; SCC: squamous cell carcinoma; NET: neuroendocrine tumor; BRCA1-KO: breast cancer 1 knockout; CEA: carcinoembryonic antigen.
Cell culture conditions
HEK293 cells were from ATCC (Manassas, Virginia, USA). BRCA1-KO fibroblasts were established as described previously.26 Cells were maintained in recommended culture medium until 30% confluence, at which point, they were treated with DMEM-F12 medium (Wisent, Québec, Canada) supplemented with 10% v/v of either cancer patient or control serum filtered through 0.2-μm filters. Half of the media was changed every second day. When the cells reached 80% confluence, they were passaged (one in six) using trypsin/ethylenediaminetetraacetic acid (Wisent). To confirm that there was no contamination or carryover of blood-derived cells, aliquots of the culture medium were placed in a culture plate and incubated at 37°C, 5% carbon dioxide.
Population doubling level calculation
Cells were considered at population doubling zero at the first time they were exposed to patient serum-containing culture medium. At every passage, cell number was determined and population doubling level (PDL) was calculated using the following formula: PDL = log(Nh/Ni)/log2, where Nh is the number of cells harvested at the end of the incubation time and Ni is the number of cells inoculated at the beginning of the incubation time. Cumulative PDL was calculated by adding the previously calculated PDL.
In vivo tumor growth
Five-week-old female nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Jackson Laboratory, Bar Harbor, Maine, USA) were used in compliance with McGill University Health Centre Animal Compliance Office (protocol 2012–7280). Cells growing in log phase were harvested and washed twice with Hank’s balanced salt solution (HBSS). Mice were injected subcutaneously with 2.106 cells in 200 μl HBSS/Matrigel mixture. Mice were followed-up daily for any sign of discomfort and tumor growth monitoring. Four weeks postinjection, mice were euthanized and tumor size recorded with a caliper.
Histological analysis
Mice xenotransplants were collected, fixed in 10% buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin according to standard protocols or processed for immunohistochemistry. Briefly, 5-μm tissue sections were dewaxed in xylene and rehydrated with distilled water. After antigen unmasking, and blocking of endogenous peroxidase (3% hydrogen peroxide), the slides were incubated with primary antibodies specific for tumor markers as described previously.25,26 Labeling was performed using iView DAB Detection Kit (Ventana, Oro Valley, Arizona, USA) on the Ventana automated immunostainer. Sections were counterstained lightly with hematoxylin before mounting. Histological analyses were performed by a certified pathologist who was blinded to the type of cells from which the cancerous masses, which formed in mice, had been derived.
Statistical analysis
For in vitro cell growth and viability, statistical differences were analyzed using an analysis of variance followed by the Scheffé post hoc test for multiple comparisons. Given that samples distribution was not normal, we applied the nonparametric Wilcoxon rank sum significance test to compare xenotumor sizes obtained with the sera of metastatic and nonmetastatic patients (regardless of groups; screening, control, or confirmed metastatic). To analyze data of the screening test, we ranked all data (tumor volumes) and determined the 0.95 percentile, which helped, set a cutoff. This was set at 0.13 cm3, and this value was used as a threshold for metastasis prediction in the screening group. For this purpose, we used 2 × 2 table and Fisher’s exact test. For all these tests, a p value <0.05 was considered statistically significant.
Results
Cancer patient sera did not affect either cell proliferation or cell viability during in vitro exposure
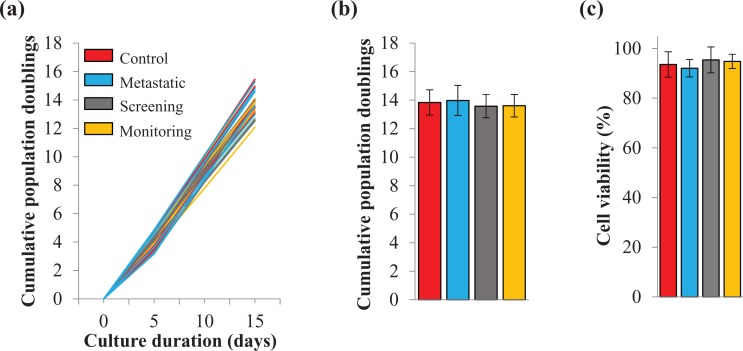
Our previous studies had shown that serum from metastatic cancer patients increased the proliferation and induced the transformation of primed human cells.25,26 In the present study, we tested the effectiveness of this discovery to be used to develop a biological platform that could be used as a cancer screening tool in healthy individuals and as a way to predict which patients might develop metastases after primary tumor resection. We verified that treated cells had the same growth potential and the same viability at the time when they were injected in NOD/SCID mice to test for their tumorigenic potential. For this purpose, we treated SOM cells for 2 weeks with daily medium refreshment. Every 5 days, cells were collected, counted, and passaged. At every passage, we calculated the PDL in each condition (Figure 1(a) and (b)). In addition, we determined cell viability (Figure 1(c)). Independently of the serum used, neither the cumulative PDL nor cell viability varied significantly. These data suggest that treated cells had the same proliferation potential at the end of the in vitro exposure regimen.
Figure 1.
Cell growth and viability were not affected during in vitro treatment duration. Cells were cultured for 2 weeks in human serum as stated in the legend. (a) and (b) Cell growth was analyzed by counting the number of viable cells at every passage (5 days duration for every passage). (a) Line graph shows the population doublings capability and (b) column graph represents cumulative population doublings at the end of the in vitro treatment periods. The ordinate axis are the same in (a) and (b). (c) Cell viability was calculated as the percentage of viable cells over total counted cells using trypan blue exclusion dye. Data are mean ± SD (p > 0.05).
Validation of the efficacy of the SOM cell platform as cancer screening test
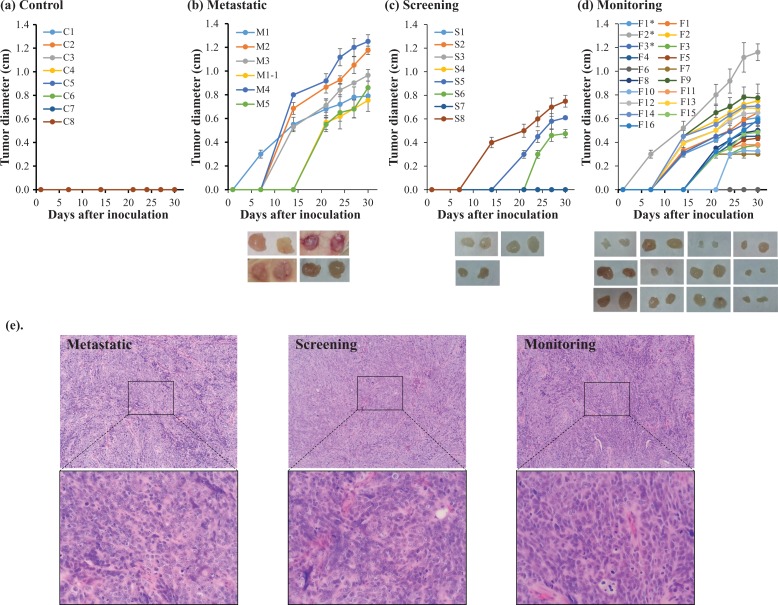
To validate the accuracy of our SOM cell-based blood test, we recruited nine patients with clinical suspicion of cancer and we enrolled them in the screening group. Eight cases were tested on HEK293 cells (cases S1–S8), and two patients (cases S8 and S9) were tested on BRCA1-KO fibroblasts. One patient was tested with both types of cells (case S8). Fifteen patients (cases M1–M5 in the HEK293 group and cases M6–M15 in the BRCA1-KO fibroblast group) who had undergone resection of primary cancer and readmitted for treatment of metastatic disease were used as positive controls. One patient was tested twice at an interval of about 3 years (case M1 and M1-1 in the HEK293 group). At the end of the in vitro treatments, cells were injected subcutaneously in NOD/SCID mice and tumor growth was monitored. Once tumors were palpable, their diameters were measured (Figure 2(b) and (c)). In addition, tumor volumes were calculated at euthanasia (Figure 3).
Figure 2.
Time course of xenotransplants growth. (a) to (c) HEK293 cells were cultured for 2 weeks in human serum as stated in the figure. Cells were injected subcutaneously in NOD/SCID mice and tumor growth was monitored every second day. Once tumors were palpable, their diameters were measured (a) to (d), and their volumes at euthanasia were calculated (see Figure 3). Values are mean ± SD (n = 2–3 mice per group). Representative pictures of tumors obtained are shown. (e) Formalin-fixed paraffin-embedded xenotransplant samples were processed for H&E staining. HEK293: human embryonic kidney 293; NOD/SCID: nonobese diabetic/severe combined immunodeficiency; H&E: hematoxylin and eosin.
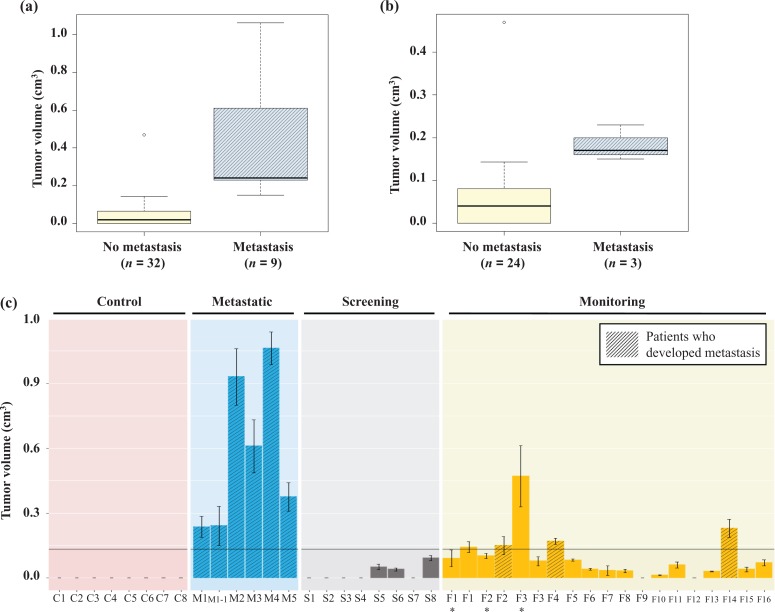
Figure 3.
Effect of patient serum on the tumorigenicity of HEK293 cells. HEK293 cells were cultured for 2 weeks in human serum as stated in the figure. Cells were injected subcutaneously in NOD/SCID mice. Four weeks after injection, growing tumors were excised and their volumes were calculated. (a) Whisker plot for the size of the xenotransplants obtained with cells treated with the sera of patients who developed metastasis and those who did not, regardless of the groups (control, metastatic, screening, or monitoring). Note that metastatic patients’ sera gave rise to tumor significantly greater than those obtained with the sera of nonmetastatic patients (p < 0.01). (b) Whisker plot for the size of the xenotransplants obtained when the analysis was done using only xenotransplants obtained with cells treated with the sera of patients in the monitoring group. Note that metastatic patients’ sera gave rise to tumor significantly greater than those obtained with the sera of nonmetastatic patients (p < 0.01). (c) Barres plot showing xenotransplants obtained for each individual involved in the study. Values are mean ± SD (n = tumors obtained with 2–3 mice per group). The line represents the threshold for metastasis prediction (set at 0.13 cm3). *Blood collected pre-op. HEK293: human embryonic kidney 293; NOD/SCID: nonobese diabetic/severe combined immunodeficiency.
Four of nine patients that were screened with the SOM cell platform transformed the SOM cells into cancer (cases S5, S6, S8, and S9). Upon further investigations, case S9 was found to have elevated carbohydrate antigen 19-9 (CA 19-9) and pancreatic cancer. Case S6 was found to have a large sessile polyp in the colon and normal carcinoembryonic antigen (CEA). Pathological analysis of the resected colon showed a T1N1 colon cancer. Case S5 on computed tomography (CT) scan was found to have large pancreatic cysts and normal CA 19-9. Because of the size of the cyst, despite normal tumor markers, the patient underwent a subtotal pancreatectomy, whose pathological analysis showed pancreatic cancer in situ. Case S8 upon further investigations was found to have a mass in the sigmoid colon and normal CEA. The pathology analysis of the resected specimen showed a T3N2 colon cancer. In summary, four of four patients that were positive at the SOM cell platform test were found to have cancer despite normal tumor markers. The other five patients who were negative at the SOM cell test were still investigated to rule out neoplastic disease. While three of them (cases S1–S3) were also negative when screened with conventional tests (serum values, imaging tests), the other two patients were found to have a thyroid nodule concerning for cancer (case S4) and a liver mass (case S7). Upon further investigations, the thyroid nodule was found to be benign as predicted from the SOM platform. The concerning features of the liver mass prompted a surgical resection of the liver whose pathology showed a benign atypical hemangioma, confirming the accuracy of the SOM platform results.
Altogether, these findings suggest that SOM cells are capable to sense cancer-produced factors even when tumor markers are negative and suggest that this in vitro blood serum-based platform might be used as a screening test to rule out neoplastic disorders.
SOM cells accurately respond to the presence of circulating cancer factors in metastatic disease and transform into cancer
All sera from patients with metastases transformed the SOM cells as confirmed by tumor formation following transplantation in NOD/SCID mice (Fisher’s exact test, p < 0.01; Tables 1 and 2, Figures 2(b), 3, and 4). Histopathological analyses of the HEK293 cells derived tumors showed that they had identical histological appearances and the types of tumors grown were not dependent of the patient’s type of cancer. The histology confirmed that most tumors were poorly differentiated carcinomas with a high mitotic index (over 90%) and small foci of necrosis (Figure 2(e)). Attempts to characterize these tumors immunohistochemically failed to show any more differentiating features.
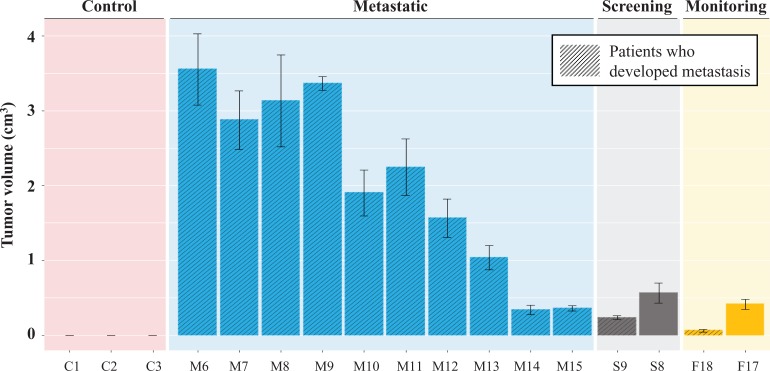
Figure 4.
Effect of patient serum on the tumorigenicity of BRCA1-deficient fibroblasts. Cells were cultured for 2 weeks in human serum as stated in the figure. Cells were injected subcutaneously in NOD/SCID mice. Four weeks after injection, growing tumors were excised and their volumes were calculated. Barres plot display xenotransplants obtained for each individual involved in the study. Values are mean ± SD (n = tumors obtained with 2–3 mice per group). NOD/SCID: nonobese diabetic/severe combined immunodeficiency; BRCA1: breast cancer 1.
On the other hand and strikingly enough, histopathological analyses of the BRCA1-KO fibroblasts showed full differentiation of the cells into colon and pancreatic cancer when they were exposed to colon cancer serum and pancreatic cancer serum, respectively, proving the hypothesis that SOM cells are able to integrate cancer factors in their genome.26
The in vitro blood test was able to predict patients at risk for metastases
Based on the evidence that the sera of metastatic patients gave rise to tumor significantly greater than those obtained with the sera of nonmetastatic patients (Wilcoxon rank sum test, p < 0.01; Figure 3(a)) and given the fact that the same trend was observed when we analyzed the xenotransplants obtained with cells treated with the sera of patients enrolled for the monitoring study (Wilcoxon rank sum test, p < 0.01; Figure 3(b)), we asked whether the size of the xenotransplants could be utilized as a predictor for future metastatic recurrence.
We ranked all the xenotransplants according to the tumor volumes and determined the 0.95 percentile. This allowed the determination of a cutoff value that was set at 0.13 cm3. This value was used as a threshold for metastasis prediction in patients enrolled in the monitoring group.
In this cohort of patients, we tested our in vitro cell-based platform with sera of 18 patients (cases F1–F18; Tables 1 and 2). Three of 15 patients (cases F1–F3) underwent the HEK293 test with blood drawn prior and after surgical resection of the tumor. The sera of all tested patients, but one (case F9), still retained their oncotransforming abilities even after 5 years from primary tumor resection. When we measured the size of the xenotransplants, we noticed that sera of patients who had no signs of metastasis generated xenotransplants with sizes smaller than 0.13 cm3 (0.04 ± 0.02 cm3; range 0.01–0.08 cm3; Figure 3(b) and (c)).
In contrast, in the HEK293 group, the sera of five patients (cases F1–F4 and F14) gave rise to xenotransplants larger than 0.13 cm3 (0.18 ± 0.12 cm3; range 0.14–0.47 cm3; Figure 3(b) and (c)). Four of them displayed metastatic disease (cases F1, F2, F4, and F14). Statistical analysis showed that xenotransplants volume and metastasis are dependent variables (Figure 3(b), Fisher’s exact test, p < 0.01). Three of the five patients had serum drawn also prior to surgical resection (cases F1–F3). When we compared the size of the xenotransplants, in these patients, we noticed that the size of the xenotransplants had not decreased after surgical resection but it had actually increased in two of them (cases F1 and F2). One of them (case F2) developed metastases at 7 months after the resection. The other patient (case F1) has been found to have CEA values higher than normal and is currently monitored every 3 months with CT scans to rule out metastatic recurrence (Table 1). Case F3 had the test repeated 6 months after surgery and it was noted that the size of the xenotransplants obtained with the serum this time was below the metastatic risk threshold compatible with her T3N0 stage.
Interestingly, the serum of one patient (case F12) who was diagnosed with metastatic lesions in the liver induced only a slight transformation of HEK293 cells with size of the xenotransplants not typical for metastatic disease but more in keeping with nonactive neoplastic disease. After surgical resection, the pathology of the liver revealed that the lesion in the liver was not malignant as predicted by the HEK293 test (Table 1 and Figure 3(c)).
Together, these data indicate that SOM cells are capable to sense cancer-produced factors even after the primary tumors have been removed and suggest a possible use of this biological platform as a way to identify patients at risks to develop metastases after primary cancer resection.
Discussion
Recently, our group reported that SOM cells turn into malignant cells when exposed to sera of patients with metastatic cancer. The above effect was not seen if SOM cells were exposed to sera coming from healthy patients.25,26 In the present study, our goal was to test the hypothesis that cells with a single oncosuppressor mutation might be used to identify cancer factors circulating in the blood and explore the potential to incorporate this discovery into a serum-based platform that could be used to both screen healthy patients for cancer disease and to predict which patients might develop metastases after primary tumor resection. To confirm the validity of this hypothesis and the accuracy of this in vitro blood test, we used the sera of four different cohorts of patients: patients with established metastatic diagnosis, healthy donors with no history or sign of cancer, patients with clinical suspicion of cancer, and patients with resected cancer disease.
The results of the tests in the screening group proved the effectiveness of this SOM cell platform to detect cancer-circulating material even at early stages and regardless of the presence of positive tumor markers. The ability of the test to predict pancreatic cancer in situ and early colon cancer seems promising since none of the blood tests available nowadays has shown such sensitivity. The negative pathology results shown on the two patients who were taken to surgery for strongly suspicious lesions strengthen the evidence that this test has great potentials to be also highly specific. For these reasons, it seems that the ability of SOM cells to “sense” neoplastic factors in the blood might help redefining new diagnostic approaches to cancer disease incorporating high sensitivity and high specificity in a single test effective at detecting different types of cancers, in different organs.
We called this novel test Metastatic And Transforming Elements Released Discovery platform (MATER-D platform) to highlight the newly found evidence that SOM cells are truly able to detect cancer factors circulating in the serum of patients, integrate them in their genome, and undergo malignant transformation.25,26
The observation that sera of patients who had cancer resection and are cancer free still retains its oncotransforming ability on the SOM cells, even after 5 years, paves the way to fascinating hypotheses. Speculations can be made that cancer cells or circulating mutated oncogenes are still present in the body even after resection of the primary tumor, as already shown in other studies27,28 but the oncogenic potential of these factors is counterbalanced by homeostatic processes such as the immune system, with mechanisms still unknown to us. The size of the cancerous xenografts could be viewed as a reflection of the efficiency of these mechanisms, with larger xenografts representing failure of the protective mechanisms to control the proliferation of the cancer cells. In our small study, the formation in the HEK293 group of malignant xenografts larger than 0.13 cm3 was correlated with a higher risk to develop metastases after surgical resection regardless of the TNM stage of the tumor. The evidence that in one patient (case F3) the size of the xenografts were above the metastatic threshold to then fall down below it after surgery, strengthen our belief that the size of the xenografts might be a valid representative of the effectiveness of the treatment and an indicator of the interaction between cancer and the body defensive mechanisms.
We couldn’t extrapolate any metastatic risk value in the BRCA1-KO fibroblast group due to the small number of patients enrolled in this cohort. We are currently running parallel experiments with both groups of SOM cells to verify the presence of any statistically significant value in the BRCA1-KO fibroblast group.
Our recent discovery that BRCA1-KO fibroblasts fully differentiate into colon and pancreatic cancers when exposed to sera of patients with colon and pancreatic cancer, respectively, strengthens the notion proved herein that SOM cells are able to incorporate genetic material in their genome and undergo malignant transformation. Identification of the oncogenic factors responsible for the malignant transformation of the SOM cells would be the next natural step to better understand the role of these hypothetical factors in cancer disease and metastatic recurrence. Putative factors that might be implicated in the observed effects and which are being currently investigated, in order to develop new diagnostic tests, are circulating cell-free DNA, or molecules packed in circulating microvesicles.19,22–24 A growing body of evidence demonstrates that cancer cells are capable of generating microvesicles in vivo, whose number and production increase with cancer stages.11,29–33 In keeping with that, high levels of circulating microvesicles have been associated with poorly differentiated tumors and shorter disease overall survival in patients with colorectal cancer and the degree of malignancy in ovarian cancer.33,34 We reported recently that oncosuppressor-deficient target cells were significantly more prone to internalize cancer patients’ serum-derived exosomes, when compared to wild-type target cells.26 This finding suggests that the efficacy of our platform seems to be dependent on the same principle behind the liquid biopsy tests: the detection of mutated genes carried in the blood either as free circulating material or packed in exosomes or microvesicles.11,12,20,21,35–39
Although the results of our experiments are striking, several limitations can be seen in this study such as the small number of patients enrolled so far and the prevalence of mainly colorectal cancer cases. These limitations can be overcome by a proper designed clinical trial, with an adequate sample size and more heterogeneous cohorts of cancer patients.
If the validity of the MATER-D platform to detect transfecting material in the serum, of both healthy patients and primary cancer-treated patients, is proven with a larger and properly designed trial, primary and tertiary prevention might be incorporated in a single test, making the secondary prevention efficacious at its best. Furthermore, this cell-based platform might have strong potential to help identifying those circulating biomarkers that can be detected early during cancer formation and possibly during metastatic recurrence. The discovery of these factors would eventually lead to the creation of antibody-based laboratory tests, which might hold strong promise for early detection of cancer in healthy patients and for supporting cancer patient management and monitoring.
Acknowledgment
We are grateful to Ayat Salman for her assistance with the Ethical Committee approvals.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was financially supported by Giuseppe Monticciolo. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ specific colonization. Nat Rev Cancer 2009; 9: 274–284. [DOI] [PubMed] [Google Scholar]
- 3. Danova M, Torchio M, Mazzini G. Isolation of rare circulating tumor cells in cancer patients: technical aspects and clinical implications. Expert Rev Mol Diagn 2011; 11(5): 473–485. [DOI] [PubMed] [Google Scholar]
- 4. Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013; 59(1): 110–118. [DOI] [PubMed] [Google Scholar]
- 5. Cohen SJ, Punt CJ, Iannotti N, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol 2009; 20: 1223–1229. [DOI] [PubMed] [Google Scholar]
- 6. Rahbari NN, Aigner M, Thorlund K, et al. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology 2010; 138: 1714–1726. [DOI] [PubMed] [Google Scholar]
- 7. Tewes M, Kasimir-Bauer S, Welt A, et al. Detection of disseminated tumor cells in bone marrow and circulating tumor cells in blood of patients with early-stage male breast cancer. J Cancer Res Clin Oncol 2015; 141(1): 87–92. [DOI] [PubMed] [Google Scholar]
- 8. Lalmahomed ZS, Mostert B, Onstenk W, et al. Prognostic value of circulating tumour cells for early recurrence after resection of colorectal liver metastases. Br J Cancer 2015; 112: 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mikulova V, Cabinakova M, Janatkova I, et al. Detection of circulating tumor cells during follow-up of patients with early breast cancer: clinical utility for monitoring of therapy efficacy. Scand J Clin Lab Invest 2014; 74(2): 132–142. [DOI] [PubMed] [Google Scholar]
- 10. Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 2003; 3(6): 453–458. [DOI] [PubMed] [Google Scholar]
- 11. Skoj J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10: 1470–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abdel-Mageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells 2014; 32: 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Venugopal C, Wang XS, Manoranjan B, et al. GBM secretome induces transient transformation of human neural precursor cells. J Neurooncol 2012; 109: 457–466. [DOI] [PubMed] [Google Scholar]
- 14. Runz S, Keller S, Rupp C, et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol Oncol 2007; 107: 563–571. [DOI] [PubMed] [Google Scholar]
- 15. Gaiffe E, Pretet JL, Launay S, et al. Apoptotic HPV positive cancer cells exhibit transforming properties. PLOS ONE 2012; 7: e36766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 2011; 71: 3792–3801. [DOI] [PubMed] [Google Scholar]
- 17. Peinado H, Alekovi M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a prometastatic phenotype through MET. Nat Med 2012; 18: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011; 2: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta 2007; 1775(1): 181–232. [DOI] [PubMed] [Google Scholar]
- 20. Harris DA, Patel SH, Gucek M, et al. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS One 2015: 10(3): e0117495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014; 3: 24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garca-Olmo D, Garca-Olmo DC, Ontanon J, et al. Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol Histopathol 1999; 14: 1159–1164. [DOI] [PubMed] [Google Scholar]
- 23. García-Olmo DC, Domínguez C, García-Arranz M, et al. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res 2010; 70: 560–567. [DOI] [PubMed] [Google Scholar]
- 24. Trejo-Becerril1 C, Perez-Cardenas E, Taja-Chayeb L, et al. : Cancer progression mediated by horizontal gene transfer in an in vivo model. PLOS One 2012; 7(12): e52754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abdouh M, Zhou S, Arena V, et al. : Transfer of malignant trait to immortalized human cells following exposure to human cancer serum. J Exp Clin Cancer Res 2014; 33(1): 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamam D, Abdouh M, Gao Z-H, et al. Transfer of malignant trait to BRCA1 deficient human fibroblasts following exposure to serum of cancer patients. J Exp Clin Cancer Res 2016: 35: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindforss U, Zetterquist H, Papadogiannakis N, et al. Persistence of K-ras mutations in plasma after colorectal tumor resection. Anticancer Res 2005; 25: 657–661. [PubMed] [Google Scholar]
- 28. Ryan BM, Lefort F, McManus R, et al. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut 2003; 52: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergsmedh A, Szeles A, Henriksson M, et al. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci U S A 2001; 98: 6407–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Al-Nedawi K, Meehan B, Rak J. Microvesicles: Messengers and mediators of tumor progression. Cell Cycle 2009; 8: 2014–2018. [DOI] [PubMed] [Google Scholar]
- 31. Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One 2009; 4: e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for premetastatic niche formation: old sayings and new thoughts. Semin Cancer Biol 2011; 21: 139–146. [DOI] [PubMed] [Google Scholar]
- 33. Silva J, Garcia V, Rodriguez M, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 2012; 51: 409–418. [DOI] [PubMed] [Google Scholar]
- 34. Taylor DD. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008; 110:13–21. [DOI] [PubMed] [Google Scholar]
- 35. Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol 2012: 83(11): 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014; 30:255–289. [DOI] [PubMed] [Google Scholar]
- 37. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002: 2: 569–279. [DOI] [PubMed] [Google Scholar]
- 38. Ogorevc E, Kralj-Iglic V, Veranic P. The role of extracellular vesicles in phenotypic cancer transformation. Radiol Oncol 2013; 47(3): 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Candelario KM, Steindler DA. The role of extracellular vesicles in the progression of neurodegenerative disease and cancer. Trends Mol Med 2014; 20(7): 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]