Abstract
Microglia are the primary innate immune cell type in the brain, and their dysfunction has been linked to a variety of central nervous system disorders. Human microglia are extraordinarily difficult to obtain for experimental investigation, limiting our ability to study the impact of human genetic variants on microglia functions. Previous studies have reported that microglia-like cells can be derived from human monocytes or pluripotent stem cells. Here, we describe a reproducible relatively simple method for generating microglia-like cells by first deriving embryoid body mesoderm followed by exposure to microglia relevant cytokines. Our approach is based on recent studies demonstrating that microglia originate from primitive yolk sac mesoderm distinct from peripheral macrophages that arise during definitive hematopoiesis. We hypothesized that functional microglia could be derived from human stem cells by employing BMP-4 mesodermal specification followed by exposure to microglia-relevant cytokines, M-CSF, GM-CSF, IL-34, and TGF-β. Using immunofluorescence microscopy, flow cytometry, and reverse transcription polymerase chain reaction, we observed cells with microglia morphology expressing a repertoire of markers associated with microglia: Iba1, CX3CR1, CD11b, TREM2, HexB, and P2RY12. These microglia-like cells maintain myeloid functional phenotypes including Aβ peptide phagocytosis and induction of pro-inflammatory gene expression in response to lipopolysaccharide stimulation. Addition of small molecules BIO and SB431542, previously demonstrated to drive definitive hematopoiesis, resulted in decreased surface expression of TREM2. Together, these data suggest that mesodermal lineage specification followed by cytokine exposure produces microglia-like cells in vitro from human pluripotent stem cells and that this phenotype can be modulated by factors influencing hematopoietic lineage in vitro.
Keywords: neurodegenerative disease, microglia, stem cells, gliogenesis, dementia and neurological disorders
Introduction
A rapidly growing body of evidence suggests that microglia, central nervous system (CNS) tissue-resident innate immune cells, are active participants in the pathogenesis of neurodegeneration. Variants in microglia/macrophage genes such as triggering receptor expressed on myeloid cells 2 (TREM2) or colony-stimulating factor 1 receptor (CSF-1R) are sufficient to cause nearly fully penetrant neurodegenerative diseases (Paloneva et al., 2000, 2002; Rademakers et al., 2012) or lead to increased risk of AD (Guerreiro et al., 2013; Jonsson et al., 2013). Genome wide association studies have highlighted the role of innate immune genetic variation in conferring risk for Alzheimer disease (Heneka et al., 2015; International Genomics of Alzheimer's Disease, 2015; Villegas-Llerena et al., 2016). These studies provide strong evidence in support of the hypothesis that myeloid cell dysfunction can drive pathogenesis in CNS disorders. Therefore, understanding the molecular pathways regulating normal and abnormal microglia function in the context of disease holds promise to identify targetable pathways for disease intervention. In vitro study of patient-derived microglia expressing disease risk variants is a potential avenue to elucidate these pathogenic mechanisms.
Human autopsy tissue captures the heterogeneity of cell phenotype and the consequence of progressive neurological disease at end stage, but is cannot be used in experimental systems to test hypotheses of disease pathogenesis. Murine models provide powerful tools to study disease, and observe how aging, environment, and the interplay between multiple organ systems influence disease pathogenesis. However, murine systems are limited by the differences between murine and human genome and molecular evolution of the immune response. Therefore, a significant need has arisen for approaches amenable to the experimental study of human microglia cells. While human microglia can be cultured from the fetal CNS, access to this tissue is limited and unreliable. Furthermore, these primary cultures have several key limitations including but not limited to the inability to control their environmental exposures prior to culture, underlying genetic diversity, early developmental state, and lack of expedient means to modulate of gene expression. The ability to generate cells derived from a stem cell population that function similarly to fully differentiated, adult microglia would greatly enhance our ability to study the function of human microglia in disease model systems. Techniques for human stem cell differentiation into CNS myeloid cells have been reported in the context of a three-dimensional (3-D) multicellular model where microglia are derived from mesoderm (Schwartz et al., 2015). A recently reported method to differentiate human microglia-like cells directly from embryoid bodies (EBs) bypassed an exogenous molecular mesodermal specification step and employed defined media containing cytokines to drive acquisition of a microglial phenotype (Muffat et al., 2016) while two more recent approaches have differentiated microglia-like cells directly from stem cell-derived hematopoietic progenitors (Abud et al., 2017; Pandya et al., 2017).
Several reports have described in vitro tools for generating microglia-like cells from murine stem cells through a heterogeneous CNS organoid culture intermediate state (Tsuchiya et al., 2005; Napoli et al., 2009; Beutner et al., 2010). While an obvious strength of this approach is the maintenance of a neural environment during microglia cell derivation, it is unclear whether this approach can be replicated using human pluripotent stem cells or whether the resulting cells will recapitulate key features of human microglia in vitro. Furthermore, the distinct role of differentiating/specification factors in human cells versus murine cells suggests the need to assess the species specific efficiency of differentiation protocols (Kelly and Hirschi, 2009). In addition, since the original publication of differentiation protocols for murine embryonic stem (ES) cell-derived microglia, multiple novel molecular markers of microglia differentiation have been reported (Hickman et al., 2013; Butovsky et al., 2014). Thus, it remains to be determined if the strategy of initiating neural differentiation to generate microglia-like cells will consistently yield cells with a microglia signature across species.
Microglia have a distinct transcriptional pattern that can be distinguished from the transcriptome of circulating monocytes (Hickman et al., 2013; Butovsky et al., 2014; Crotti and Ransohoff, 2016). In addition to a cell-type-specific transcriptome, microglia have a unique ontogeny that distinguishes them from other myeloid cells. Fate mapping studies demonstrate that microglia derive from blood islands in the embryonic yolk sac, migrating to brain at mouse embryonic Days 7 to 8 and are a component of “primitive” hematopoiesis (Ginhoux et al., 2010). This developmental ontogeny is distinct from the later “definitive” hematopoiesis, which eventually gives rise to tissue macrophages of other organs as well as circulating monocytes (Slukvin, 2013; Prinz and Priller, 2014). The elucidation of distinct microglia ontogeny has suggested one feature that may underlie their distinct physiological roles in human neurological disease. In this context, protocols for differentiating microglia-like cells from human pluripotent stem cells that recapitulate the unique ontogenic experience of microglia may be a preferred means of generating an in vitro approach for the study of human microglia. Both ES and induced pluripotent stem (iPS) cells are currently employed for CNS differentiations; both confer advantages. iPS cells can be created directly from patient cells, thus allowing for association between disease phenotype in vivo and cellular phenotype in vitro. Existing human ES lines are commercially available and have been engineered to express inducible CRISPR/Cas 9 systems (Gonzalez et al., 2014) which provide a powerful platform for systematic knockout and knockin strategies.
We sought to develop a method to derive microglia from ES and induced pluripotent stem (iPS) cells in two-dimensional (2-D) culture that draws upon current understanding of microglial ontogeny. We further aimed to reproducibly generate viable defined populations that can be employed in functional studies relevant to human neurodegenerative disease. Human ES and iPS cells were first specified toward mesoderm using previously reported approaches (Winnier et al., 1995) and then exposed to a cocktail of cytokines previously demonstrated to promote microglia differentiation and maturity (Etemad et al., 2012; Ohgidani et al., 2014). The resulting cells expressed molecular markers associated with microglia and demonstrated physiological responses including the induction of NFkB responsive genes after exposure to lipopolysaccharide (LPS) and the capacity to phagocytose aggregated amyloid beta peptide (Aβ). To further assess how recapitulation of myeloid ontogeny influences the molecular features of microglia in vitro, we assessed the impact of extracellular signals involved in definitive hematopoiesis. We observed that exposure to concurrent Wnt activation and activin inhibition, signals identified as promoting definitive hematopoiesis (Sturgeon et al., 2014) resulted in decreased expression of the microglia associated surface protein TREM2 (Schmid et al., 2002; Takahashi et al., 2005; Hickman and El Khoury, 2014). These findings align with a recent human stem cell microglia (ScMglia) differentiation protocol incorporating activin signaling which results in high efficiency (Abud et al., 2017). Taken together, the results reported here demonstrate that recapitulation of early embryonic mesodermal specification can be employed to generate human microglia-like cells derived from human ES and iPS cells.
Materials and Methods
Pluripotent Stem Cell Culture
Skin fibroblasts from healthy control subjects were collected through institutional review board (IRB) approval with the University of Washington Alzheimer Disease Research Center. Fibroblasts were reprogrammed, through approval of the University of Washington IRB and in compliance with institutional ethical standards, into pluripotent stem cells using retrovirally delivered vectors (modified from reprogramming vectors available at Addgene) based on published protocols (Park et al., 2008): pmxs-oct4-ires-gfp, pmxs-sox2-ires-gfp, pmxs-klf4-ires-gfp, pmxs-myc-ires-gfp, and pmxs-glis1-ires-gfp and isolated after 10 days of mouse embryonic feeder (MEF) layer culture at 37℃ with 5% CO2 in medium consisting of Dulbecco’s modified Eagle’s Medium with F12 nutrient mix (1:1, DMEM/F12, Invitrogen), 20% knockout serum replacement (Invitrogen, Carlsbad CA), 20 ng/ml basic fibroblast growth factor (bFGF, Peprotech, #100-18B), 1% non-essential amino acids (NEAA, Invitrogen), 1% penicillin/streptomycin (P/S, Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, St. Louis MO), and 2 mM L-glutamine (Invitrogen). iPS and H1 ES cells were subsequently transferred to culture on Matrigel (Corning) in mTeSR1 medium (StemCell Technologies). H1 ES cells (WA01, WiCell Research Institute) were fed daily and cells were passaged either 1:2 or 1:3 approximately every 6 days using 1 mg/ml dispase (Sigma-Aldrich) in the case of ES cells. Prior to differentiation experiments, iPSC lines were passaged at the same frequency, but grown in culture on MEF cells. To dissociate iPSC colonies from the MEF feeder layer, iPSC were dissociated first with Collagenase IV (Gibco Cat#17104-019) washed off the feeder layer with media then transferred to a 15 ml conical tube. To separate iPSC from MEF single cells, the tube rested unperturbed for 30 min at 37°, allowing the iPS colonies to settle to the bottom while single-cell MEF cells remained in the supernatant.
Stem Cell Differentiation
EB Generation: On Day 0, human stem cells were lifted with Dispase (Gibco) from one well of a six-well plate and split into four wells of a 96-well V-bottom culture plate (Corning), centrifuged at 800 × g for 3 min, and incubated overnight at 37℃ in DMEM/F12 plus 1% P/S, 1% L-glutamine, 20 ng/ml bFGF, 1 × N2 supplement (Invitrogen), 1 × B27 supplement (Invitrogen), 0.05% bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO, A1595-50 mL), 10 µM Y-27632, 0.4% polyvinyl alcohol (Figure 1). Cells from one well of a six-well plate produced approximately four to eight EBs.
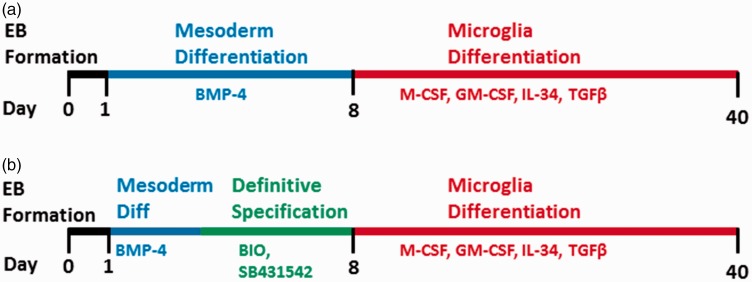
Figure 1.
Schematic representation of microglia differentiation from human pluripotent stem cells (hPSC). (a) Microglial differentiation methods began with BMP-4 to direct stem cells down a mesodermal lineage during nonadherent embryoid body (EB) culture by Day 1. After introduction of EBs to adherent culture on poly-l-ornithine and fibronectin, differentiation cues were provided by M-CSF, GM-CSF, IL-34, and TGF-β. EBs were manually lifted and replated into new wells every 4 to 6 days, starting on Day 24. (b) For experiments involving differentiation toward a definitive mesoderm lineage, BMP-4-containing differentiation medium was replaced from Day 3 to Day 8 with differentiation medium fortified with the small molecules SB431542 and BIO, which inhibit TGFβ signaling through activin receptor-like kinases and promote Wnt signaling through inhibition of GSK-3, respectively.
Pelleted EBs were transferred the following day (Day 1) into 96-well ultra-low attachment plates (Corning, #7007) containing 200 µl of DMEM/F12 plus 1% P/S, 1% L-glutamine, 20 ng/ml bFGF, 10 ng/ml bone morphogenic protein 4 (R&D Systems, #314-BP-010), and 10% fetal bovine serum (FBS). EBs were transferred to new ultra-low attachment wells containing fresh differentiation medium on Day 3. Induction toward a definitive mesodermal lineage was begun on Day 3 by transferring EBs into new wells containing 200 µl of DMEM/F12 plus 1% P/S, 1% L-glutamine, 20 ng/ml bFGF, 4 µM of Wnt pathway activator 6-bromoindirubin-3’-oxime (BIO, StemCell Technologies, Vancouver, BC, Canada), and 2 µM of activin pathway inhibitor SB431542 (Figure 1(b)).
Six-well culture plates were adsorbed with 0.01% poly-L-ornithine (PLO, Sigma-Aldrich) for at least 2 hr, washed three times with phosphate-buffered saline (PBS, Invitrogen, Carlsbad CA), and adsorbed with 1 µg/ml fibronectin (Fn, Sigma-Aldrich, St. Louis, MO, #F1141) for at least 2 hr and washed three times with PBS. On Day 7, EBs were transferred to coated plates (Passage 0, P0) and fed with a 50%/50% mix of Microglia Medium (ScienCell Research, #1901) and specification medium (DMEM/F12 with 1% P/S, 1% L-glutamine, 20 ng/ml bFGF, 1 × B27, 1% FBS [Atlanta Biologicals], 10 ng/ml human granulocyte macrophage colony-stimulating factor [GM-CSF, R&D Systems, Minneapolis MN, #215-GM-050/CF], 10 ng/ml human macrophage colony-stimulating factor [M-CSF, R&D Systems, #216-MC-025], 10 ng/ml interleukin 34 [IL-34, R&D Systems, #5265-IL-010/CF], and 2 ng/ml TGFβ-1 [R&D Systems, #240-B-002]). For all experiments employing flow cytometry analysis or sorting for functional assays, three EBs were cultured for differentiation in one well of a six-well dish prior to collection of cells for assay. For experiments analyzing gene expression of microglia markers (Figure 3), individual EBs were plated and differentiated independently in one well each of a 12-well dish. Cells were fed with this medium mix every 2 to 3 days for at least 20 days and until experimentation. After Day 23, EBs were manually lifted using a P1000 pipette and transferred to a new PLO/Fn-coated plate every 4 to 6 days, increasing EB passage number by one each time. Quantification for surface marker expression, mRNA expression, and phagocytosis were performed using samples up to and including P3.
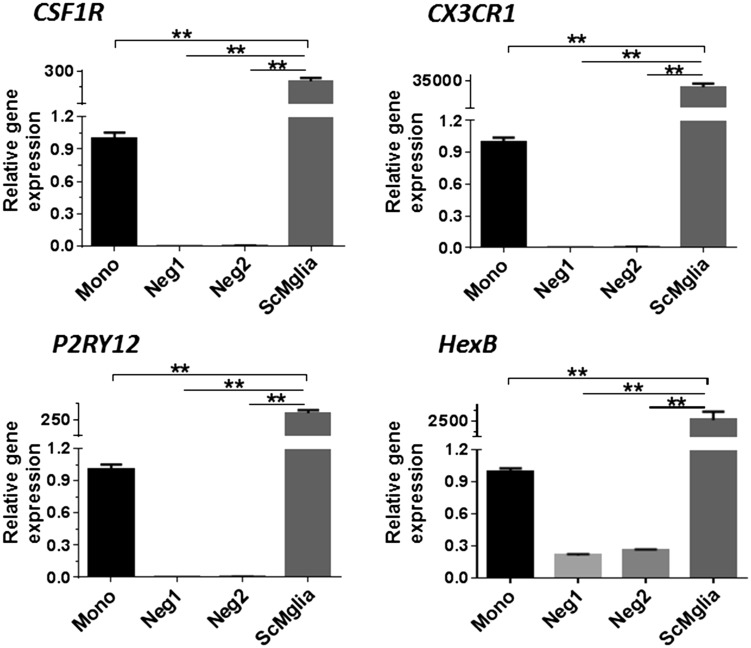
Figure 3.
ES-derived ScMglia express increased levels of genes associated with microglia. Individual EBs from one ES line were differentiated in independent wells. Gene expression for CSF1R, CX3CR1, P2RY12, and HexB in wells demonstrating microglia-like cells as assessed by morphology (ScMglia) were compared with wells in which no microglia-like cells were observed (Neg1, Neg2) and undifferentiated human circulating monocytes (Mono). RNA was collected from individual differentiation wells (n = 3 technical replicates, analyzed by one-way analysis of variance and Tukey’s post hoc test **p < .01).
Immunofluorescence Staining
Cells were washed with PBS and fixed with 4% paraformaldehyde (Electron Microscopy Science, Hatfield PA) for 15 min. Cells were washed with PBS containing 1 mg/ml magnesium chloride (Sigma-Aldrich) and 0.1 mg/ml calcium chloride (Sigma-Aldrich) three times and then blocked and permeabilized with blocking buffer (PBS with 0.5 mg/ml sodium azide, 0.1% BSA, 1 mM EDTA, 0.5% Triton X-100, and 10% goat serum (Sigma-Aldrich)) for 1 hr at room temperature (RT). Cells were then stained for 1 hr at RT with rabbit anti-Iba1 (Wako Pure Chemical Industries, Richmond, VA, 019-19741) diluted in Antibody Buffer (PBS with 0.5 mg/ml sodium azide, 0.1% BSA, 1 mM EDTA, 0.5% Triton X-100). Cells were washed three times with PBS containing MgCl2 and CaCl2 then stained for 20 min with Alexa Fluor 594 goat anti-rabbit secondary antibody (1:200 dilution, Life Technologies, Carlsbad CA) in antibody buffer. Cells were washed three times with PBS containing MgCl2 and CaCl2 then counterstained with 4’,6-diamidino-2-phenylindole for 5 min at RT. Cells were washed once more with PBS and imaged using an inverted Zeiss Axiovert 200 M fluorescent microscope.
Flow Cytometry
Flow cytometry beginning with 1000 to 5000 cells was performed by trypsinizing cells after manual removal of EBs, centrifuging at 300 × g for 10 min at 4℃, resuspending in 200 µl fluorescence-activated cell sorting (FACS) Medium (HBSS plus 1% 1 M [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] [HEPES, Life Technologies, #15630-080] pH 7.3 and 10% FBS). Cells were blocked with 4 µl Human Fc block (BD Pharmingen, #564219) per ml of FACS medium and incubated on ice for 5 min. Cells were then labeled with either conjugated antibodies or isotype controls (list antibodies) for 45 min on ice. Cells were washed three times with cold FACS medium, centrifuging at 500 × g for 4 min each time before being resuspended in 350 µl of FACS medium and analyzed on either an LSRII flow cytometer (BD Biosciences) for surface marker quantification or a FACSAria III cell sorter (BD Biosciences) using FACSDiva software. Antibodies used for surface marker expression quantification were rat anti-human CX3CR1-APC (Clone 2A9-1, eBioscience, Cat#17-6099-42), monoclonal rat anti-human CD11b-DyLight550 (Clone M1/70.15, Novus Biologicals, #NB600-1327R), and monoclonal rat anti-human TREM2-Alexa Fluor 700 (R&D Systems, #FAB17291N), each at 1:200 dilution. Compensation was performed using unstained stem cell-derived microglia-like cells and single-stained controls while gates were set using fluorescence-minus-one controls. Data were analyzed using FCS Express software (De Novo Software).
Cells sorted based on surface marker expression were either collected in FACS medium, pelleted, and lysed with QIAzol or sorted directly into culture plates for use in the microglial activation assays.
Aβ Phagocytosis
Aβ 1-42 (American Peptide Company, #H-1368) was resuspended to 100 µM in PBS and allowed to fibrillize at 37℃ for 48 hr (Reed-Geaghan et al., 2009). pHrodo (Life Technologies, P36600) was dissolved in dimethyl sulfoxide at 100 µg dye per 10 µl solvent. Aβ was thawed and centrifuged at 14,000 rpm for 10 min, supernatant was aspirated, and the pellet was resuspended in 110 µl 0.1 M NaHCO3; 16.5 µl of pHrodo solution was added, and the mixture was vortexed for 1 hr at RT; 500 µl of Hank’s balanced salt solution (HBSS, Invitrogen) was added, and the solution was centrifuged for 1 min at 14,000 rpm. The solution was washed once in 100% methanol and then three more times in HBSS before being resuspended at 100 µM.
Stem cells were differentiated as described earlier and following EB removal, cells were incubated with 1 µM pHrodo-conjugated Aβ at 4℃ or 37℃ for 6 hr (modified from Li et al., 2015). Cells were then washed with cold PBS, trypsinized on ice, and prepared for analysis by flow cytometry, using anti-human TREM2-Alexa Fluor 700 antibody. TREM2+ stem cell-derived microglia were identified by gating based on unstained stem cell-derived microglia that had been treated with pHrodo-conjugated Aβ. The geometric mean of the TREM2+ population’s Aβ-pHrodo signal (585 nm peak, measured using a 567-597 nm bandpass detector) was compared between groups incubated at 4℃ and 37℃ with labeled Aβ protein. Results were compared with similarly treated HEK293T human tumor cells as a negative control. Significance was exerted at p < .05, as calculated by paired, one-way paired Student’s t-test, and error bars represent standard error.
Microglia Marker Gene expression
CD14 + human monocytes were collected through University of Washington IRB approval from healthy subjects. Fifty milliliter of whole blood was collected in EDTA tubes, and CD14 + cells were isolated with the MACS cell separation system and CD14 MicroBeads (Miltenyi Biotec). Adherent ES- and iPSC-derived ScMglia derived from differentiations where one EB was independently differentiated in 1 well of a 12-well dish, or cells from wells in which differentiation in the same single EB/well format did not yield microglia-like cells as determined by morphology (“negative” differentiations) were rinsed with PBS before lysing with QIAzol. RNA was extracted using miRNeasy Micro Kit (QIAgen) and transcribed to cDNA (High Capacity cDNA RT Kit, Applied Biosystems). Taqman Assay on Demand probe primer sets (Applied Biosystems) for the following genes were used: CSF1R (Hs00911250_m1), CX3CR1 (Hs01922583_s1), TMEM119 (Hs01938722 u1), HEXB (Hs01077594_M1), and P2RY12 (Hs01881698_s1). Relative gene expression was determined by calculating delta CT with respect to beta actin expression followed by delta-delta Ct to primary monocytes. Statistical significance of gene expression differences between the populations was determined by one-way analysis of variance followed by Tukey’s post hoc test.
Inflammatory Gene Expression
Differentiated ScMglia cells expressing TREM2 were sorted into PLO/Fn-coated 96-well plates by FACS and allowed to rest for 24 hr in serum-free specification medium. Cells were then treated with serum-free specification medium containing either 0 ng/ml or 50 ng/ml of LPS for 24 hr. Wells were then washed with PBS and lysed with QIAzol reagent (QIAgen). mRNA was extracted using a miRNeasy Micro Kit (QIAgen) then amplified and transcribed to cDNA (Ovation Pico WTA System V2 kit, NuGEN) before being analyzed for expression of interleukin-1 beta (IL-1β, Applied Biosystems, #Hs00174097_m1), monocyte chemoattractant factor-1 (MCP-1 or CCL2, Applied Biosystems, #Hs00234140_m1), tumor necrosis factor (TNFα, Applied Biosystems, #Hs01113624_g1), interleukin-6 (IL-6, Applied Biosystems, #Hs00985639_m1), tumor necrosis factor alpha-induced protein 3 (TNFAIP3, Applied Biosystems, #Hs00234713_m1), and cluster of differentiation 68 (CD68, Applied Biosystems, #Hs02836816_g1). Relative gene expression in LPS-treated samples was determined by calculating delta Ct with respect to beta actin expression and then by calculating delta-delta Ct with respect to the paired control (0 ng/ml LPS) sample. Significance was exerted at p < .05, as calculated by paired, one-tailed paired or unpaired Student’s t-test, and error bars represent standard error in all figures.
Results
Microglia-Like Cells can be Derived From Pluripotent Stem Cells
To determine if cells with microglia features can be derived from stem cell-derived EBs exposed to drivers of mesoderm specification, we assessed the evolution of morphological phenotype as well as microglial marker protein expression through the differentiation process. Following initial EB plating (P0), we observed cells with microglia morphological features migrating out from the EB within a heterogeneous “skirt” of cells surrounding the EB. Similar to previous reports (Muffat et al., 2016), we observed that EBs with cystic structures (Figure 2(a)) were more likely to yield a “skirt” containing cells with microglia-like morphology than dense, non-cystic EBs (Figure 2(b)). This outward migration began as early as one day after plating and continued over the next 3 weeks. Between Day 15 and Day 24, increasing numbers of adherent cells that migrated away from the EB and its “skirt” exhibited morphologies some similar to cultured primary murine microglia (Caldeira et al., 2014).
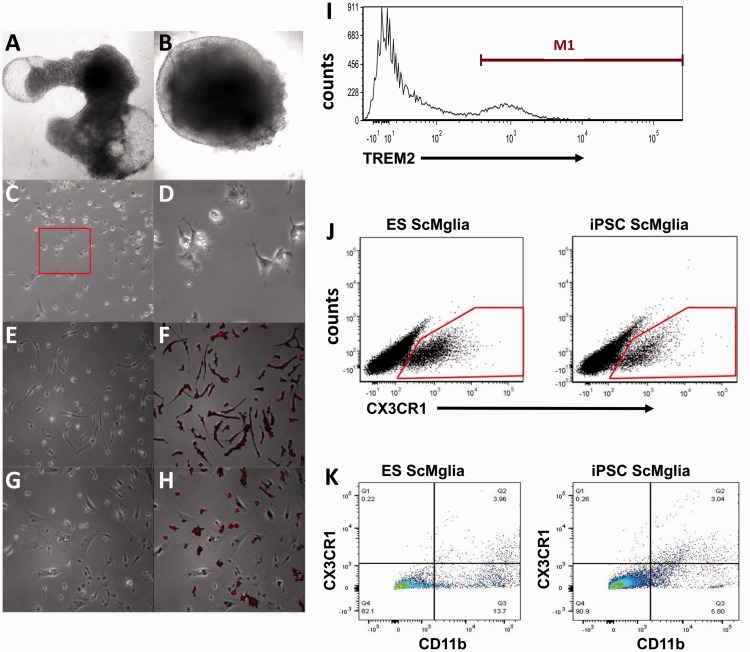
Figure 2.
Microglia-like cells (ScMglia) differentiated from hPSC develop microglia-like morphology and express microglia/myeloid markers. (a, b) Representative images (taken with 4 × objective) of cystic (a) and dense, noncystic (b) EBs in culture at Day 10. (c) Representative phase contrast images at 10 × of differentiating cells at Day 21 and (d) magnified 32 × images of microglial-like precursor cells at Day 21 exhibiting rounded morphologies with microglia-like processes (red box inset in (c)). (e) Phase image (32 × objective) of microglia-like cells (ScMglia) differentiated from iPS cells at Day 28, showing ramified morphology and even spacing among cells that have migrated away from the EB. (f) Most cells with this morphology also stained positively for microglial marker Iba1 (red). (g) Phase imaging showing fewer ScMglia were observed after extended culture (8 to 10 days after P1 plating) and (h) Iba1 immunolabeling demonstrates iPS-derived microglia-like cells continued to be Iba1 immunoreactive. Some microglia-like cells began to exhibit a more rounded morphology. (i) Representative flow cytometry analysis of ScMglia derived from an iPSC line showing 20.02% of gated cells express TREM2 (the M1 population); a total of 4,924 cells (events) were counted. (j) Flow cytometry analysis of ScMglia derived from both iPS and ES cells demonstrates both ES and iPSC–derived microglia-like cells express CX3CR1. (k) Differentiated ScMglia show double immunolabeling for CD11b and CX3CR1. Both ES and iPSC-derived cells have propensity for microglia differentiation using this protocol (representative FACS plots from at least n = 3 separate experiments).
EBs lifted and replated in new wells (P1) demonstrated new outgrowth of the EB “skirt” and migration of microglia-like cells away from the EB over a shorter timescale. Microglia-like cells were most apparent 4 to 6 days after P1 replating. After this time, the proportion of cells with microglia-like morphologies decreased over time until approximately 10 days after passage, when few ramified or rounded cells were observed. Immunofluorescent labeling for the marker Iba1 demonstrated that cells with microglia-like morphology expressed this myeloid protein while other cells in the EB skirt without a microglia-like morphology were not immunoreactive for Iba1 (Figure 2(g) and (h)). In subsequent experiments aimed at isolating microglia-like cells (here after designated as ScMglia) from the heterogeneous EB skirt, cells were harvested 4 to 6 days after passage.
EBs could be replated successfully as many as five times; however, the number of ScMglia decreased with increasing passage number. Therefore, we assessed the generation of ScMglia from EBs of one normal control iPSC line and one H1 ES line at Stages P0 to P5 to establish the optimal passage number. In these experiments, we quantified ScMglia numbers using FACS analysis for the cell surface marker, TREM2. We observed that both in iPSC and ES lines, the highest percent of TREM2 positive cells were collected in the P1 population (14.6% and 28.3% in the iPSC and the ES line, respectively, n = 3 for each line, representative flow cytometry histogram shown 2I) compared with either the P0 or late stage (P4-5) passages which ranged from 3% to 8%. These experiments revealed that EB’s derived from an iPSC or an ES line demonstrated the capacity to differentiate into TREM2 expressing microglia-like cells in response to this differentiation protocol. Furthermore, we determined that there is a maximal “peak” passage during differentiation and that prolonged time in culture eventually led to relative decrease in the proportion of TREM2 and Iba1 expressing cells within the differentiation well.
To further characterize the range of ScMglia surface protein expression and heterogeneity, ScMglia from P0 to P3 cultures containing three EBs in each differentiation well derived from either an iPSC or an ES line were analyzed by flow cytometry with a repertoire of established microglia/myeloid cell surface markers. Similar to what was observed with TREM2 expression, both the iPSC and the ES line produced CD11b and CX3CR1 expressing cells ranging from 6% to 10% and approximately 3% to 4% doubly expressing cells in both iPSC and ES lines as measured over three independent differentiation experiments (representative scatter plots shown in Figure 2(j) and (k)). To investigate whether three microglia specific surface markers are expressed by overlapping or distinct populations, we quantified triple labeled cytometry in three ES differentiations and found cells CD11b, CX3CR1, TREM2+ triple label to be 3% (n = 3). These observations suggest that microglia surface marker expression varies with a larger population expressing only one or two of the markers assayed. Furthermore, the finding supports the hypothesis that the stem cell differentiation approach employed here yields a heterogeneous population of microglia-like cells in multiple states of maturation.
We found one contributor to relatively low efficiency in production of cells expressing surface microglial markers was that not every EB eventually produced microglia-like cells. Our initial experiments involved culturing multiple EBs in the same well in order to generate a sufficient cell number for flow sorting isolation. While this approach was sufficiently robust to generate the number of ScMglia to be flow sorted for functional assays as described later, we set out to determine if differentiating each EB in individual wells would then allow a more direct comparison of marker expression in those populations which appeared to produce cells with microglia morphology (ScMglia) compared with those EBs which did not. Those differentiation populations which had not adopted a microglia morphology were termed as “negative.” In general, out of 12 EBs, two to four EBs would result in the majority of the cells in the well manifesting microglia morphology. These wells were then employed as the “ScMglia” population. The remainder of wells, considered negative, produce a heterogeneous population of cells with varying morphologies suggestive of other lineages. Using real-time reverse transcription polymerase chain reaction, mRNA for HexB, CX3CR1, CSF1R, P2Ry12, and TMEM119 was measured in the ScMglia population, the “negative” populations, and human circulating undifferentiated monocytes. The ScMglia population demonstrated significantly increased expression of microglia markers CX3CR1, P2Ry12, HEXB compared with negative EB differentiations as well as compared with undifferentiated monocytes (Figure 3). TMEM119, a recently described microglia specific protein (Bennett et al., 2016), was not measurably expressed by any of the cell types in this culture system (data not shown), suggesting that not all molecular features associated with mature microglia are recapitulated in these ScMglia. Furthermore, these studies suggested that there is heterogeneity in the propensity of each EB to yield microglia-like cells, consistent with what has recently been reported (Muffat et al., 2016).
Stem Cell-Derived Microglia-Like Cells Phagocytose Aβ
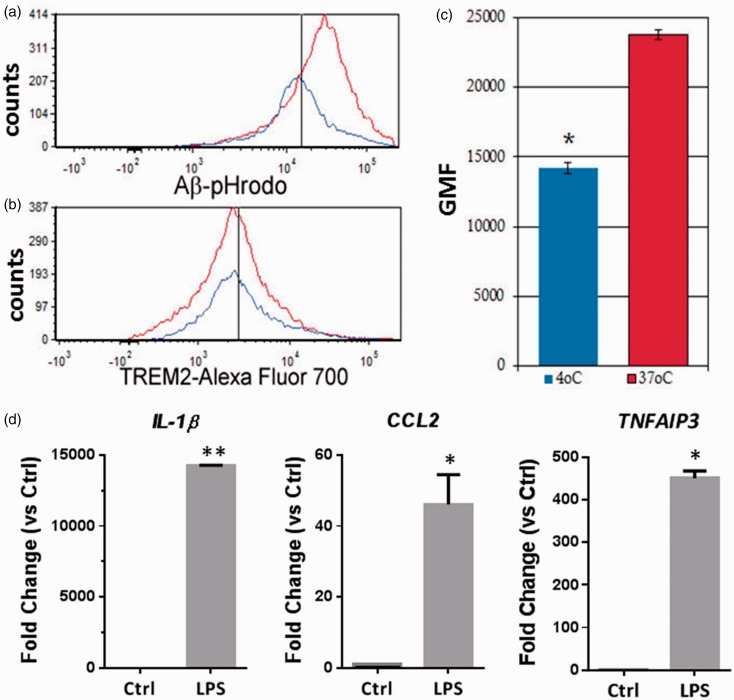
Microglia are thought to influence the pathogenesis of AD as mediators of Aβ phagocytosis and degradation. Thus, to validate the utility of the ScMglia in an in vitro functional assay, we measured the capacity of ScMglia to internalize a pH sensitive Aβ molecule that is fluorescent upon acidification within the phagosome. When treated with 1 µM pHrodo-labeled Aβ1-42 for 6 hr at either 4℃ or 37℃, TREM2 expressing ScMglia showed a statistically significant increase in pHrodo signal (n = 3, p < .0005, Figure 4(a) and (c)), demonstrating that ScMglia show increased internalization of Aβ1-42 at 37℃. Because we analyzed flow cytometry based on TREM2 expression on samples that were differentially incubated at 4° or 37℃, we asked whether temperature impacted cell-surface TREM2 expression itself. When the cell surface expression of TREM2+ cells (which had been treated with Aβ) was compared between samples incubated at 4℃ and those incubated at 37℃, no statistical difference was found, using a one-way Student’s t-test (Figure 4(b)). Thus, TREM2 surface expression does not differ by temperature and is unlikely to influence measurement of ScMglia aggregated Aβ1-42 phagocytosis.
Figure 4.
ScMglia expressing TREM2 internalize Aβ and respond to LPS. (a) Phagocytosis of Aβ was determined by a shift in cellular signal at 37℃ versus signal at 4℃. After gating data based on TREM2 expression, H1 ES-derived microglia-like cells show a positive shift in Aβ fluorophore signal at 37℃ compared with the same cells at 4℃, suggesting the presence of phagocytic internalization of Aβ in TREM2+ differentiated cells. (b) No significant difference was observed with respect to TREM2 expression between Aβ-pHrodo-treated microglial-like cells at 4℃ and those incubated with labeled Aβ at 37℃. Black vertical lines in (a) and (b) represent approximations of the average of the geometric means of fluorescence for cells incubated at 4℃. (c) Quantification of shift in fluorescent signal, indicating phagocytosis of Aβ-pHrodo proteins by TREM2+ H1 ES-derived microglia-like cells. Bars indicate average of geometric means of fluorescence from three independent experiments. (d) Expression of NFkB responsive genes, IL-1β, CCL2, and TNFAIP3 is induced in TREM2+ ES cell-derived ScMglia cells after treatment with LPS. ScMglia were first sorted for TREM2 expression, cultured for 24 hr followed by 24 hr incubation with 50 ng/ml LPS (Student’s t-test, ** p < .01; * p < .05).
Stem Cell-Derived Microglia-Like Cells Respond to Inflammatory Stimuli
Functional characterization of stem cell-derived microglia should include assessment of their ability to respond to inflammatory stimuli. To assess the functional capacity of ScMglia, we first determined whether the ScMglia generated using this protocol could be isolated by FACS for surface TREM2 expression and retain viability 24 hr after sorting. Isolated ScMglia demonstrated normal microglia morphology 24 h following FACS isolation and remained viable after stimulation with 50 ng/ml LPS for 24 hr (data not shown). After LPS treatment, ScMglia gene expression was evaluated for known downstream targets of toll-like receptor 4 (TLR4) signaling including the canonical pro-inflammatory gene IL-1β, the late NFκB responsive gene TNFAIP3, and the LPS inducible chemokine, CCL2 (MCP-1). In the TREM2+ ScMglia population, expression of IL-1β, CCL2, and TNFAIP3 were all significantly induced by LPS stimulation compared with untreated controls (Figure 4(d)). These data demonstrate that multiple genes in the TLR4/NFκB pathway are inducible in ScMglia derived using this approach, supporting the conclusion that ScMglia have the capacity to respond to an inflammatory stimulus.
Signals Promoting Definitive Hematopoiesis Negatively Influence TREM2 Expression
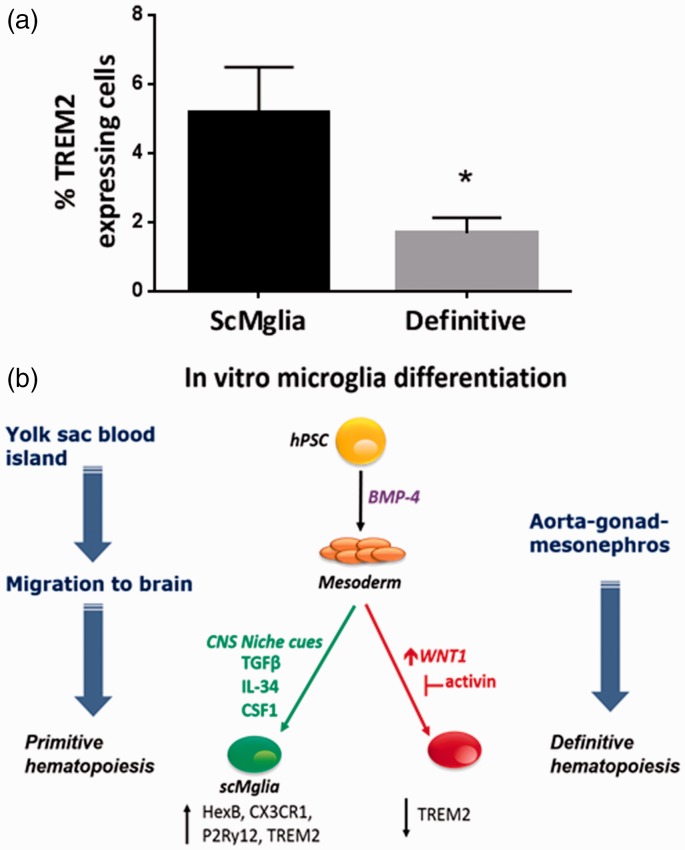
Microglia differ from other myeloid cells in developmental ontogeny. Detailed fate-mapping of mammalian developmental hematopoiesis in mice suggest that yolk sac-derived microglia precursors are generated in primitive mesoderm prior to the second wave of “definitive” hematopoiesis (Ginhoux et al., 2010). We hypothesized that EBs producing microglia-like cells are in a developmental state recapitulating primitive mesodermal specification. Therefore, we asked if exposure to small molecules driving the next wave of definitive hematopoietic specification would negatively impact the efficiency of microglia-like differentiation. Wnt signaling is the proximal driver of definitive specification during hematopoiesis while activin signaling sustains primitive specification (Sturgeon et al., 2014). To our knowledge, there have been no prior studies comparing signals for primitive hematopoiesis to factors associated with definitive hematopoiesis in their impact on the microglia-like features of stem cell-derived myeloid cells. Thus, we leveraged modulators of Wnt and activin signaling to manipulate developmental specification in culture. We introduced the Wnt signal activating compound BIO and activin pathway inhibitor, SB431542, to drive an IPSC line toward definitive hematopoiesis (Sturgeon et al., 2014) on Day 3 to 7 of the differentiation protocol, and sought to assess the relative expression of relevant microglia cell surface markers. TREM2 is more highly expressed in resident microglia and some tissue macrophage populations compared with myeloid progenitors or circulating monocytes (Turnbull et al., 2006). We observed that in iPSC cell-derived EBs (n = 5), pharmacological Wnt activation, and activin inhibition, signals promoting definitive specification resulted in significantly decreased number of cells demonstrating surface expression of TREM2 (Figure 5a).
Figure 5.
Definitive specification of hPSC decreases cell surface expression of resident macrophage/microglia marker TREM2. Comparison of BMP-4-only mesodermal differentiation method (“ScMglia”) and definitive mesodermal differentiation (“definitive”) achieved through incubation with SB431542 and BIO between Days 3 to 7 of stem cell-derived microglia-like cell differentiation protocol. Expression of TREM2 is approximately threefold higher in cells derived using the BMP-4-only ScMglia method compared with definitive (n = 5, Student’s t-test, *p < .05). (b) Select cues influencing microglia differentiation during embryonic development can be recapitulated in vitro. Formation of the embryonic body allows for development of all three germinal layers. Within yolk sac blood islands, primitive mesoderm is formed which can be simulated in culture through use of small molecules such as BMP-4. During development, primitive mesodermal precursors exit yolk sac blood islands and migrate to brain where under developmental and brain environmental cues they differentiate into microglia. Exogenous brain molecular cues, such as TFGβ and CSF1R, can be employed to mimic those encountered by primitive microglial precursors.
These data suggest that definitive specification impairs the development of ScMglia supporting the hypothesis that human microglia arise from yolk sac-derived primitive mesoderm as has been demonstrated in murine system (Ginhoux et al., 2010). Thus, the human ScMglia developed using this approach may be similar to primitive mesodermal-derived microglia-like cells that migrate into the CNS early in development and can be distinguished from mesoderm-derived peripheral macrophages that develop during definitive hematopoiesis (Figure 5(b)).
Discussion
We report here that stimulation of human ES and iPS lines with mesodermal specification cues followed by cytokines associated with the neuroglial niche produces cells exhibiting microglia-like morphology, expressing genes and cell-surface proteins, as well as functional behavior consistent with a microglial phenotype. In this study, we further demonstrated that definitive specification by small molecules used to manipulate Wnt and activin signaling can drive the cellular phenotype of differentiated cells away from what is expected for the microglia phenotype. These findings show that a relatively simple, 2-D culture protocol with BMP4-guided mesodermal differentiation produces a cell population with microglia-like morphology, surface protein, and gene expression as well as disease relevant microglia functions. This approach, similar to the recently described methods (Muffat et al., 2016; Pandya et al., 2017), demonstrates that these in vitro methods recapitulate aspects of microglial ontogeny. We show that factors known to drive definitive hematopoietic specification leads to decreased TREM2 surface expression in ScMglia, a surface marker associated with microglia maturation. This suggests that in vitro differentiation approaches such as these have the potential to capture developmental cues known to influence in vivo microglial development and remain useful candidates in in vitro disease modeling methods.
Tsuchiya et al. (2005) were among the first to report an in vitro method of generating microglia from murine stem cells using an approach modified from one designed for neuronal differentiation from murine ES cells. Following that initial report, new methods were developed describing a microglia differentiation method (Napoli et al., 2009) and further detailed in Beutner et al. (2010) based on isolation of microglial precursors after induction of neuronal differentiation in ES cells. In this protocol, driving neural differentiation gives rise to a heterogeneous population of cells with some expressing myeloid markers including CD11b, CD11c, and CD36. These myeloid cells were designated ES cell-derived microglial precursors (ESdMs). They observed that ESdMs demonstrated chemokine dependent migration, bead phagocytosis, and adoption of activation states similar to primary microglia (Beutner et al., 2010). Interestingly, cells isolated through this reported method are proliferative and can be passaged multiple times, suggesting that there are distinct features of ESdMs that are not observed in primary microglia. Further validation of this method more recently demonstrated that murine ES-derived microglia share transcriptomic and activation features with primary murine microglia (Beins et al., 2016); however, similar comparisons between human ES-derived and primary human microglia have not been reported.
Recently, microglia derived from human stem cells in the context of a 3-D neural environment were shown to express cell surface CD11b and CD14 and to phagocytose yeast particles (Schwartz et al., 2015). Stem cell-derived microglia cultured within the 3-D structure expressed several microglia associated genes including TREM2, CD11b, CD68, and Iba1. In contrast to previous methods initiating microglia differentiation from a neuronal precursor population, microglia/myeloid cells were first differentiated directly from mesodermal precursor cells and then incorporated into a neural environment. The protocol we report here, and a recently described hematopoietic progenitor-like iPSC differentiation protocol (Pandya et al., 2017), share the developmental cues to drive microglia differentiation from mesoderm state, and may also offer a tractable method for laboratories seeking to test specific phenotypes in a 2-D in vitro model.
Our finding that individual EBs demonstrated different efficiencies at yielding microglia is consistent with recent reports suggesting that EBs with a “cystic” appearance were more likely to yield microglia-like cells (Muffat et al., 2016). This principle significantly impacts ScMglia differentiation protocols as a mixed differentiation culture with multiple EBs led to markedly decreased efficiency in our hands. Identifying EBs more likely to produce microglia early, such as those demonstrating a cystic phenotype, will be valuable to incorporate into future differentiation methods. Of note, the creation of EBs prior to generating microglia precursor cells may not be absolutely critical to microglia generation. Rapidly evolving protocols have provided further insight into the in vitro tools that can be employed to produce microglia-like cells with differing methods. Pandya et al. (2017) describe the isolation of CD34 + hematopoietic progenitor-like cells differentiated directly from human iPSC cultures which can then be cocultured with astrocytes to yield a microglia-like cell population. This report suggests that microglia-like cells can be isolated following varying degrees of mesodermal specification and recapitulation of developmental structure and cues prior to microglial differentiation. It remains to be determined if there are significant phenotypic differences between the various methods, and if mesodermal specification influences the propensity for ScMglia to develop specific features associated with microglia differentiation.
We observed cell surface expression of CD11b, CX3CR1, and TREM2 in the differentiated ScMglia described here. Further, we found a heterogeneous population of cells that were singly, doubly, or triply immunoreactive for the select microglia markers. The contribution of differentiation method to the resulting diversity in microglia phenotype needs further exploration. Interestingly, similar challenges regarding heterogeneity and maturity have arisen in the efforts to differentiate stem cell-derived neuronal cell types (Dolmetsch and Geschwind, 2011) which underscores the complexity of CNS stem cell tools and the need to be clear about experimental paradigms and caveats. Regardless, the diversity of microglia function and transcriptomic signatures are well described (Schmid et al., 2009; Gertig and Hanisch, 2014; Bachstetter et al., 2015; Ritzel et al., 2015; Grabert et al., 2016). Indeed, the regional and age-related heterogeneity in microglia characteristics may reflect the plasticity of microglia cells and their ability to adapt to different environmental cues.
While the approach we utilized to generate stem cell-derived microglia does not lead to a homogenous population of microglia as determined by surface marker expression, determination of whether the stem cell-derived microglia perform disease relevant cellular functions is a key criteria for assessing their utility. To that end, we found that ScMglia cells did indeed demonstrate capacity to phagocytose Aβ peptide, a functional assay that can be utilized in studying dysfunction of innate immune mechanisms in AD. Therefore, we believe ScMglia can be employed to quantitatively evaluate the impact of disease associated genetic variants on the capacity of microglia to phagocytose a disease-relevant molecule. Induction of immune gene expression in response to a proinflammatory stimulus is another common assessment of microglia function in vitro. Consistent with a myeloid response, ScMglia demonstrate increased expression of canonical inflammatory genes, TNFAIP3, CCL2 (MCP-1), and IL-1β, in response to LPS. Expression of TNFα and IL-6 was not increased after 24 hr of exposure to LPS, which is in contrast to observations in murine primary microglia (Jayadev et al., 2010). Thus, ScMglia may be less sensitive to sole LPS stimulation, a finding suggested by previous reports comparing murine ES-derived microglia to primary microglia (Beins et al., 2016). Factors that may influence inflammatory responses include the maturity of ScMglia and inherent differences between human and murine-cultured microglia. However, the presence of a transcriptional response to proinflammatory stimulation validates these cells as usable models for specific questions relating to the influence of genetic variation on proinflammatory signaling pathways in human stem cell-derived microglia.
Given the lack of expression of TMEM119, which is expressed postnatally in mouse (Bennett et al., 2016), we suspect that ScMglia are not fully differentiated toward a mature microglia phenotype. The incomplete maturation may reflect the need for additional time after induction of differentiation or additional differentiation cues during the differentiation period. While the heterogeneity of microglia and contribution of environment in determining that spectrum of phenotype has been described (Schmid et al., 2009), the specific environmental cues experienced by microglial precursors in humans have not been fully delineated. Thus, it is not possible to utilize all factors that promote microglia maturation in in vitro microglia development models. One mechanism by which environmental cues influence microgliogenesis, phenotypic diversity, and functional heterogeneity is through epigenetic regulation of enhancer activity (Gosselin et al., 2014; Lavin et al., 2014). For instance, comparing gene expression profiles in microglia relative to peritoneal macrophage have identified cell subtype specific enhancer signatures that are regulated by the immediate local environment (Gosselin et al., 2014). Therefore, assessing the impact of various exogenous cues on enhancer activity may be an additional means for assessing the fidelity of a “microglia”-like state in vitro. Indeed, identification of what is the “true” microglia signature express is hampered by species differences, degree of manipulation once dissected from the brain, and likely plasticity of microglia once isolated suggesting that the cell analyzed ex vivo remains to some degree an imperfect representative of its in vivo counterpart.
We show here that after driving mesodermal specification in vitro, factors promoting specification to definitive hematopoiesis that give rise to circulating monocytes result in a significantly smaller population of cells expressing a key microglia-associated marker. Based on these results, we hypothesized that the yield of microglia-like cells could be increased by activin signaling activation or Wnt inhibition at a time point after BMP4-mediated mesodermal specification has been achieved. Indeed, a recently published robust microglia differentiation method supports the efficacy of incorporating activin signaling to differentiate human stem cells (Abud et al., 2017). Manipulation of Wnt signaling to promote primitive rather than definitive myeloid differentiation is stage and timing specific (Sturgeon et al., 2014); thus, future studies aimed at determining when mesodermal specification has proceeded for a sufficient time period to enable suppression of definitive hematopoiesis will likely enable further refinement of the differentiation protocol employed here toward one that generates a larger portion of cells expression microglia specific lineage markers. Approaches such as ours and in recent reports highlight the complexity of in vitro manipulation of extracellular developmental cues to recapitulate embryological differentiation patterns.
In summary, we demonstrate that developmental cues which guide differentiation of microglia-like cells promote microglia-like morphology, gene and protein expression, as well as basic microglia functions. We describe a reproducible method for generating microglia-like cells by first deriving mesoderm with BMP-4 followed by exposure to microglia relevant cytokines. Our approach was based on recent studies that defined microglia ontogeny from primitive yolk sac mesoderm as distinct from peripheral macrophages that arise during definitive hematopoiesis. We demonstrate that Wnt and activin signaling can be employed to manipulate the development of microglia-like cells. This suggests that methods which leverage our understanding of microglial ontogeny may lead to more phenotypically “accurate” microglia-like cells. We believe therefore that understanding the molecular signals involved in microglia ontogeny will further refine stem cell to microglia differentiation approaches with the goal of developing in vitro human disease models to study the role of human microglia in the cellular and molecular pathogenesis of human neurological disease.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH grants: K02 AG04447 (S. J.), University of Washington ADRC Pilot Grant (P. J. A.) K08DK082783 (A. R.), R01AG053002 (S. J., G. A. G.), U01HL099993, and P50 AG05136.
References
- Abud, E. M., Ramirez, R. N., Martinez, E. S., Healy, L. M., Nguyen, C. H. H., Newman, S. A., Yeromin, A. V., Scarfone, V. M., Marsh, S. E., Fimbres, C., Caraway, C. A., Fote, G. M., Madany, A. M., Agrawal, A., Kayed, R., Gylys, K. H., Cahalan, M. D., Cummings, B. J., Antel, J. P., Mortazavi, A., Carson, M. J., Poon, W. W., & Blurton-Jones, M. (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron, 94(2), 278–293, e279. [DOI] [PMC free article] [PubMed]
- Bachstetter A. D., Van Eldik L. J., Schmitt F. A., Neltner J. H., Ighodaro E. T., Webster S. J., Patel E., Abner E. L., Kryscio R. J., Nelson P. T. (2015) Disease-related microglia heterogeneity in the hippocampus of Alzheimer's disease, dementia with Lewy bodies, and hippocampal sclerosis of aging. Acta Neuropathol Commun 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beins E., Ulas T., Ternes S., Neumann H., Schultze J. L., Zimmer A. (2016) Characterization of inflammatory markers and transcriptome profiles of differentially activated embryonic stem cell-derived microglia. Glia 64: 1007–1020. [DOI] [PubMed] [Google Scholar]
- Bennett M. L., Bennett F. C., Liddelow S. A., Ajami B., Zamanian J. L., Fernhoff N. B., Mulinyawe S. B., Bohlen C. J., Adil A., Tucker A., Weissman I. L., Chang E. F., Li G., Grant G. A., Hayden Gephart M. G., Barres B. A. (2016) New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113: E1738–E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutner C., Roy K., Linnartz B., Napoli I., Neumann H. (2010) Generation of microglial cells from mouse embryonic stem cells. Nat Protoc 5: 1481–1494. [DOI] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M. P., Moore C. S., Cialic R., Lanser A. J., Gabriely G., Koeglsperger T., Dake B., Wu P. M., Doykan C. E., Fanek Z., Liu L., Chen Z., Rothstein J. D., Ransohoff R. M., Gygi S. P., Antel J. P., Weiner H. L. (2014) Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 17: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldeira C., Oliveira A. F., Cunha C., Vaz A. R., Falcao A. S., Fernandes A., Brites D. (2014) Microglia change from a reactive to an age-like phenotype with the time in culture. Front Cell Neurosci 8: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti A., Ransohoff R. M. (2016) Microglial physiology and pathophysiology: Insights from genome-wide transcriptional profiling. Immunity 44: 505–515. [DOI] [PubMed] [Google Scholar]
- Dolmetsch R., Geschwind D. H. (2011) The human brain in a dish: The promise of iPSC-derived neurons. Cell 145: 831–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad S., Zamin R. M., Ruitenberg M. J., Filgueira L. (2012) A novel in vitro human microglia model: Characterization of human monocyte-derived microglia. J Neurosci Methods 209: 79–89. [DOI] [PubMed] [Google Scholar]
- Gertig U., Hanisch U. K. (2014) Microglial diversity by responses and responders. Front Cell Neurosci 8: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M. F., Conway S. J., Ng L. G., Stanley E. R., Samokhvalov I. M., Merad M. (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Zhu Z., Shi Z. D., Lelli K., Verma N., Li Q. V., Huangfu D. (2014) An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Link V. M., Romanoski C. E., Fonseca G. J., Eichenfield D. Z., Spann N. J., Stender J. D., Chun H. B., Garner H., Geissmann F., Glass C. K. (2014) Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K., Michoel T., Karavolos M. H., Clohisey S., Baillie J. K., Stevens M. P., Freeman T. C., Summers K. M., McColl B. W. (2016) Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19: 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R., et al. (2013) TREM2 variants in Alzheimer's disease. N Eng J Med 368: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M. T., et al. (2015) Neuroinflammation in Alzheimer's disease. Lancet Neurol 14: 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S. E., El Khoury J. (2014) TREM2 and the neuroimmunology of Alzheimer's disease. Biochem Pharmacol 88: 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S. E., Kingery N. D., Ohsumi T. K., Borowsky M. L., Wang L. C., Means T. K., El Khoury J. (2013) The microglial sensome revealed by direct RNA sequencing. Nat Neurosci 16: 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Genomics of Alzheimer's Disease Consortium (2015). Convergent genetic and expression data implicate immunity in Alzheimer's disease. Alzheimers Dement, 201511: 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayadev S., Case A., Eastman A. J., Nguyen H., Pollak J., Wiley J. C., Moller T., Morrison R. S., Garden G. A. (2010) Presenilin 2 is the predominant gamma-secretase in microglia and modulates cytokine release. PloS One 5: e15743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T., et al. (2013) Variant of TREM2 associated with the risk of Alzheimer's disease. N Eng J Med 368: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M. A., Hirschi K. K. (2009) Signaling hierarchy regulating human endothelial cell development. Arterioscler Thromb Vasc Biol 29: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. (2014) Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159: 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Melief E., Postupna N., Montine K. S., Keene C. D., Montine T. J. (2015) Prostaglandin E2 receptor subtype 2 regulation of scavenger receptor CD36 modulates microglial Abeta42 phagocytosis. Am J Pathol 185: 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J., Li Y., Yuan B., Mitalipova M., Omer A., Corcoran S., Bakiasi G., Tsai L. H., Aubourg P., Ransohoff R. M., Jaenisch R. (2016) Efficient derivation of microglia-like cells from human pluripotent stem cells. Nature Med 22: 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I., Kierdorf K., Neumann H. (2009) Microglial precursors derived from mouse embryonic stem cells. Glia 57: 1660–1671. [DOI] [PubMed] [Google Scholar]
- Ohgidani M., Kato T. A., Setoyama D., Sagata N., Hashimoto R., Shigenobu K., Yoshida T., Hayakawa K., Shimokawa N., Miura D., Utsumi H., Kanba S. (2014) Direct induction of ramified microglia-like cells from human monocytes: Dynamic microglial dysfunction in Nasu-Hakola disease. Sci Rep 4: 4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J., Kestila M., Wu J., Salminen A., Bohling T., Ruotsalainen V., Hakola P., Bakker A. B., Phillips J. H., Pekkarinen P., Lanier L. L., Timonen T., Peltonen L. (2000) Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet 25: 357–361. [DOI] [PubMed] [Google Scholar]
- Paloneva J., Manninen T., Christman G., Hovanes K., Mandelin J., Adolfsson R., Bianchin M., Bird T., Miranda R., Salmaggi A., Tranebjaerg L., Konttinen Y., Peltonen L. (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet 71: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H., Shen M. J., Ichikawa D. M., Sedlock A. B., Choi Y., Johnson K. R., Kim G., Brown M. A., Elkahloun A. G., Maric D., Sweeney C. L., Gossa S., Malech H. L., McGavern D. B., Park J. K. (2017) Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci 20: 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I. H., Lerou P. H., Zhao R., Huo H., Daley G. Q. (2008) Generation of human-induced pluripotent stem cells. Nat Protoc 3: 1180–1186. [DOI] [PubMed] [Google Scholar]
- Prinz M., Priller J. (2014) Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat Rev Neurosci 15: 300–312. [DOI] [PubMed] [Google Scholar]
- Rademakers R., et al. (2012) Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet 44: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed-Geaghan E. G., Savage J. C., Hise A. G., Landreth G. E. (2009) CD14 and toll-like receptors 2 and 4 are required for fibrillar A{beta}-stimulated microglial activation. J Neurosci 29: 11982–11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzel R. M., Patel A. R., Pan S., Crapser J., Hammond M., Jellison E., McCullough L. D. (2015) Age- and location-related changes in microglial function. Neurobiol Aging 36: 2153–2163. [DOI] [PubMed] [Google Scholar]
- Schmid C. D., Melchior B., Masek K., Puntambekar S. S., Danielson P. E., Lo D. D., Sutcliffe J. G., Carson M. J. (2009) Differential gene expression in LPS/IFNgamma activated microglia and macrophages: in vitro versus in vivo. J Neurochem 109(Suppl 1): 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C. D., Sautkulis L. N., Danielson P. E., Cooper J., Hasel K. W., Hilbush B. S., Sutcliffe J. G., Carson M. J. (2002) Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J Neurochem 83: 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. P., Hou Z., Propson N. E., Zhang J., Engstrom C. J., Santos Costa V., Jiang P., Nguyen B. K., Bolin J. M., Daly W., Wang Y., Stewart R., Page C. D., Murphy W. L., Thomson J. A. (2015) Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc Natl Acad Sci U S A 112: 12516–12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slukvin (2013) Hematopoietic specification from human pluripotent stem cells: current advances and challenges toward de novo generation of hematopoietic stem cells. Blood 122: 4035–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon C. M., Ditadi A., Awong G., Kennedy M., Keller G. (2014) Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat Biotechnol 32: 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Rochford C. D., Neumann H. (2005) Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med 201: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Park K. C., Toyonaga S., Yamada S. M., Nakabayashi H., Nakai E., Ikawa N., Furuya M., Tominaga A., Shimizu K. (2005) Characterization of microglia induced from mouse embryonic stem cells and their migration into the brain parenchyma. J Neuroimmunol 160: 210–218. [DOI] [PubMed] [Google Scholar]
- Turnbull I. R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., Colonna M. (2006) Cutting edge: TREM-2 attenuates macrophage activation. J Immunol 177: 3520–3524. [DOI] [PubMed] [Google Scholar]
- Villegas-Llerena C., Phillips A., Garcia-Reitboeck P., Hardy J., Pocock J. M. (2016) Microglial genes regulating neuroinflammation in the progression of Alzheimer's disease. Curr Opin Neurobiol 36: 74–81. [DOI] [PubMed] [Google Scholar]
- Winnier G., Blessing M., Labosky P. A., Hogan B. L. (1995) Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev 9: 2105–2116. [DOI] [PubMed] [Google Scholar]