Abstract
The indicating FTA elute micro card™ has been developed to collect and stabilize the nucleic acid in biological samples and is widely used in human and veterinary medicine and other disciplines. This card is not recommended for protein analyses, since surface treatment may denature proteins. We studied the ability to analyse proteins in human plasma and vaginal fluid as applied to the indicating FTA elute micro card™ using the sensitive proximity extension assay (PEA). Among 92 proteins in the Proseek Multiplex Oncology Iv2 panel, 87 were above the limit of detection (LOD) in liquid plasma and 56 among 92 above LOD in plasma applied to FTA cards. Washing and protein elution protocols were compared to identify an optimal method. Liquid-based cytology samples showed a lower number of proteins above LOD than FTA cards with vaginal fluid samples applied. Our results demonstrate that samples applied to the indicating FTA elute micro card™ are amendable to protein analyses, given that a sensitive protein detection assay is used. The results imply that biological samples applied to FTA cards can be used for DNA, RNA and protein detection.
Keywords: FTA Elute Card, Vaginal Fluid, Plasma, Proximity Extension Assay
1. Introduction
A number of sampling devices have been developed to collect blood or plasma/serum for protein analyses, ranging from solid phase matrices to the recent use of nanocapillary technology [1]. The collection of blood spots, buccal cells and saliva on filter paper is a common sampling method, used for the screening of newborns for inherited diseases, in forensic medicine and for many other purposes. Specific surface treatments have been developed to stabilize nucleic acids applied onto filter paper. Whatman FTA technology consists of two distinct chemistries, both of which have the ability to lyse cells on contact, denature proteins and protect DNA and RNA from degradation. The FTA elute card™ (GE Healthcare, United Kingdom) contains a chaotropic salt and proteins remain tightly bound while DNA is eluted from the matrix. The indicating FTA elute micro card™ also includes a purple dye that turns white when a clear sample is applied. This colour change is useful for verifying that the biological sample has been deposited on the card surface, specifically when the card is used for self-collection of samples. The indicating FTA elute micro card™ allows for a simple elution of DNA and RNA from the surface of the card, and is extremely suitable for genotyping, sequencing and real-time PCR applications [2, 3].
The indicating FTA elute micro card™ is presently used for the self-collection of cervico-vaginal fluid (CVF) samples in cervical cancer screening programmes [4–10]. It will therefore be of interest to determine if samples collected onto the FTA card™ can also be used for analyses of protein biomarkers. The FTA elute card is not recommended for protein analyses, since surface treatment may denature proteins and to the best of our knowledge, no study has been performed to determine whether proteins applied to the FTA elute micro card™ can be detected. The aim of this study was to examine the potential for using the indicating FTA elute micro card™ for protein analyses by employing the very sensitive and highly specific protein extension assay (PEA) [11, 12]. PEA is based on the use of two antibodies towards the target protein, providing high specificity. Each antibody has a DNA tail attached and if the DNA tails on the two antibodies are within reach of each other, they can act as a substrate for DNA polymerase. Protein detection is achieved through a real-time PCR, resulting in high analytical sensitivity. The PEA has previously been used to measure protein in biomarker studies [13, 14]. If protein detection is indicated as feasible in samples applied to the indicating FTA elute micro card™, it will offer a quick, cost-effective, easy-to-store option for sample collection, both for diagnostic purposes and population screening.
2. Samples and Methods
2.1 Samples and preparation
CVF samples were collected from women participating in the organized cervical cancer screening using the Rover Viba brush (Rover Medical Devices B.V., Oss, The Netherlands) and applied to the indicating FTA elute micro card™ as described earlier [15].
All FTA card samples were selected from HPV-negative women. One sample, which had been stored for two weeks, was selected for a first buffer-test. Twenty-eight CVF samples were gathered representing a storage time on the card of 1–29 months prior to analysis. Twelve samples had been stored for one month, six samples for seven months, six samples for 19 months and four samples were stored for 29 months. Five additional samples, with storage times of less than two weeks, were selected for the testing protocols for washing.
Plasma samples were obtained from five women. For each woman, 10 μl plasma aliquots were applied to an indicating FTA elute micro card™; the card surfaces were dried and stored at room temperature for either 24 h or 72 h. All samples applied to the FTA cards were processed using the 3 mm Ø Harris micro punch (Whatman, Inc., Clifton, NJ) for collecting punches as previously described [15]. The following washing procedures were compared: I) no wash prior to elution; II) 1 × 5 min. wash in 100 μl water; III) 2 × 5 min. wash in 100 μl water. Proteins were eluted from the indicating FTA elute micro card™ by placing the punch in either a 20 μl Radioimmunoprecipitation assay buffer (RIPA) or 20 μl Phosphate-buffered saline (PBS)-Tween 20 (0.05 %) buffer for 60 to 90 minutes.
We also investigated samples from 28 women that were collected using the procedure applied in liquid-based cytology. These samples were collected using a cytobrush and dissolved in a 20 ml ThinPrep storage solution (Hologic, Inc. San Diego, USA), and stored at room temperature for 3–10 days prior to freezing. From this solution, 1 μl was used for the PEA analysis.
2.2 Protein analysis
One μl of either liquid plasma, elution from FTA card, or liquid-based cytology sample was analysed using the proximity extension assay (PEA) and the Proseek Multiplex Oncology I v2 panel (Olink Bioscience AB, Uppsala, Sweden). This panel includes 92 proteins selected on the basis of their reported associations with cancer. The protein analysis is reported as normalized protein expression levels (NPX), which are Ct values normalized by the subtraction of values for extension control, as well as an interplate control; the scale is shifted using a correction factor (normal background noise) and reported in Log 2 scale. All assay characteristics including detection limits and measurements of assay performance and validations are available from the manufacturer's webpage (http://www.olink.com/products/proseek-multiplex/downloads/).
2.3 Statistical analysis
All analyses and illustrations were performed in R [16]. Spearman correlations were used throughout. The significance of differences in distributions was calculated using the double-sided Wilcoxon signed-rank test. Resulting P-values were corrected for multiple testing using either Bonferroni or FDR correction and considered significant if the q-value < 0.05. Linear modelling for estimating the correlation between the NPX measurement and storage time was performed with the lm function and the ANOVA function.
3. Results
3.1 Elution buffer optimization
We first compared two elution buffers, PBS-Tween 20 (0.05 %) and RIPA, for their efficiency in terms of recovering proteins from the indicating FTA elute micro card™. A single punch from one FTA sample was eluted in 20 μl PBS-Tween 20 (0.05 %) or RIPA buffer and analysed using the Proseek Multiplex Oncology I v2 panel. For both buffers, 80 of the 92 proteins in the panel had levels above the assay background (Spearman's rho, R2=0.92, p-value < 2.2e-16) (Figure 1). With a cut-off of >1 NPX above background, the PBS-Tween 20 buffer resulted in the detection of 43 proteins and the RIPA buffer in 33 proteins. Additionally, 60 proteins showed higher NPX values for the sample FTA elution card than a blank FTA elution card for PBS-Tween 20 buffer and 52 proteins for the sample eluted in RIPA buffer. Thus, the PBS-Tween 20 buffer performed better than the RIPA buffer and was therefore used in the following analyses.
Figure 1.
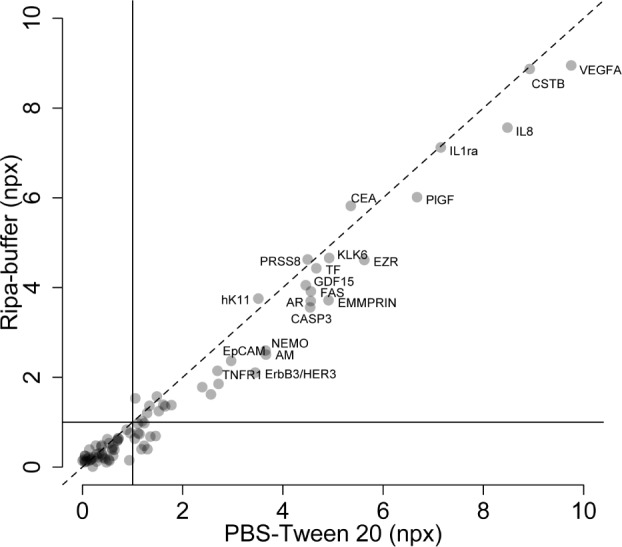
Comparison of the NPX values for the 92 proteins in the Proseek Multiplex Oncology Iv2 panel from analysis of a sample applied to the indicating FTA elute micro card™ and eluted with either RIPA (y-axis) or PBS-Tween 20 buffer (x-axis). PBS-Tween reports slightly higher NPX-values for the majority of the 92 proteins.
3.2 Proteins are measurable from the FTA cards
We next compared liquid plasma samples with plasma applied to the indicating FTA elute micro card™ and stored for either 24 h or 72 h. One punch from each FTA card was eluted in 20 μl PBS-Tween 20 buffer and analysed using the Proseek Multiplex Oncology I v2 panel. As expected from the elution protocol, the plasma sampled and applied to the FTA showed generally lower protein amounts than the liquid plasma samples. In total, 87 of the 92 proteins in the panel were detected in all liquid plasma samples and 56 and 52 proteins were detected in the samples on FTA cards stored for 24 h and 72 h, respectively (Table 1).
Table 1.
List of proteins and percentage of samples over limit of detection (LOD) for each protein when using liquid plasma (left column) plasma on indicating FTA elute micro card™ stored for 24h (middle column) and 72h (right column)
| Protein | Plasma (% of samples over LOD) | 24 h (% of samples over LOD) | 72 h (% of samples over LOD) |
|---|---|---|---|
| IL-8 | 100% | 100% | 100% |
| VEGF-A | 100% | 100% | 100% |
| AM | 100% | 100% | 100% |
| GDF-15 | 100% | 100% | 100% |
| PlGF | 100% | 100% | 100% |
| CSF-1 | 100% | 100% | 100% |
| CSTB | 100% | 100% | 100% |
| MCP-1 | 100% | 100% | 100% |
| KLK6 | 100% | 100% | 100% |
| TRAIL-R2 | 100% | 100% | 100% |
| LAP-TGF-beta-1 | 100% | 100% | 100% |
| TF | 100% | 100% | 100% |
| TNF-R1 | 100% | 100% | 100% |
| PDGF-subunit-B | 100% | 100% | 100% |
| PARK7 | 100% | 100% | 100% |
| CXCL11 | 100% | 100% | 100% |
| VE-statin | 100% | 100% | 100% |
| IL-7 | 100% | 100% | 100% |
| SCF | 100% | 100% | 100% |
| CXCL9 | 100% | 100% | 100% |
| TNF-R2 | 100% | 100% | 100% |
| TNFSF14 | 100% | 100% | 100% |
| GH | 100% | 100% | 100% |
| FasL | 100% | 100% | 100% |
| FAS | 100% | 100% | 100% |
| CCL19 | 100% | 100% | 100% |
| EMMPRIN | 100% | 100% | 100% |
| CXCL10 | 100% | 100% | 100% |
| Ep-CAM | 100% | 100% | 100% |
| HGF | 100% | 100% | 100% |
| LITAF | 100% | 100% | 100% |
| CXCL5 | 100% | 100% | 100% |
| MK | 100% | 100% | 100% |
| U-PAR | 100% | 100% | 100% |
| CDH3 | 100% | 100% | 100% |
| LYN | 100% | 100% | 100% |
| Flt3L | 100% | 100% | 100% |
| HB-EGF | 100% | 100% | 100% |
| CD69 | 100% | 100% | 100% |
| TR-AP | 100% | 100% | 100% |
| CDKN1A | 100% | 100% | 100% |
| REG-4 | 100% | 100% | 100% |
| VEGF-D | 100% | 100% | 100% |
| HE4 | 100% | 100% | 100% |
| CXCL13 | 100% | 100% | 100% |
| CA-125 | 100% | 100% | 100% |
| PRSS8 | 100% | 100% | 100% |
| PECAM-1 | 100% | 100% | 100% |
| FR-alpha | 100% | 100% | 100% |
| TIE2 | 100% | 100% | 80% |
| MIC-A | 100% | 100% | 80% |
| IL-6 | 100% | 100% | 60% |
| ICOSLG | 100% | 100% | 60% |
| FS | 100% | 100% | 60% |
| NTRK3 | 100% | 100% | 20% |
| ILT-3 | 100% | 80% | 80% |
| VEGFR-2 | 100% | 80% | 80% |
| ITGA1 | 100% | 80% | 80% |
| ErbB3-HER3 | 100% | 60% | 80% |
| MIA | 100% | 60% | 80% |
| PTPN22 | 100% | 60% | 60% |
| SELE | 100% | 60% | 40% |
| FUR | 100% | 60% | 40% |
| AR | 100% | 60% | 20% |
| eIF-4B | 100% | 40% | 40% |
| EZR | 100% | 40% | 20% |
| PRL | 100% | 40% | 20% |
| IL-12 | 100% | 20% | 40% |
| MMP-1 | 100% | 20% | 40% |
| ErbB2-HER2 | 100% | 20% | 40% |
| TNFRSF4 | 100% | 20% | 40% |
| IL-6RA | 100% | 20% | 20% |
| BAFF | 100% | 20% | 20% |
| ErbB4-HER4 | 100% | 20% | 20% |
| NEMO | 100% | 20% | 20% |
| FADD | 100% | 20% | 0% |
| IFN-gamma | 100% | 20% | 0% |
| VIM | 100% | 0% | 40% |
| CASP-3 | 100% | 0% | 20% |
| CD40-L | 100% | 0% | 0% |
| IL-1ra | 100% | 0% | 0% |
| hK11 | 100% | 0% | 0% |
| CAIX | 100% | 0% | 0% |
| TGF-alpha | 100% | 0% | 0% |
| EGFR | 100% | 0% | 0% |
| THPO | 100% | 0% | 0% |
| IL-17RB | 100% | 0% | 0% |
| CEA | 80% | 20% | 20% |
| MYD88 | 80% | 0% | 20% |
| EPO | 80% | 0% | 0% |
| IL-2 | 60% | 80% | 100% |
| TNF | 40% | 100% | 100% |
The correlation between all observations for all proteins between 24 h and 72 h was high (Spearman's rho, R2=0.89, p-value < 2.2e-16) (Figure 2). In a per-protein analysis, replacing values below the limit of detection with protein specific LOD-values, five proteins (CSF-1, NTRK3, ICOSLG, SCF and PECAM-1) were found to have nominally significant differences between the two time-points (p < 0.05, Wilcox test). None of these associations remained significant after multiple hypothesis corrections. Only two proteins showed a mean difference of more than 1 NPX unit between samples stored for 24 h and 72 h (CSF-1 and TNF-R1, both of which had lower NPX values at 72 h).
Figure 2.
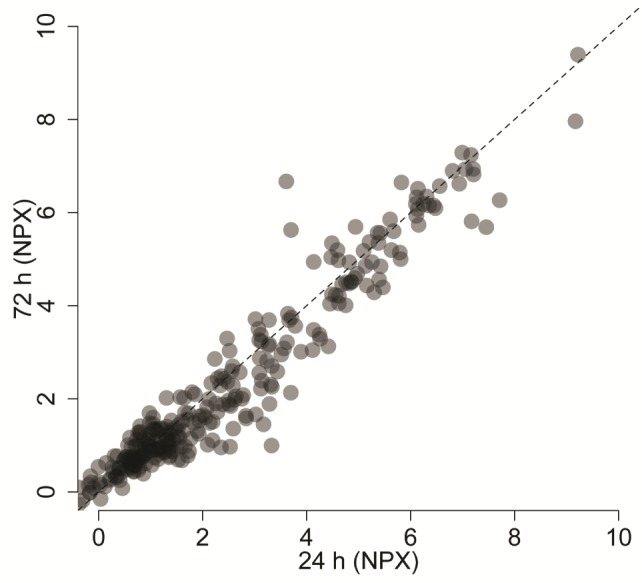
Correlation between NPX values for all observations with measurements above the limit of detection at two time-points from cervicovaginal fluid samples stored on FTA cards for 24 h (x-axis) and 72 h (y-axis), Spearman's rho, R2=0.89, p-value < 2.2e-16
3.3 Washing optimization
There was a large overlap between the proteins that were detectable in liquid plasma and on the FTA cards. Only six proteins were detectable in liquid plasma and on FTA cards stored for 24 h, but not on FTA cards stored for 72 h (Figure 3). Twelve out of 15 plasma samples on the FTA cards did not pass the Proseek internal controls for incubation, extension and detection. Nonetheless, 56 of 92 proteins were detected over LOD in all samples for FTA cards stored 24 h and 51 of 92 proteins were detected in all samples for FTA cards stored for 72 h. We reasoned that components in the surface chemistry of the FTA elute cards™ had a negative effect on the assay.
Figure 3.
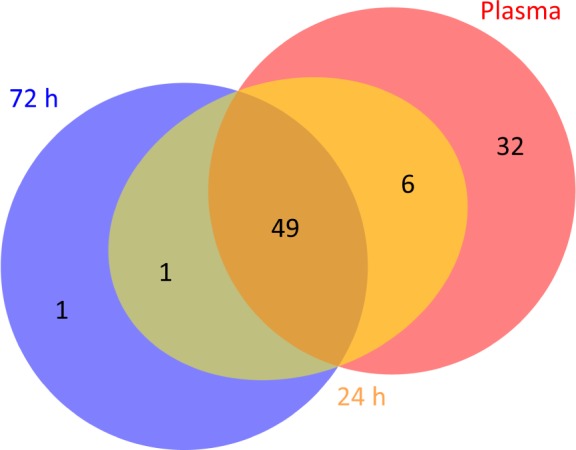
The overlap in proteins above LOD in 100% of the samples when analysing liquid plasma (plasma), plasma samples applied to the indicating FTA elute micro card™ and stored for 24 h (t24) and 72 h (t72) prior to analysis
In order to reduce the problem regarding failed internal controls, we tested three different protocols for washing punches prior to protein elution. One punch from each of 28 indicating FTA elute micro cards™ with a vaginal fluid sample was analysed using one of three different protocols: I) no wash prior to elution; II) 1 × 5 min. wash in 100 μl water; III) 2 × 5 min. wash in 100 μl water. Samples were then eluted in 20 μl PBS-Tween 20 buffer and analysed using the Proseek Multiplex Oncology I v2 panel.
With method I (no wash), eight samples showed signs of inhibition of internal controls, compared to one sample with method II and zero samples with method III. In total, 57 of 92 proteins had levels over LOD in all 28 samples using method I and 73 proteins were detected in 80% of the samples. With method II, 29 of 92 proteins were detectable in all 28 samples and 56 proteins were detected in 80% of the samples. With method III, 21 of 92 proteins had levels over LOD in all 28 samples and 45 proteins were detected in 80% of the samples. The number of proteins detected using the different extraction protocols as a function of the fraction of individuals with protein levels over LOD is shown in Figure 4.
Figure 4.
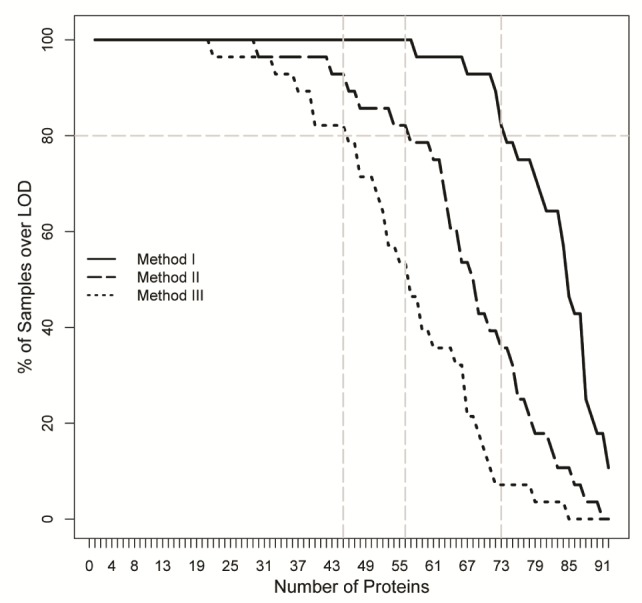
The number of proteins detected over limit of detection (LOD) for the three washing methods as a function of the percentage of samples studied with a NPX value over the LOD
3.4 Detectable proteins remain stable on the FTA-cards
The 28 samples were stored for up to 29 months. No significant effect of storage time on protein amounts was observed using Bonferroni correction (p > 0.065). With FDR correction, 19 proteins showed an effect due to storage time (p < 0.05). The urokinase plasminogen activator surface receptor (UPAR) protein had the largest difference between samples stored for one month and samples stored for 29 months, with a mean difference of 3.59 NPX (Supplementary Table S1).
3.5 Adding more material does not improve detectability
The additional wash improved the analysis, but may also have resulted in loss of protein. We therefore investigated whether increasing the input material, i.e., the number of punches from a sample, would result in higher protein amounts. To this end one, two, three or four punches were obtained from an FTA card with CVF sample from five women and washed for 2 × 5 min. in 100 μl water (method III), eluted in a 20 μl PBS-Tween 20 buffer and analysed using the Proseek Multiplex Oncology I v2 panel. The samples using one punch passed all the assay controls, while the samples using two punches or more did not pass the incubation control. Extension and detection controls were not affected, regardless of the number of punches used.
Independent of the number of punches used, roughly 50% of proteins in the panel showed levels above the LOD in 100% of the women (47/92 with one punch, 47/92 with two punches, 46/92 with three punches and 46/92 with four punches). When considering proteins that were measurable in more than 50% of individuals, 62 and 58 proteins were above LOD, using one and four punches, respectively. Thus, increasing the number of punches did not increase the number of proteins to a level above the LOD when using the presently used wash and elusion protocol.
3.6 Comparison with liquid cytology samples
For comparison we also investigated samples collected with the procedure used in liquid-based cytology (Thin-Prep transport solution). A total of 28 cytology samples were analysed using the Proseek Multiplex Oncology I v2 panel. Two of the samples did not pass the internal assay controls and were removed from the analysis. Only seven proteins were above the LOD in 100% of the 26 cytology samples, compared to 21 proteins that were above the LOD in 100% of the FTA samples (method III) (Table 2).
Table 2.
List of proteins and percentage of samples over the limit of detection (LOD) among the indicating FTA elute micro card™ samples (left column) and the Liguid-based cytology samples (right column); 2 of the 28 Liguid-based cytology samples did not pass the quality controls and were excluded. 7 proteins were detected in all 26 Liguid-based cytology samples, compared to 21 proteins that were detected in the 28 FTA samples.
| Protein | FTA elute micro cardTM (% of samples over LOD) | Protein | Liquid cytology samples (% of samples over LOD) |
|---|---|---|---|
| IL-8 | 100% | IL-8 | 100% |
| VEGF-A | 100% | VEGF-A | 100% |
| AM | 100% | IL-1ra | 100% |
| PlGF | 100% | CSTB | 100% |
| EZR | 100% | U-PAR | 100% |
| IL-1ra | 100% | HE4 | 100% |
| CSTB | 100% | PRSS8 | 100% |
| MCP-1 | 100% | MCP-1 | 96% |
| TNF-R1 | 100% | TNF-R1 | 96% |
| PARK7 | 100% | CEA | 96% |
| FADD | 100% | EZR | 92% |
| FAS | 100% | hK11 | 92% |
| EMMPRIN | 100% | Ep-CAM | 92% |
| LITAF | 100% | CXCL5 | 92% |
| CASP-3 | 100% | TF | 88% |
| CDKN1A | 100% | FAS | 88% |
| TGF-alpha | 100% | EMMPRIN | 88% |
| HE4 | 100% | CASP-3 | 88% |
| eIF-4B | 100% | FR-alpha | 88% |
| CA-125 | 100% | GDF-15 | 85% |
| PRSS8 | 100% | MK | 85% |
| GDF-15 | 96% | TGF-alpha | 85% |
| TF | 96% | CXCL13 | 85% |
| CXCL9 | 96% | VIM | 85% |
| PTPN22 | 96% | PlGF | 81% |
| CXCL10 | 96% | IL-6 | 81% |
| CXCL5 | 96% | CXCL10 | 77% |
| MK | 96% | HGF | 77% |
| U-PAR | 96% | AR | 77% |
| LYN | 96% | CXCL9 | 73% |
| CXCL13 | 96% | ErbB3-HER3 | 73% |
| CEA | 96% | TNF-R2 | 65% |
| FUR | 93% | TR-AP | 65% |
| Ep-CAM | 93% | KLK6 | 62% |
| AR | 93% | TRAIL-R2 | 62% |
| NEMO | 93% | FADD | 62% |
| KLK6 | 89% | LITAF | 62% |
| hK11 | 89% | NEMO | 62% |
| ErbB3-HER3 | 89% | eIF-4B | 62% |
| IL-6 | 82% | CA-125 | 58% |
| LAP-TGF-beta-1 | 82% | LAP-TGF-beta-1 | 50% |
| FasL | 82% | CSF-1 | 46% |
| CD69 | 82% | PARK7 | 46% |
| VIM | 82% | FUR | 46% |
| FR-alpha | 82% | CAIX | 42% |
| HGF | 79% | ErbB4-HER4 | 42% |
| ICOSLG | 79% | CD69 | 31% |
| CSF-1 | 71% | MIC-A | 31% |
| ErbB2-HER2 | 71% | AM | 27% |
| MIC-A | 71% | ErbB2-HER2 | 27% |
| VE-statin | 68% | Flt3L | 27% |
| CDH3 | 64% | CCL19 | 23% |
| Flt3L | 57% | PTPN22 | 23% |
| PECAM-1 | 57% | LYN | 23% |
| CCL19 | 54% | PECAM-1 | 23% |
| MYD88 | 54% | SCF | 15% |
| ILT-3 | 46% | HB-EGF | 15% |
| TNF-R2 | 46% | CXCL11 | 12% |
| PDGF-subunit-B | 39% | BAFF | 12% |
| TNF | 39% | TNFRSF4 | 12% |
| IL-7 | 36% | IL-12 | 8% |
| TNFSF14 | 36% | FasL | 8% |
| ErbB4-HER4 | 36% | MYD88 | 8% |
| ITGA1 | 36% | MIA | 8% |
| TRAIL-R2 | 32% | CDKN1A | 8% |
| CXCL11 | 32% | EGFR | 8% |
| SCF | 21% | ITGA1 | 8% |
| FS | 21% | FS | 8% |
| REG-4 | 18% | ILT-3 | 4% |
| HB-EGF | 14% | TIE2 | 4% |
| MMP-1 | 11% | PDGF-subunit-B | 4% |
| TIE2 | 7% | IL-2 | 4% |
| BAFF | 7% | IL-7 | 4% |
| MIA | 7% | IL-6RA | 4% |
| TNFRSF4 | 7% | MMP-1 | 4% |
| TR-AP | 7% | TNFSF14 | 4% |
| THPO | 7% | GH | 4% |
| NTRK3 | 7% | CDH3 | 4% |
| SELE | 4% | REG-4 | 4% |
| IL-12 | 4% | VEGF-D | 4% |
| PRL | 4% | IL-17RB | 4% |
| GH | 4% | CD40-L | 0% |
| CAIX | 4% | SELE | 0% |
| EGFR | 4% | EPO | 0% |
| CD40-L | 0% | VE-statin | 0% |
| EPO | 0% | PRL | 0% |
| IL-2 | 0% | TNF | 0% |
| IL-6RA | 0% | ICOSLG | 0% |
| VEGFR-2 | 0% | VEGFR-2 | 0% |
| IFN-gamma | 0% | IFN-gamma | 0% |
| VEGF-D | 0% | THPO | 0% |
| IL-17RB | 0% | NTRK3 | 0% |
4. Discussion
This study shows that plasma and cervico-vaginal fluid samples applied to the indicating FTA elute micro card™ are amendable to protein analysis. The PBS-Tween (0.05 %) elution is the preferable elution buffer. The majority of proteins in the Proseek Multiplex Oncology I v2 panel that were detectable in liquid plasma were also above LOD in samples applied to the indicating FTA elute micro card™. The NPX values were generally lower in the FTA card samples, although some proteins showed high NPX values. This was to some extent expected because of the elution process, since for FTA card samples, the 10 μl of plasma in a punch was eluted in a 20 μl of buffer, resulting in half of the concentration of proteins entering the assay, compared to the liquid plasma samples.
Our comparison of washing methods showed that although extensive washing may results in a loss of protein, adding a second wash to the punches from FTA cards reduced the number of samples that did not pass the internal controls. The results showed that the proteins on the FTA card appear stable. The protein profile in the vaginal fluid samples on FTA cards from 28 women stored for up to two years showed an effect on 19 proteins, with FDR correction (p < 0.05); however, this effect was not seen using Bonferroni correction (p > 0.065). We also studied whether the number of proteins above LOD detected in the FTA card samples could be increased by adding more starting material, i.e., more punches in the washing and elution steps. However, this did not result in additional proteins appearing above LOD, but only increased the number of samples failing internal control. An alternative would be to use a protocol that selectively removes inhibitors from the FTA cards' surface prior to the elution of the proteins. The liquid cytology samples showed a slightly higher abundance of some proteins compared to samples from the indicating FTA elute micro card™, but the number of proteins with levels above LOD was higher for the FTA cards. The lower number of detectable proteins in the liquid cytology samples may have been due to the procedure for sample collection and storage, and in particular, the substantial dilution of the original sample in the transport solution.
Our study demonstrates the feasibility of measuring proteins from vaginal fluid and plasma collected onto the indicating FTA elute micro card™. This opens up the possibility of studying both DNA, RNA and protein biomarkers in a single sample applied to the indicating FTA elute micro card™. The indicating FTA elute micro card™ is extremely suitable where biobanking is concerned, since these cards can be stored at room temperature and do not require large storage space or the subzero conditions needed for liquid cytology samples.
5. Compliance with Ethical Research Standards
Daniel Ekman is employed at Olink Bioscience AB, Uppsala. All other authors declare no conflicts of interest.
6. Acknowledgements
This study was supported by the Swedish Cancer Foundation and the Swedish Foundation for Strategic Research (SSF). BioCARE is a national strategic cancer research program at the University of Gothenburg. The PEA measurements were performed by Olink Bioscience AB, Uppsala, Sweden.
References
- [1].Chen JQ, Wakefield LM, Goldstein DJ. Capillary nano-immunoassays: advancing quantitative proteomics analysis, biomarker assessment, and molecular diagnostics. J Transl Med. 2015;13:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang DY, Chang CW, Lagace RE, Oldroyd NJ, Hennessy LK. Development and validation of the AmpFlSTR(R) Identifiler (R) Direct PCR Amplification Kit: a multiplex assay for the direct amplification of single-source samples. Journal of forensic sciences. 2011;56(4):835–45. [DOI] [PubMed] [Google Scholar]
- [3].da Cunha Santos G, Liu N, Tsao MS, Kamel-Reid S, Chin K, Geddie WR. Detection of EGFR and KRAS mutations in fine-needle aspirates stored on Whatman FTA cards: is this the tool for biobanking cytological samples in the molecular era? Cancer Cytopathol. 2010;118(6):450–6. [DOI] [PubMed] [Google Scholar]
- [4].Gustavsson I, Lindell M, Wilander E, Strand A, Gyllensten U. Use of FTA card for dry collection, transportation and storage of cervical cell specimen to detect high-risk HPV. J Clin Virol. 2009;46(2): 112–6. [DOI] [PubMed] [Google Scholar]
- [5].Phongsavan K, Gustavsson I, Marions L, Phengsavanh A, Wahlstrom R, Gyllensten U. Detection of human papillomavirus among women in Laos: feasibility of using filter paper card and prevalence of high-risk types. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2012;22(8):1398–406. [DOI] [PubMed] [Google Scholar]
- [6].Gyllensten U, Gustavsson I, Lindell M, Wilander E. Primary high-risk HPV screening for cervical cancer in post-menopausal women. Gynecol Oncol. 2012;125(2):343–5. [DOI] [PubMed] [Google Scholar]
- [7].Santos CR, Franciscatto LG, Barcellos RB, Almeida SE, Rossetti ML. Use of FTA elute card impregnated with cervicovaginal sample directly into the amplification reaction increases the detection of human papillomavirus DNA. Brazilian journal of microbiology 2012;43(1):389–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Catarino R, Vassilakos P, Bilancioni A, Vanden Eynde M, Meyer-Hamme U, Menoud PA, et al. Randomized Comparison of Two Vaginal Self-Sampling Methods for Human Papillomavirus Detection: Dry Swab versus FTA Cartridge. PloS one. 2015;10(12):e0143644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang SM, Hu SY, Chen F, Chen W, Zhao FH, Zhang YQ, et al. Clinical evaluation of human papillomavirus detection by care HPV test on physician-samples and self-samples using the indicating FTA Elute(R) card. Asian Pac J Cancer Prev. 2014;15(17): 7085–9. [DOI] [PubMed] [Google Scholar]
- [10].Wang SM, Hu SY, Chen W, Chen F, Zhao FH, He W, et al. Feasibility and accuracy evaluation of three human papillomavirus assays for FTA card-based sampling: a pilot study in cervical cancer screening. BMC Cancer. 2015;15:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39(15):e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PloS one. 2014;9(4):e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Enroth S, Bosdotter Enroth S, Johansson A, Gyllensten U. Effect of genetic and environmental factors on protein biomarkers for common non-communicable disease and use of personally normalized plasma protein profiles (PNPPP). Biomarkers. 2015;20(6–7):355–64. [DOI] [PubMed] [Google Scholar]
- [14].Helseth R, Weiss TW, Opstad TB, Siegbahn A, Solheim S, Freynhofer MK, et al. Associations between circulating proteins and corresponding genes expressed in coronary thrombi in patients with acute myocardial infarction. Thrombosis research. 2015;136(6):1240–4. [DOI] [PubMed] [Google Scholar]
- [15].Gustavsson I, Sanner K, Lindell M, Strand A, Olovsson M, Wikstrom I, et al. Type-specific detection of high-risk human papillomavirus (HPV) in self-sampled cervicovaginal cells applied to FTA elute cartridge. J Clin Virol. 2011;51(4):255–8. [DOI] [PubMed] [Google Scholar]
- [16].R Core Team. R: A Language and Environment for Statistical Computing.: R Foundation for Statistical Computing; 2014.


